Significance
The diabetes drugs metformin and phenformin have interesting anticancer properties including the selective inhibition of cancer stem cells (CSCs). We show that these drugs (i) have remarkably similar metabolic profiles, (ii) reduce the tricarboxylic acid cycle and selected glyolytic intermediates during transformation, providing physiological evidence that mitochondrial complex I is a target, and (iii) have very different effects during transformation and in CSCs. These observations provide insight into the metabolic effects of these drugs in cancer contexts and their selective effects in CSCs that underlie potential cancer treatments.
Keywords: glycolysis, metabolism, cancer metabolism, metabolic profiling
Abstract
Metformin, a first-line diabetes drug linked to cancer prevention in retrospective clinical analyses, inhibits cellular transformation and selectively kills breast cancer stem cells (CSCs). Although a few metabolic effects of metformin and the related biguanide phenformin have been investigated in established cancer cell lines, the global metabolic impact of biguanides during the process of neoplastic transformation and in CSCs is unknown. Here, we use LC/MS/MS metabolomics (>200 metabolites) to assess metabolic changes induced by metformin and phenformin in an Src-inducible model of cellular transformation and in mammosphere-derived breast CSCs. Although phenformin is the more potent biguanide in both systems, the metabolic profiles of these drugs are remarkably similar, although not identical. During the process of cellular transformation, biguanide treatment prevents the boost in glycolytic intermediates at a specific stage of the pathway and coordinately decreases tricarboxylic acid (TCA) cycle intermediates. In contrast, in breast CSCs, biguanides have a modest effect on glycolytic and TCA cycle intermediates, but they strongly deplete nucleotide triphosphates and may impede nucleotide synthesis. These metabolic profiles are consistent with the idea that biguanides inhibit mitochondrial complex 1, but they indicate that their metabolic effects differ depending on the stage of cellular transformation.
Altered metabolism is a hallmark of malignantly transformed cells. Cancer risk is linked to metabolic syndrome, a disease state that includes obesity, type 2 diabetes, high cholesterol, and atherosclerosis. Retrospective studies of type 2 diabetes patients treated with metformin, the most widely prescribed antidiabetic drug, show a strong correlation between drug intake and reduced tumor incidence or reduced cancer-related deaths (1–4).
In the breast lineage, metformin inhibits growth of cancer cell lines (5–7), blocks transformation in a Src-inducible cell system (8, 9), and selectively inhibits the growth of cancer stem cells (CSCs) (8). As a consequence of its selective effects on CSCs, combinatorial therapy of metformin and standard chemotherapeutic drugs (doxorubicin, paclitaxel, and cisplatin) increases tumor regression and prolongs remission in mouse xenografts (8, 10). In addition, metformin can decrease the chemotherapeutic dose for prolonging tumor remission in xenografts involving multiple cancer types (10).
Phenformin, a related biguanide and formerly used diabetes drug, acts as an anticancer agent in tumors including lung, lymphoma, and breast cancer with a greater potency than metformin. Phenformin mediates antineoplastic effects at a lower concentration than metformin in cell lines, a PTEN-deficient mouse model, breast cancer xenografts, and drug-induced mitochondrial impairment (11–14). The chemical similarities of these biguanides, as well as their similar effects in diabetes and cancer, have led to the untested assumption that phenformin is essentially a stronger version of metformin.
In a Src-inducible model of cellular transformation and CSC formation, multiple lines of evidence suggest that metformin inhibits a signal transduction pathway that results in an inflammatory response (15). In the context of atherosclerosis, metformin inhibits NF-κB activation and the inflammatory response via a pathway involving AMP kinase (AMPK) and the tumor suppressor PTEN (16, 17). As metformin alters energy metabolism in diabetics, we speculated that metformin might block a metabolic stress response that stimulates the inflammatory pathway (15). However, very little is known about the metabolic changes that inhibit the inflammatory pathway.
Previous studies on metformin-induced metabolic effects in cancer have focused on single metabolic alterations or pathways in already established cancer cell lines. Metformin leads to activation of AMPK, which plays a key role in insulin signaling and energy sensing (18). Metformin can reduce protein synthesis via mTOR inhibition (19). In addition, metformin may directly impair mitochondrial respiration through complex I inhibition and has been described to boost glycolysis as a compensation mechanism (14, 20). In this regard, lactic acidosis can be a side effect of metformin and phenformin treatment of diabetic patients, presumably because inhibition of complex I prevents NADH oxidation, thereby leading to a requirement for cytosolic NADH to be oxidized by the conversion of pyruvate to lactate. There is some knowledge about the metabolic effects of metformin (21, 22), but very little is known about the specific metabolic alterations linking biguanides to inhibition of neoplastic transformation.
