Significance
Stomata are widespread in aerial part of plants as passages exchanging gas and water with environment. Therefore, stomata are crucial for photosynthesis as well as global carbon and water circulation. Auxin, as the first identified phytohormone, participates in many aspects of plant growth and development, but whether auxin regulates stomatal development is unknown. This study establishes that auxin negatively regulates stomatal development through MONOPTEROS (MP) repression of mobile peptide gene STOMAGEN expression in mesophyll cells, which is mediated by direct binding of MP to auxin response elements in the STOMAGEN promoter. This study advances our knowledge about the roles of auxin and the versatile regulator MP in plant growth and development.
Abstract
Plants, as sessile organisms, must coordinate various physiological processes to adapt to ever-changing surrounding environments. Stomata, the epidermal pores facilitating gas and water exchange, play important roles in optimizing photosynthetic efficiency and adaptability. Stomatal development is under the control of an intrinsic program mediated by a secretory peptide gene family—namely, EPIDERMAL PATTERNING FACTOR, including positively acting STOMAGEN/EPFL9. The phytohormone brassinosteroids and environment factor light also control stomatal production. However, whether auxin regulates stomatal development and whether peptide signaling is coordinated with auxin signaling in the regulation of stomatal development remain largely unknown. Here we show that auxin negatively regulates stomatal development through MONOPTEROS (also known as ARF5) repression of the mobile peptide gene STOMAGEN in mesophyll. Through physiological, genetic, transgenic, biochemical, and molecular analyses, we demonstrate that auxin inhibits stomatal development through the nuclear receptor TIR1/AFB-mediated signaling, and that MONOPTEROS directly binds to the STOMAGEN promoter to suppress its expression in mesophyll and inhibit stomatal development. Our results provide a paradigm of cross-talk between phytohormone auxin and peptide signaling in the regulation of stomatal production.
Auxin is the first identified phytohormone, which exerts multifaceted influences on plant growth and development, such as embryonic root initiation (1, 2), shoot apical meristem function (3), and floral primordia initiation (4). As a “molecular glue,” auxin facilitates the formation of its coreceptor complexes comprising F-box proteins (TIR1/AFBs) and AUXIN/INDOLE-3-ACETIC ACID proteins (AUX/IAAs), and subsequent AUX/IAAs ubiquitination and degradation by 26S proteasome, thus releasing auxin response factors (ARFs) from AUX/IAAs repression to regulate auxin-responsive gene expression by either activation or repression (2, 3, 5–11). Although many physiologic processes are reported to be regulated by auxin (1–4, 6, 12), the full understanding of the functions of this versatile phytohormone has not been reached.
Stomata, the pores flanked by a pair of guard cells, mainly constitute the epidermis of plant leaves together with trichomes and neighboring pavement cells that separate stomata to maintain the one-cell spacing rule (13, 14). As a gas and water passage between external environment and internal plant tissue, stomata play important roles in photosynthesis and global carbon and water circulation (15). Stomatal generation undergoes several stages, including meristemoid mother cell, meristemoid, guard mother cell, and guard cells, which is modulated by an intrinsic program (14) mainly involving putative peptide ligands [EPIDERMAL PATTERNING FACTOR (EPF) family] (16–20), membrane proteins (receptor-like protein TMM and receptor-like kinase ERECTA family) (21–23), MAPK cascades (protein kinase YDA, MKK4/5/7/9, and MPK3/6) (24–26), and transcription factors (bHLH and MYB type) (23, 27–31). EPF factors, the small secretory peptides, are proposed to act at the top of this hierarchical signaling pathway (16–20). Interestingly, the EPF family is comprised of members with completely opposite functions (32), such as negatively acting EPF1 and EPF2 (16, 17, 20) and positively acting STOMAGEN/EPFL9 (18). EPF1 and EPF2 are expressed in the epidermis, and their encoding peptides were recently shown to be ligands of ERECTA and TMM, negatively regulating stomatal development (16, 17, 19, 20). In contrast, STOMAGEN is expressed in mesophyll, and its encoding peptide then migrates to the epidermis where it is proposed to promote stomatal development by competitively inhibiting TMM-mediated signaling (18). Modulation of EPF expression could drastically alter stomatal development (16–18, 20), which might be due to cascade amplification from the top signals. Thus, it was proposed that EPFs are a novel class of peptide hormones (32, 33). Although intrinsic program regulating stomatal development has been well characterized, how the top signals from EPFs are regulated remains elusive.
