Abstract
Pluripotent stem cells transition between distinct naive and primed states that are controlled by overlapping sets of master regulatory transcription factors. In this issue of Cell Stem Cell, Buecker et al. (2014) and Factor et al. (2014) demonstrate that alternate enhancer usage, regulated by state-specific binding partners of master regulators, defines these pluripotent state transitions.
Master regulatory transcription factors direct tissue-specific expression patterns and unique cellular responses to signaling pathways by targeting cell-type-specific enhancer regions (Mullen et al., 2011). Oct4, for example, functions as a master transcription factor critical for pluripotency maintenance in embryonic stem cells (ESCs), and the levels of Oct4 are tightly regulated to control cellular transitions either toward pluripotency or toward embryonic lineage commitment (Radzisheuskaya et al., 2013). Recent evidence shows that pluripotency can be defined by two distinct phases: naive, a preim-plantation developmental ground state; or primed, a postimplantation pluripotent state prepared for lineage specification and commitment (Nichols and Smith, 2009). Though naive and primed pluripotent cells have distinct gene expression profiles, both preim-plantation- and post-implantation- derived ESCs express the same core pluripotency transcription factors, Oct4, Sox2, and Nanog (Tesar et al., 2007), raising questions about the molecular characteristics that distinguish these two states. In this issue of Cell Stem Cell, Factor et al. and Buecker et al. profile enhancer and transcription factor occupancy landscapes in naive and primed stem cells, providing mechanistic insight into the dynamic regulation of key overlapping factors across distinct pluripotent states.
Tesar and colleagues performed transcriptome profiling in mouse ESCs and epiblast stem cells (EpiSCs), representative of naive and primed pluripotent states, respectively. This analysis showed small-scale differences in transcriptional output, yet these changes are mirrored by dramatic alterations in chromatin profiles at enhancers (Factor et al., 2014). In particular, nearly all genes preferentially expressed in pluripotent cells are marked by differential enhancer usage between naive and primed states. Enhancers used specifically in naive ESCs, termed naive-dominant enhancers, are characterized by high levels of enhancer histone signals H3K4me1 and H3K27ac, and these enhancers and their associated marks are lost following transition of naive ESCs into primed EpiSCs. In contrast, enhancers used exclusively in EpiSCs and not naive cells contain low but detectable levels of enhancer histone signals in ESCs. This suggests that these regulatory elements, termed “seed enhancers,” may function as placeholders in precursor cells to ensure proper enhancer usage in subsequent, differentiated cell types (Factor et al., 2014). Seed enhancers were further shown to be significantly enriched for H3K27ac in subsequent embryonic and adult tissues, and in many cases they expand into multienhancer clusters, recently described as stretch enhancers or superenhancers, which are important for maintaining cellular identity (Hnisz et al., 2013; Parker et al., 2013).
In contrast to naive-dominant enhancers, seed enhancers rarely interact with pluripotency gene promoters and are comparatively depleted for factors such as the mediator and cohesin complexes that promote enhancer-promoter interactions in mouse ESCs. The reorganization of enhancer usage during the transition to primed EpiSCs suggests that regulatory transcription factors, such as Oct4, must be redirected from decommissioned naive-dominant enhancers to newly functional seed enhancers. To examine the behavior of these common regulatory factors during the transition between naive and primed pluripotency, Swigut, Wysocka, and colleagues (Buecker et al., 2014) performed a careful analysis of transcription factor binding. The authors performed genome-wide profiling of Oct4 binding using an in vitro system modeling preimplantation to postimplantation differentiation. Consistent with the dramatic changes in enhancer usage and chromatin profiles described above, Oct4 occupancy shows substantial reorganization between naive and primed stem cell states. Naive state-specific Oct4 binding correlates with downregulation of associated transcripts following transition into a primed stem cell state, whereas primed state-specific Oct4 binding is associated with transcriptional upregulation. Changes in Oct4 localization are further mirrored by alterations in p300 binding and H3K27ac levels, suggesting that Oct4 indeed localizes to state-specific active enhancers.
The levels of Oct4 expression are comparable between naive and primed states despite clear differences in binding landscapes. However, by comparing the protein-protein interaction profiles between Oct4 and other factors across these two pluripotent states, Buecker et al. provide further insight into the mechanisms driving Oct4 reorganization. Chromatin remodeling complexes, protein modification enzymes, and several transcription factors show differential interaction with Oct4 in naive versus primed stem cells. For example, Esrrb, Klf5, and Tcf3 interact with Oct4 specifically in naive ESCs, and Otx2, Zic2, Zfp281, and others were identified as interacting proteins specifically in primed epiblast-like stem cells. De novo motif identification of Oct4 binding sites also revealed differential enrichments for DNA recognition motifs corresponding to Esrrb and Klf4/5, or to Otx2 and Zic2, consistent with biochemically identified binding partners in naive and primed states, respectively. The differential protein interaction and DNA interaction profiles suggest that cooperative binding by Oct4 with cell-state-specific regulatory partners may be responsible for driving reorganization of enhancer usage during differentiation (Figure 1).