Here, we perform a metabolomic analysis on the effects of metformin and phenformin in a Src-inducible model of transformation and in CSCs. This inducible model permits an analysis of the transition from nontransformed to transformed cells in an isogenic cell system and hence differs from analyses of already established cancer cell lines. We studied CSCs to address why this population, which is resistant to standard chemotherapeutics and hypothesized to be a major reason for tumor recurrence, is selectively inhibited by metformin. Our results indicate the metabolic effects of metformin and phenformin are remarkably similar to each other, with only a few differences. Both biguanides dramatically decrease tricarboxylic acid (TCA) cycle intermediates in the early stages of transformation, and they inhibit the boost in select glycolytic intermediates that normally occurs with transformation along with increases in glycerol 3-phosphate and lactate, which are metabolites branching from glycolysis. Unexpectedly, in CSCs, biguanides have only marginal effects on glycolytic and TCA cycle metabolites, but they severely decrease nucleotide triphosphates. These detailed metabolic analyses provide independent support for the idea that metformin inhibits mitochondrial complex 1 (14, 20), and they indicate that the metabolic effects of biguanides depend on the stage of the cellular transformation.
Results
Phenformin Inhibits Morphological Transformation of ER-Src Cells at a Lower Concentration Than Metformin.
We previously showed metformin inhibits cellular transformation using an inducible breast cancer model (8, 9). This model involves a derivative of the spontaneous immortalized breast epithelial cell line MCF-10A (23) expressing an ER-Src fusion gene that consists of the v-Src oncogene and the ligand-binding domain of the estrogen receptor. Activation of Src via tamoxifen results in morphological transformation and the ability to form colonies in anchorage-independent growth assays (9, 24).
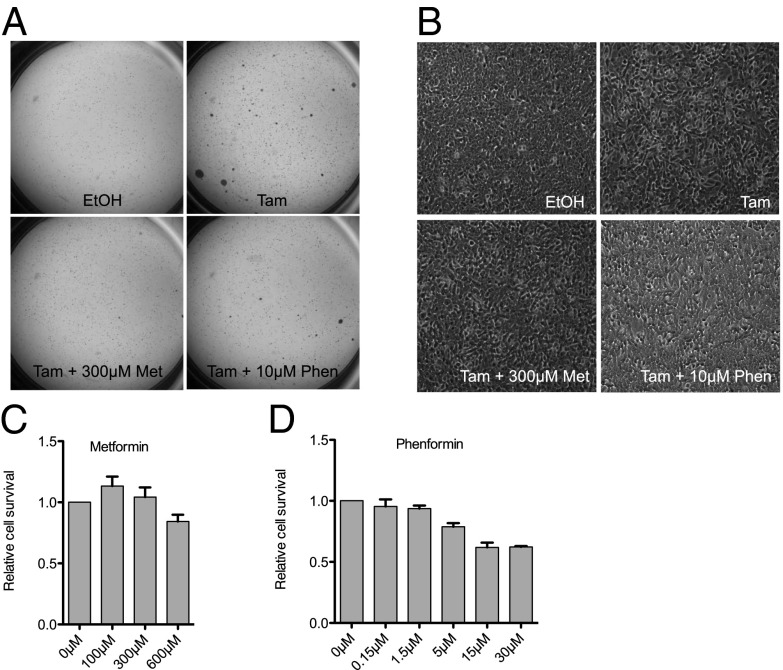
As phenformin appears to be a more potent anticancer drug than metformin in various cell types (11, 12, 25), we first asked whether the related biguanide phenformin could achieve this same effect with increased potency. Indeed, soft agar assays showed that treatment with metformin or phenformin for 24 h during tamoxifen-induced Src activation reduces the number of colonies to that of cells treated only with vehicle (Fig. 1A). Additionally, morphologic transformation due to loss of contact inhibition is suppressed by both biguanides. Phenformin shows a comparable, and perhaps stronger, effect, even though it is used at a 30-fold lower concentration than metformin (Fig. 1B). In accordance with the clinical data for diabetes treatment, phenformin is both more potent and more toxic than metformin. At the effective concentration, phenformin (10 µM) shows slightly reduced cell viability, whereas metformin (300 µM) does not affect cell survival (Fig. 1 C and D).
Fig. 1.
Metformin and phenformin block malignant transformation. ERSrc cells were treated with EtOH, tamoxifen, tamoxifen + metformin, or tamoxifen + phenformin for 24 h, and soft agar assays (A) and morphology assays (B) were performed. Cell viability via MTT was measured after 24-h treatment of ERSrc cells with different concentrations of metformin (C) or phenformin (D). Error bars indicate SEM.
Induction of Cellular Transformation Is Associated with Metabolic Changes Typical of Cancer Cells.