Phytohormones and external stimuli, such as brassinosteroids (BRs), light, and carbon dioxide, are also involved in modulating stomatal production (13, 14, 34–36). Here we show that nuclear receptor-mediated auxin signaling negatively regulates stomatal development, and that ARF5/MONOPTEROS (MP) is involved in regulating this process. MP directly associates with the STOMAGEN promoter and represses STOMAGEN expression in an auxin response element (AuxRE)-dependent manner. The regulation of STOMAGEN by MP occurs in mesophyll, where photosynthesis mainly takes place, providing a possibility of specifically manipulating auxin signaling in mesophyll to coordinate stomatal development with photosynthesis without disturbing the whole-body plan.
Results
Nuclear Receptor-Mediated Auxin Signaling Inhibits Stomatal Development.
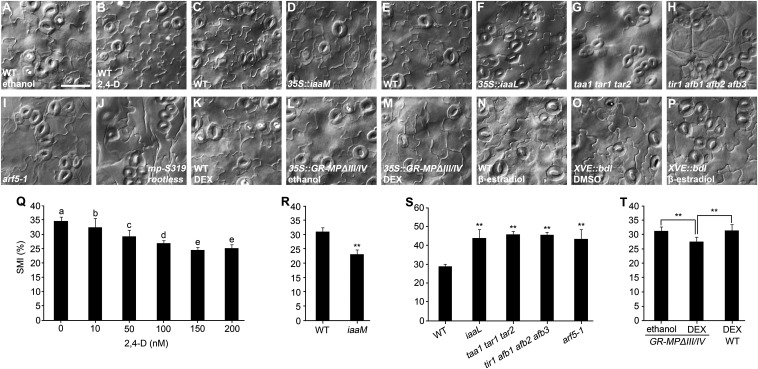
To explore the role of auxin in stomatal development, we treated germinating seeds with various concentrations of auxin analog 2,4-D for 8 d and found that the stomata and meristemoids indexes (SMIs; stomata plus meristemoids per total epidermal cells) in the abaxial epidermis of cotyledons are progressively reduced with the increasing concentration of 2,4-D, with 150 nM being the minimal concentration tested for the maximal inhibition (Fig. 1 A, B, and Q), indicating that auxin inhibits stomatal development in a dose-dependent manner. Moreover, the 35S::iaaM transformants, which produce elevated levels of auxin in vivo (37), display significantly reduced SMIs compared with wild type (Fig. 1 C, D, and R). These results indicate that excess auxin from either exogenous application or genetic manipulation inhibits stomatal development. We further examined transgenic and mutant plants with deficiencies in endogenous auxin biosynthesis or signaling pathways. It is known that taa1 tar1 tar2 triple mutant is defective in the main route for auxin biosynthesis in vivo (38–41), and that the 35S::iaaL transformants have reduced free auxin levels due to the transformation of active auxin to inactive auxin–lysine conjugate (42). Both taa1 tar1 tar2 mutant and 35S::iaaL seedlings exhibit clustered stomata and significantly increased SMIs in the abaxial epidermis of cotyledons (Fig. 1 E–G and S), indicating that reduced auxin promotes excess stomatal production and abnormal stomatal patterning. Taken together, these results demonstrate that auxin inhibits stomatal development.
Fig. 1.
Auxin negatively regulates stomatal development. (A–P) Differential interference contrast microscopy images of abaxial cotyledon epidermis of 8-dpg seedlings with indicated genotypes. Comparisons should be made in the same group, such as A and B; C and D; E–J; K–M; and N–P. The concentration of 2,4-D used in B is 150 nM. GR-MPΔIII/IV could be conditionally nucleus-localized on DEX (M) but not ethanol induction (L). MPΔIII/IV lacks domain III/IV, which mediates dimerization with AUX/IAA proteins. (O and P) XVE::bdl line expresses stabilized bdl on β-estradiol induction (P) rather than on solvent DMSO induction (O). (Q) SMIs obtained from various concentrations of 2,4-D treatment. Values indicated by distinct letters are significantly different (P < 0.01; Tukey’s least significant difference, n = 10). (R–T) SMIs obtained from C and D; E–I; and K–M, respectively. Data are means ± SDs (n = 10 for R and T; n = 9 for S) with Student t test (**P < 0.001). (Scale bar: 50 μm.)
To determine how the auxin signal is transduced to influence stomatal development, we examined the auxin receptors’ quadruple mutant tir1 afb1 afb2 afb3 and observed prominent stomatal clusters and increased SMI (Fig. 1 E, H, and S), indicating that nuclear receptor-mediated auxin signaling plays a pivotal role in the regulation of stomatal development.