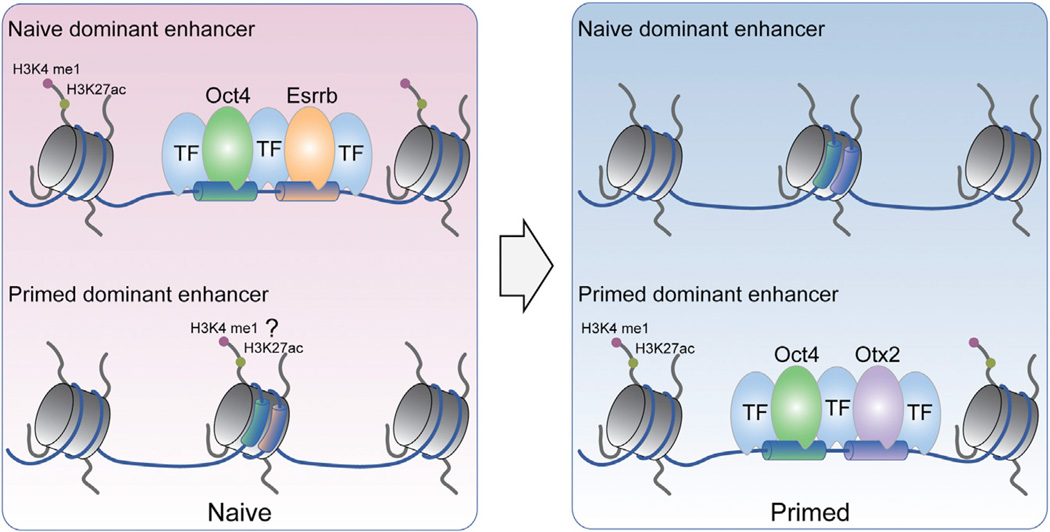
Figure 1. Dynamic Enhancer Organization across Naive and Primed Stem Cell States.
Epigenome profiling by Buecker et al. and Factor et al. reveal differential enhancer usage between naive ESCs (left) and primed EpiSCs (right) by genes preferentially expressed in pluripotent stem cells. Enhancers identified exclusively in ESCs (Naive-dominant enhancers, top) are characterized by strong enhancer histone signals H3K4me1 and H3K27ac in the naive state, whereas primed-dominant enhancers (bottom) show strong enrichment for H3K4me1 and H3K27ac in EpiSCs. Similarly, Oct4 binding at promoter-distal enhancers is significantly reorganized between naive and primed stem cells. Oct4 localization is directed by state-specific regulatory partners: Oct4 interacts and cobinds with specific transcription factors in naive ESCs, such as Esrrb, and with other distinct factors in primed EpiSCs, such as Otx2. Whereas naive-dominant enhancers are lost in transition to primed stem cells, Factor et al. report low but detectable enhancer histone signals at primed-dominant “seed enhancers” in the naive state.
Indeed, Otx2 expression levels and genomic binding events were further shown to be significantly enriched in primed stem cells, and primed state-specific Otx2 binding sites represent regions cobound by Oct4 that were previously inaccessible to this master regulatory factor in naive ESCs (Buecker et al., 2014). The Fgf5 locus, for example, is activated in the postimplantation epiblast and is regulated by a cluster of distal enhancers bound by Otx2 and Oct4 specifically in the primed stem cell state. Similar to seed enhancers, this enhancer cluster is reminiscent of large enhancer regulatory domains or superenhancers, and it appears active specifically after the transition from naive to primed pluripotency. However, in contrast to seed enhancers, nuclease sensitivity and enhancer histone signals were not detected at these sites in ESCs, and this appears to be the case among most primed-specific Otx2/Oct4 binding sites. Whether Otx2/Oct4-bound enhancers are distinct from seed enhancers, or whether this discrepancy is an artifact arising from differences in experimental systems or methodologies in data analysis, is not currently evident.
Nonetheless, these data shed light on a novel mechanism underlying cell-state-specific regulatory circuitries important for defining pluripotency and lineage specification and commitment. When considered in combination with additional recent reports, this mechanism likely represents a fundamental paradigmfor cell-type-specific expression patterns and cellular responses to signaling pathways. Genome-wide mapping of enhancer activity in Drosophila, for example, revealed tissue-specific localization patterns for the ecdysone receptor (EcR) in response to hormone signaling in distinct cell types (Shlyueva et al., 2014). Similar to results for Oct4, differential EcR partner motifs defined cell-type-specific target enhancers that, in most cases, represent previously inaccessible chromatin sites. Meanwhile, large-scale comparisons of DNA-binding and protein interactions across distinct human cell lines similarly revealed tissue-specific colocalization patterns dynamically regulated across conditions and cell types (Xie et al., 2013). The mechanisms that regulate protein-protein interaction networks to effect changes in cooperative transcription factor binding, as well as understanding how inaccessible regions of the genome are made accessible or otherwise regulated, are central questions for future research, and the answers to these questions have important consequences for our understanding of the regulation of pluripotent states.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Cell Stem Cell. 2014;14 doi: 10.1016/j.stem.2014.04.003. this issue, ■ ■ ■ – ■ ■ ■. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Factor D, Corradin O, Zentner GE, Saiakhova A, Song L, Chenoweth JG, McKay RD, Crawford GE, Scacheri PC, Tesar PJ. Cell Stem Cell. 2014;14 doi: 10.1016/j.stem.2014.05.005. this issue, ■ ■ ■ – ■ ■ ■. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, vanBueren KL, Chines PS, Narisu N, Black BL, et al. NISC Comparative Sequencing Program; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors; NISC Comparative Sequencing Program Authors. Proc. Natl. Acad. Sci. USA. 2013;110:17921–17926. [Google Scholar]
- Radzisheuskaya A, Chia Gle B, dos Santos RL, Theunissen TW, Castro LF, Nichols J, Silva JC. Nat. Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stelzer C, Gerlach D, Yáñez-Cuna JO, Rath M, Boryń LM, Arnold CD, Stark A. Mol. Cell. 2014;54:180–192. doi: 10.1016/j.molcel.2014.02.026. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Xie D, Boyle AP, Wu L, Zhai J, Kawli T, Snyder M. Cell. 2013;155:713–724. doi: 10.1016/j.cell.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]