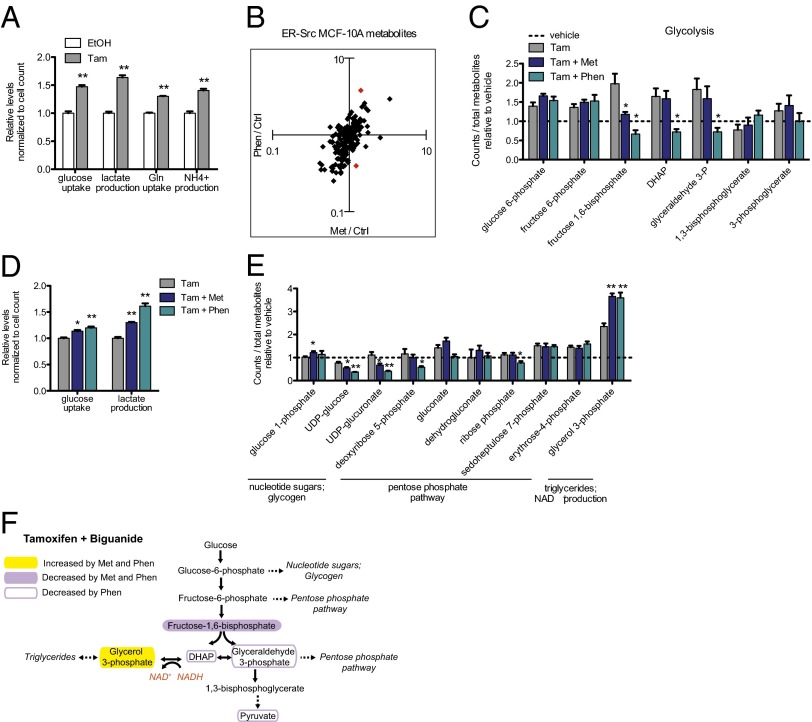
Fully transformed cancer cells commonly display the Warburg effect, characterized by a high rate of glucose consumption and lactate production (26). Additionally, many transformed cells consume high amounts of glutamine as an additional nutrient source and consequently generate a large amount of ammonium that is secreted from the cell. In accord with these observations, analysis of media from induced ER-Src cells reveals a significant increase in glucose and glutamine uptake 24 h after Src activation (Fig. 2A). In addition, ammonium and lactate production are increased following Src induction (Fig. 2A). This switch to typical tumor cell metabolism that occurs within only 24 h of Src activation validates our inducible model for metabolic analysis of cellular transformation.
Fig. 2.
Metformin and phenformin alter the metabolic state of the transformation process, specifically preventing an increase in some glycolytic intermediates. Twenty-four hours after Src induction by tamoxifen (Tam) or control treatment with ethanol (EtOH), glucose and glutamine uptake and lactate and ammonium production were measured in the media of MCF10A ERSrc cells (A), n = 3. To identify metformin vs. phenformin differences, fold changes of both drugs for all LS-MS/MS metabolites over the tamoxifen-only sample were determined and ratios were calculated. Red dots indicate differentially regulated metabolites outside of a 99.7% CI generated over all ratios (B). Relative levels of glycolytic intermediates measured with LC-MS/MS for tamoxifen ± metformin or phenformin and vehicle (ethanol)-treated samples (C), n = 4. Glucose uptake and lactate production were measured in the media 24 h after Src induction ± metformin (D), n = 3. Relative levels of metabolites branching from glycolysis with tamoxifen ± metformin or phenformin and vehicle-treated samples (E), n = 4. Diagram of glycolytic intermediates altered by metformin or phenformin (F) treatment. The role of glycerol 3-phosphate production in regeneration of NAD+ is indicated in orange. For all panels, *P < 0.05 and **P < 0.01 compared with control sample. Error bars indicate SEM.
Metformin and Phenformin Have Very Similar, but Nonidentical, Metabolic Profiles During Cellular Transformation.
Based on their chemical relationship and a few similar effects in diabetes and cancer, it is generally assumed that phenformin is a stronger version of metformin. To address this issue in more detail and to determine the global metabolic impact of metformin and phenformin on cells undergoing transformation, we measured more than 200 metabolites by LC/MS/MS 24 h after tamoxifen treatment in the presence or absence of biguanides (Table S1). Fold change values for each biguanide vs. tamoxifen-only treatment were determined for all metabolites (Fig. 2B). Taking into account the consistently stronger potency of phenformin over metformin, the vast majority of metabolites behave similarly with the two biguanides. Only two metabolites appear to be differentially affected (P < 0.003), namely serine and anthranilate, and a similar analysis in a stably transformed breast cancer cell line, CAMA-1, reveals five differentially affected metabolites.
Metformin and Phenformin Prevent the Tamoxifen-Induced Boost in Glycolytic Intermediates.
To identify metabolic pathways altered by biguanides during the initial stages of transformation, we focused on significantly changed metabolites in biguanide-treated samples compared with tamoxifen-only treatment (P < 0.05). As expected, levels of multiple glycolytic intermediates are increased during transformation (Fig. 2C and Fig. S1A). Interestingly, increases in glycolytic intermediates are only observed for the early part of the pathway. All intermediates preceding 1,3 bisphosphoglycerate are increased during transformation, whereas this and all subsequent intermediates including pyruvate are not.
Addition of either biguanide causes a decrease in specific glycolytic intermediates, but not in the entire pathway (Fig. 2C). With phenformin treatment, three successive intermediates in the middle of glycolysis—fructose 1,6-bisphosphate, dihydroxyacetone phosphate (DHAP), and glyceraldehyde-3-phosphate—are significantly reduced compared with tamoxifen-only treatment and even lower than the untransformed state (dotted line). With metformin treatment, fructose 1,6-biphosphate is significantly reduced, albeit to a lesser extent than with phenformin, and DHAP and glyceraldehyde-3-phosphate are slightly reduced. Neither biguanide has an effect on the earliest glycolytic intermediates that are increased during transformation nor on later glycolytic intermediates whose levels are unaffected during transformation.
The decrease in specific glycolytic intermediates is not due to a defect in glucose uptake. Analysis of cell culture media 24 h after tamoxifen treatment shows that phenformin and (to a lesser extent) metformin actually increase glucose uptake (Fig. 2D), consistent with previous reports that metformin increases the dependency on glycolysis (4). Lactate production is also increased in the presence of biguanides, again with phenformin having the stronger effect (Fig. 2D). Thus, despite promoting increased glucose consumption and lactate production, metformin and phenformin ultimately decrease specific glycolytic intermediates, suggesting rapid glucose processing that depletes intermediates from key junctions in glycolysis.
Biguanide Treatment Increases Glycerol 3-Phosphate and Lactate Production During Transformation.
We asked whether decreased glycolytic intermediates in the presence of biguanides during transformation might be due to increased partitioning to metabolites branching from glycolysis. Surprisingly, although a number of anabolic precursors of the pentose phosphate pathway, nucleotide sugars, or glycogen synthesis are depleted with biguanide treatment, glycerol 3-phosphate is increased by both metformin and phenformin (Fig. 2 E and F). Glycerol 3-phosphate, which is generated from the glycolytic intermediate DHAP, can serve as an intermediary between glucose and lipid metabolism. However, analysis of 14C-glucose incorporation into the lipid fraction reveals that biguanides instead decrease de novo lipogenesis (Fig. S1B), indicating that glycerol 3-phosphate levels are increased for an alternate purpose. As conversion of DHAP to glycerol 3-phosphate regenerates NAD+ from NADH (shown in orange, Fig. 2G), we hypothesize increased glycerol 3-phosphate levels promote NAD+ regeneration, which is required to maintain glycolysis.
Although glycerol 3-phosphate is increased with both drugs, UDP-glucose and UDP-glucuronate, branching out of glycolysis at the glucose-6-phosphate step via glucose 1-phosphate, are decreased. As UDP-glucose is a metabolic precursor for glycogen synthesis, biguanides may direct glycolytic intermediates away from glycogen synthesis, which is a nutrient storage pathway that normally occurs in cells during energy abundance (Fig. 2E). UDP-glucuronate feeds in the pentose phosphate pathway, but this pathway is not significantly affected by either biguanide (Fig. 2E).
Metformin and Phenformin Decrease the Level of TCA Cycle Intermediates.
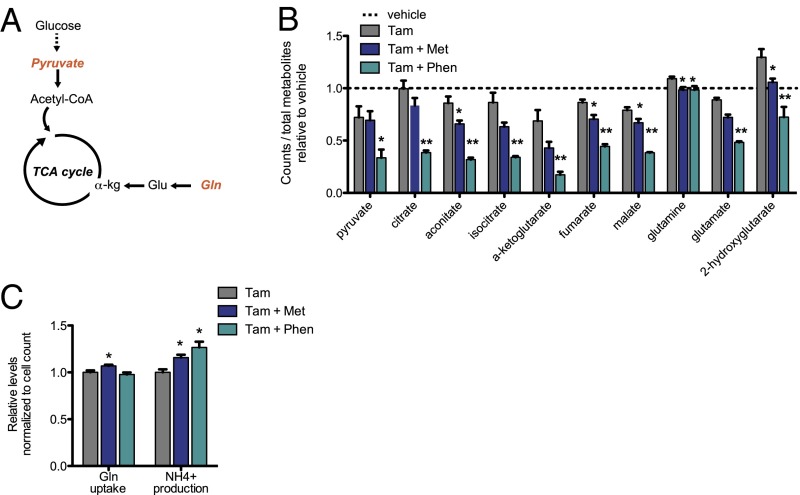
In addition to boosting glycolysis, cancer cells must allocate nutrients toward the TCA cycle to generate ATP and intermediates necessary for macromolecule biosynthesis (27). Along with glucose-derived pyruvate, glutamine flux contributes substantially to fueling the TCA cycle in many cancer cells. Strikingly, nearly all TCA cycle metabolites are strongly decreased with both metformin and phenformin (Fig. 3 A and B). Decreased levels of TCA cycle intermediates correlate with decreased pyruvate (with phenformin), increased shunting of glucose-derived carbons toward lactate, and decreased levels of glutamate and (marginally) glutamine (Fig. 3B). Glutamine uptake is not decreased by biguanides, indicating there is no defect in glutamine transport across the cell membrane (Fig. 3C). Ammonium production is increased by biguanide treatment (Fig. 3C), suggesting increased utilization of glutamine as an attempt to refuel the TCA cycle via anaplerosis.
Fig. 3.
Metformin and phenformin decrease TCA cycle intermediates. Schematic of the TCA cycle with key molecules fueling the cycle indicated in orange (A). Relative levels of TCA cycle intermediates following treatment with tamoxifen ± metformin or phenformin as measured by LC-MS/MS (B), n = 4. Glutamine uptake and NH4+ production were measured in the media 24 h after Src induction ± metformin or phenformin (C), n = 3. *P < 0.05 and **P < 0.01 compared with tamoxifen treatment alone. Error bars indicate SEM.
Our results appear to differ from a previous report in prostate cancer suggesting that metformin does not inhibit the TCA cycle but rather alters the fuel source by decreasing the oxidation of glucose-derived pyruvate and increasing glutamine anaplerosis (28). We considered the possibility that this apparent difference in TCA cycle inhibition might be due to analysis of stably transformed cancer cells as opposed to cells early in the process of transformation. However, biguanide treatment of a stably transformed breast cancer cell line (CAMA-1) leads to a decrease in TCA cycle intermediates (Fig. S2), suggesting that the metabolic reduction of the TCA cycle by biguanides may be important for inhibiting transformation.
Biguanides Induce a CSC-Specific Depletion of Nucleotide Triphosphates.
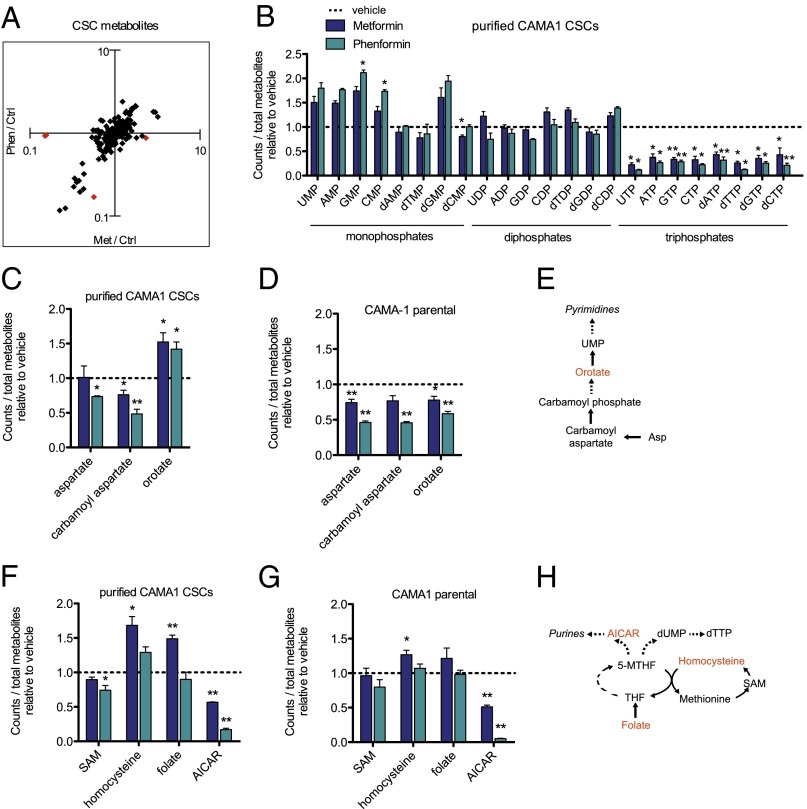
Metformin selectively kills breast CSCs and, as a consequence, can act together with standard chemotherapeutic drugs to increase tumor regression and prolong relapse in mouse xenografts (8, 10). CSCs represent a minor population of cancer cells either in primary tissue or cancer cell lines, but they are enriched in mammospheres that form when cultivated in nonadherent and nondifferentiating conditions (29, 30). We performed metabolic profiling on 7-d-old mammospheres from the transformed breast cancer cell line CAMA-1 that were treated with metformin, phenformin, or vehicle for 24 h; as a control, we analyzed the CAMA-1 parental cell line. In accordance with observations during cellular transformation (Fig. 2B), the vast majority of metabolites are similarly regulated with both drugs (Fig. 4A).
Fig. 4.
Metformin and phenformin alter the metabolic state of breast cancer stem cells and deplete NTPs. Fold change comparisons identify metabolites differently regulated in metformin vs. phenformin samples measured by LC-MS/MS after 24 h of treatment in breast CSCs. Red diamonds represent differentially altered metabolites (A). Relative levels of nucleoside monophosphates, diphosphates, and triphosphates in metformin- or phenformin-treated CAMA-1 CSCs compared with untreated CSCs (B), n = 4. Relative levels of pyrimidine precursors in CAMA-1 CSCs (C) and parental CAMA-1 (D) treated with metformin or phenformin, n = 4. Schematic of key metabolites in pyrimidine synthesis (E). Relative levels of folate metabolites in CAMA-1 CSCs (F) and parental CAMA-1 (G) treated with metformin or phenformin for 24 h, n = 4. Schematic depicting of folate and regeneration of 5-MTHF in purine and dTTP synthesis (H). *P < 0.05 and **P < 0.01 compared with vehicle control. Error bars indicate SEM.
Surprisingly, the degree of the metabolic effects induced by biguanides differs considerably between the transformation and CSC systems. CSCs treated with biguanides show only marginal effects on glycolytic and TCA cycle intermediates (Fig. S3 A and B). In contrast and unexpectedly, levels of all ribonucleotide and deoxyribonucleotide triphosphates (NTPs) are strongly decreased on biguanide treatment, with the effects of phenformin being stronger than metformin. Conversely, levels of all ribonucleotide and some deoxyribonucleotide monophosphates trend toward being increased by biguanide treatment, whereas little if any effect is seen on nucleotide diphosphates (Fig. 4B). Importantly, this depletion of NTPs by metformin and phenformin occurs specifically in CSCs and not in the parental CAMA-1 cell line (Fig. S4A). Although isolated effects on nucleotide metabolism are observed during tamoxifen-induced transformation (Fig. S4B), the magnitude and extent of NTP pool depletion are much greater in CSCs. Biguanide treatment also increases the levels of early precursors in nucleotide metabolic pathways, including orotate (Fig. 4 C and E). Although the orotate precursors aspartate and carbamoyl aspartate are similarly regulated in CSCs and parental CAMA-1 cell line, the increased orotate level is specific to CSCs (Fig. 4 C and D and Fig. S4A). These observations indicate that CSCs have distinct responses to biguanides and, in particular, appear to be defective in converting nucleotide precursors to NTPs.
Folate Metabolism and Aminoimidazole Carboxamide Ribonucleotide Levels Are Altered by Biguanides.
Analysis of metabolites that feed into purine and pyrimidine synthesis reveals that CSCs treated with metformin, but not phenformin, have a buildup of folate (Fig. 4 F and H). Folate is enzymatically reduced to tetrahydrofolate (THF) and subsequently converted to N5-methyl-THF (5-MTHF) to serve as a methyl donor for both purine and dTTP synthesis. 5-MTHF can also serve as a 1-carbon donor to homocysteine to produce methionine, and conversely, homocysteine can be regenerated from methionine via the intermediate S-adenosyl-methionine (SAM). In both CSCs and parental CAMA1 cells, we observed increased folate and homocysteine with metformin treatment, possibly indicating a defect in entry of folate into the THF-cycling pathway for nucleotide synthesis. Interestingly altered folate metabolism in the Escherichia coli food source has been implicated in metformin-mediated benefits in Caenorhabditis elegans (31). Additionally, both phenformin and metformin decrease aminoimidazole carboxamide ribonucleotide (AICAR), an intermediate required for purine synthesis (Fig. 4 F–H). As decreased AICAR is observed in both CSCs and the parental line, the CSC-specific depletion of NTPs suggests that this population may have greater NTP utilization and hence be more sensitive to AICAR levels.
Discussion
Phenformin and Metformin Have Remarkably Similar Metabolic Profiles, with Phenformin Having Increased Potency.
It has been assumed that phenformin is essentially a stronger version of metformin, but the evidence is limited to their chemical similarity and a few common effects in diabetes and cancer contexts. Our detailed metabolic analysis (>200 metabolites) indicates that the metabolic profiles of metformin and phenformin are remarkably similar, with phenformin causing stronger effects even when used at a 30-fold lower concentration. At least in part, this likely reflects the slightly greater lipophilic character of phenformin relative to metformin that facilitates drug uptake. Although both biguanides use the OCT1 transporter for cellular entry, phenformin may be more readily taken up to reach its cellular targets (32, 33). However, it is possible that phenformin may also have a stronger effect on the cellular target(s) per se.
Despite the remarkably similar metabolic profiles, a very small number of metabolites are uniquely altered by only one biguanide. Although metabolites that appear to be specifically affected by phenformin might simply reflect a quantitative difference between the two biguanides, metabolites such as anthranilate that are only affected by metformin cannot be explained in such a manner. These rare examples, which could be considered to occur from off-target effects of the drugs, have the potential to differentially affect medical outcomes. Nevertheless, the remarkably similar metabolic profiles, together with other lines of evidence, suggest that phenformin be considered as a more powerful alternative to metformin as an anti-cancer agent.
Biguanides Lead to a Depletion of Select Glycolytic and All TCA Cycle Intermediates During Cellular Transformation.
The process of neoplastic transformation creates a demand for increased synthesis of macromolecules, and this is accommodated by increased uptake of glucose and glutamine from the medium. Tamoxifen-induced transformation causes increased levels of all metabolites involved in glycolysis up to and including the step mediated by triose phosphate isomerase, presumably a consequence of increased glucose uptake. However, glycolytic intermediates after this step are not increased, even though the cells produce more lactate. It is possible that increased lactate production is not simply due to increased flux through the entire glycolytic pathway but rather involves differences in the competition for pyruvate to be converted to lactate or to citrate for entry into the TCA cycle (see below). Alternatively, the latter glycolytic intermediates may not accumulate due to rapid processing toward lactate.
Interestingly, the biguanides selectively decrease three consecutive metabolites in the middle of the glycolytic pathway. For each drug, the levels of these three metabolites are reduced to a comparable extent, an observation that could be explained by a decrease in the step that converts fructose 6-phosphate to fructose 1,6-diphosphate. Alternatively, it might reflect biguanide-induced partitioning of glucose-derived carbons toward glycerol 3-phosphate, a metabolite whose level is significantly increased by metformin and phenformin.
Both biguanides cause a quantitatively similar decrease in all TCA metabolites tested, strongly suggesting decreased flux into the TCA cycle. Reduced levels of some TCA metabolites have been observed previously with metformin treatment (22). There are two explanations, which are not mutually exclusive, to explain the effects on the TCA cycle. First, biguanides lead to decreased levels of pyruvate and increased levels of lactate production, presumably by increasing the conversion of pyruvate to lactate. As pyruvate directly leads into the TCA cycle, lowering its intracellular levels is expected to reduce the levels of all TCA metabolites. Second, biguanides decrease the levels of glutamate, a metabolite that leads directly into the TCA cycle on conversion to α-ketoglutarate. Thus, biguanides may decrease input into the TCA cycle by inhibiting precursors generated either by carbon or nitrogen metabolism (pyruvate and glutamate, respectively), and hence reduce ATP production and anabolic metabolites necessary for cell growth that are derived from the TCA cycle.
Transformation in the inducible ER-Src model is mediated by an inflammatory response that depends on NF-κB and STAT3 (9, 24), and metformin blocks this response by an unknown mechanism (15). Our results indicate that this inflammatory response is associated with increased glucose uptake and increased glycolytic intermediates, although the mechanistic connection is unknown. Further, they suggest that the biguanide-mediated effects on metabolism effectively decrease the inflammatory stimuli or signal transduction pathway that is required for transformation.
Biguanides Differently Affect the Transformation Process and CSCs, Suggesting Unique Metabolic States of These Two Systems.
As the biguanides presumably affect the same target(s) in all cells, we were surprised to find different metabolic profiles during the transformation process and in CSCs. Although TCA cycle and glycolysis were mainly affected during transformation, the biguanides more specifically affected NTP levels in the CSCs. The decreased NTP levels in CSCs are likely to limit the availability for energetics, RNA, DNA, and biosynthesis of cofactors such as FAD, NADH, and CoA. In addition, metformin causes a defect in folate utilization in CSCs, as evidenced by increased levels of folate pathway metabolites. Consistent with this observation, the folate derivative 5-formimino-tetrahydrofolate increases in metformin-treated breast cancer cell lines (34), and patients treated with metformin have a higher serum level of homocysteine, a metabolite involved in folate cycling (35).
The differential metabolic effects of biguanides strongly suggest that CSCs have a distinct metabolic state compared with other cancer cells. We speculate that CSCs might have reduced requirements for glycolysis and the TCA cycle, perhaps analogous to yeast cells growing on nonfermentative carbon sources, and increased dependence for NTPs, perhaps due to a reduced energy state. It is also tempting to speculate that the severe defect in NTP levels (and perhaps the defect in folate metabolism) underlies the increased sensitivity of CSCs to metformin treatment compared with typical cancer cells. More generally, our observations suggest that the metabolic effects of metformin may differ considerably among cancer cell types and states.
Evidence for Mitochondrial Complex 1 Being a Target of Biguanides and Future Use of Metabolic Profiles.
The direct target(s) of metformin is unknown, although mitochondrial complex I is the best candidate at present (14, 20). A defect in complex I will decrease oxidation of NADH to NAD+, a critical reaction to maintain the function of the TCA cycle, and ultimately inhibit oxidative phosphorylation leading to ATP. Several observations in this paper are consistent with the hypothesis that biguanides target complex I. First, biguanides decrease the levels of all TCA intermediates, suggesting an overall defect in TCA cycle function due to the relative inability to oxidize NADH to NAD+. Second, decreased TCA cycle function is likely to result in preferential conversion of pyruvate to lactate as opposed to entering the TCA cycle, and this is observed in biguanide-treated cells. In this regard, inhibition of complex I by rotenone boosts lactate production (36) and reduces TCA cycle intermediates (37), allowing an alternate way to produce ATP when the electron transport chain is not functional. Third, the increased lactate production and increased levels of glycerol-3-phosphate may reflect stimulation of two key reactions that effectively oxidize NADH to NAD+, thereby compensating for lower levels of NAD+ due to decreased complex I activity. Fourth, the strong decrease in NTPs in CSCs suggests a major defect in energy state necessary, likely reflecting a defect in oxidative phosphorylation.
Our detailed metabolic profiling provides independent support for the idea that complex I is a target of biguanides. As a complement to biochemical studies, examining metabolic profiles conferred by drugs with known targets (e.g., rotenone) or caused by functional inhibition or deletion of an individual gene should be very helpful for identifying the physiological target of biguanides. In addition, metabolic profiles conferred by biguanide treatment should be very useful for identifying new drugs with similar metabolic properties, and such drugs might have potential for treatment of diabetes or cancer.
Materials and Methods
Cell and culture conditions, tamoxifen-induced transformation of MCF-10A ER-Src cells, metformin (300 µM) and phenformin (10 µM) treatment (24), mammosphere culture (10), metabolic profiling by target LC MS (38), basal lipogenesis analysis (39), analysis using the BioProfile FLEX analyzer (Nova Biomedicals) for levels of glucose, lactate, glutamine, and ammonia (40), and statistical analyses were performed as described previously and in more detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Min Yuan for metabolomics technical assistance. A.J. was supported by a fellowship from the Postdoc Programme of the German Academic Exchange Service (DAAD), N.J.G. by National Science Foundation Graduate Research Fellowship Grant 1000087636, K.N.G.-H. by the Paul & Daisy Soros Fellowship for New Americans, M.C.H. by National Institutes of Health (NIH) Grant AG032375, the American Cancer Society New Scholar Award, and the Glenn Foundation for Medical Research, and K.S. by NIH Grant CA 107486.
Footnotes
The authors declare no conflict of interest.
1A.J. and N.J.G. contributed equally to this work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409844111/-/DCSupplemental.
References
- 1.Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330(7503):1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiralerspong S, et al. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol. 2009;27(20):3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowling RJ, Niraula S, Stambolic V, Goodwin PJ. Metformin in cancer: Translational challenges. J Mol Endocrinol. 2012;48(3):R31–R43. doi: 10.1530/JME-12-0007. [DOI] [PubMed] [Google Scholar]
- 4.Pollak MN. Investigating metformin for cancer prevention and treatment: The end of the beginning. Cancer Discov. 2012;2(9):778–790. doi: 10.1158/2159-8290.CD-12-0263. [DOI] [PubMed] [Google Scholar]
- 5.Alimova IN, et al. Metformin inhibits breast cancer cell growth, colony formation and induces cell cycle arrest in vitro. Cell Cycle. 2009;8(6):909–915. doi: 10.4161/cc.8.6.7933. [DOI] [PubMed] [Google Scholar]
- 6.Liu B, et al. Metformin induces unique biological and molecular responses in triple negative breast cancer cells. Cell Cycle. 2009;8(13):2031–2040. doi: 10.4161/cc.8.13.8814. [DOI] [PubMed] [Google Scholar]
- 7.Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66(21):10269–10273. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69(19):7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch HA, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17(4):348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iliopoulos D, Hirsch HA, Struhl K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res. 2011;71(9):3196–3201. doi: 10.1158/0008-5472.CAN-10-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, et al. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem J. 2008;412(2):211–221. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- 12.Appleyard MV, et al. Phenformin as prophylaxis and therapy in breast cancer xenografts. Br J Cancer. 2012;106(6):1117–1122. doi: 10.1038/bjc.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dykens JA, et al. Biguanide-induced mitochondrial dysfunction yields increased lactate production and cytotoxicity of aerobically-poised HepG2 cells and human hepatocytes in vitro. Toxicol Appl Pharmacol. 2008;233(2):203–210. doi: 10.1016/j.taap.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci USA. 2013;110(3):972–977. doi: 10.1073/pnas.1221055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li SN, et al. Metformin inhibits nuclear factor kappaB activation and decreases serum high-sensitivity C-reactive protein level in experimental atherogenesis of rabbits. Heart Vessels. 2009;24(6):446–453. doi: 10.1007/s00380-008-1137-7. [DOI] [PubMed] [Google Scholar]
- 17.Kim SA, Choi HC. Metformin inhibits inflammatory response via AMPK-PTEN pathway in vascular smooth muscle cells. Biochem Biophys Res Commun. 2012;425(4):866–872. doi: 10.1016/j.bbrc.2012.07.165. [DOI] [PubMed] [Google Scholar]
- 18.Hardie DG. Role of AMP-activated protein kinase in the metabolic syndrome and in heart disease. FEBS Lett. 2008;582(1):81–89. doi: 10.1016/j.febslet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Larsson O, et al. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci USA. 2012;109(23):8977–8982. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Mir MY, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275(1):223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 21.Scotland S, et al. Mitochondrial energetic and AKT status mediate metabolic effects and apoptosis of metformin in human leukemic cells. Leukemia. 2013;27(11):2129–2138. doi: 10.1038/leu.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakikhani M, et al. Alterations in cellular energy metabolism associated with the antiproliferative effects of the ATM inhibitor KU-55933 and with metformin. PLoS ONE. 2012;7(11):e49513. doi: 10.1371/journal.pone.0049513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–6086. [PubMed] [Google Scholar]
- 24.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosilio C, et al. The metabolic perturbators metformin, phenformin and AICAR interfere with the growth and survival of murine PTEN-deficient T cell lymphomas and human T-ALL/T-LL cancer cells. Cancer Lett. 2013;336(1):114–126. doi: 10.1016/j.canlet.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 27.Lunt SY, Vander Heiden MG. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 28.Fendt SM, et al. Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer Res. 2013;73(14):4429–4438. doi: 10.1158/0008-5472.CAN-13-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao MJ, et al. Enrichment of a population of mammary gland cells that form mammospheres and have in vivo repopulating activity. Cancer Res. 2007;67(17):8131–8138. doi: 10.1158/0008-5472.CAN-06-4493. [DOI] [PubMed] [Google Scholar]
- 30.Grimshaw MJ, et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10(3):R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabreiro F, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153(1):228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117(5):1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shitara Y, et al. Role of organic cation/carnitine transporter 1 in uptake of phenformin and inhibitory effect on complex I respiration in mitochondria. Toxicol Sci. 2013;132(1):32–42. doi: 10.1093/toxsci/kfs330. [DOI] [PubMed] [Google Scholar]
- 34.Corominas-Faja B, et al. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging (Albany, NY Online) 2012;4(7):480–498. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahin M, Tutuncu NB, Ertugrul D, Tanaci N, Guvener ND. Effects of metformin or rosiglitazone on serum concentrations of homocysteine, folate, and vitamin B12 in patients with type 2 diabetes mellitus. J Diabetes Complications. 2007;21(2):118–123. doi: 10.1016/j.jdiacomp.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Xu Q, Vu H, Liu L, Wang TC, Schaefer WH. Metabolic profiles show specific mitochondrial toxicities in vitro in myotube cells. J Biomol NMR. 2011;49(3-4):207–219. doi: 10.1007/s10858-011-9482-8. [DOI] [PubMed] [Google Scholar]
- 37.Basu SS, Blair IA. Rotenone-mediated changes in intracellular coenzyme A thioester levels: Implications for mitochondrial dysfunction. Chem Res Toxicol. 2011;24(10):1630–1632. doi: 10.1021/tx200366j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurent G, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50(5):686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.