Because TIR1/AFB-mediated auxin signaling regulates gene expression eventually through ARF transcription factors (6, 43), we sought to determine which of the 23 ARFs in Arabidopsis are involved in the regulation of stomatal development. It is proposed that the genes implicated in regulating asymmetric divisions of stomatal precursor cells may also act to regulate earlier development (e.g., embryogenesis), such as those encoding components in MAPK signaling pathway (YDA, MPK3, and MPK6) (24, 25, 44). We focused on MP based on previous demonstrations. First, prominent embryo defects are observed in the well-characterized mp mutant, but not in any of the other arf mutants available so far (45). Second, an extraembryonic cell adjacent to the embryonic cell—namely, hypophysis, which is specified to become the primary root meristem founder cell, often undergoes aberrant division in both yda and mp mutants, leading to varied degrees of rootless phenotypes (1, 2, 44). Moreover, abnormal cotyledon development is obvious in yda, mpk3 mpk6, and mp mutants (1, 25, 44). Therefore, we examined stomatal development phenotype in mp/arf5 mutants, and observed significantly increased SMI and stomatal clusters in the lethal mp allele arf5-1 seedlings (Fig. 1 E, I, and S). A weak allele mpS319 homozygote is reported to have only a small portion of seedlings with root deficit (2, 46). To determine whether mpS319 homozygotes also exhibit stomatal development variation, we examined stomatal patterning of mpS319 homozygous seedlings with and without roots, respectively, and found that only rootless seedlings (∼8.6% of progenies of mpS319 heterozygote) exhibit a prominent stomatal cluster phenotype (Fig. 1 E and J and Fig. S1).
To explore whether MP overexpression might reduce SMI, we created 35S::GR-MPΔIII/IV transgenic lines expressing glucocorticoid receptor (GR)-fused MP lacking domain III/IV (MPΔIII/IV). MPΔIII/IV is known not to interact with AUX/IAA proteins, thus possessing higher activity than the full-length of MP (47, 48). GR-MPΔIII/IV is imported into nucleus upon dexamethasone (DEX) treatment. We found that GR-MPΔIII/IV significantly reduces SMI on DEX induction (Fig. 1 K–M and T).
Because ARF proteins are shown to be inhibited by AUX/IAA proteins, we examined the effects of a stabilized and gain-of-function version of BDL—namely, bdl (49), on stomatal development. We generated transgenic plants expressing bdl under the control of a chemical-inducible promoter XVE (50). As expected, on β-estradiol induction, pXVE::bdl transgenic seedlings produce excess stomatal clusters (Fig. 1 N–P), indicating that AUX/IAA–ARF module is also involved in the regulation of stomatal development. Because BDL and MP is a well-studied IAA-ARF pair regulating embryonic root initiation, shoot apical meristem function, and floral primordia initiation (2–4), and our findings implicate MP in the regulation of stomatal development, it is probable that the BDL-MP pair is also involved in the regulation of stomatal development. Confirmation of this possibility can be made by investigating whether BDL is expressed in mesophyll and whether the ARFs other than MP regulate stomatal development. Taken together, these results indicate that MP-mediated nuclear auxin signaling inhibits stomatal development.
MP Functions to Regulate Stomatal Development at Early Seedling Developmental Stage.
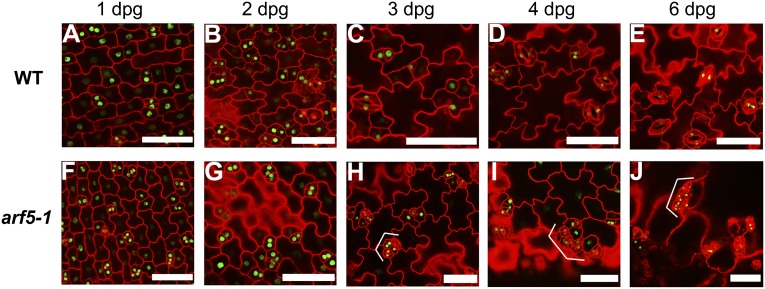
To determine how stomatal clusters are generated in the arf5-1 mutant, we tracked and compared time-course developmental progression in arf5-1 with that in WT using stomatal lineage marker TMM promoter (22). Nucleus-localized luciferase (LUC) fused with GFP driven by the TMM promoter (pTMM::LUC-NLS-GFP) was constructed to visualize stomatal lineage cells. Abnormal cell divisions appear at the third day postgermination (3 dpg) in arf5-1 with adjacent meristemoids and immature stomata highly marked with GFP signals, which indicate potential cell-division abilities (Fig. 2 A–C and F–H). The increasingly prominent stomatal clusters are generated in arf5-1 at 4 dpg and 6 dpg (Fig. 2 D, E, I, and J). These results indicate that, as early as 3 dpg, the role of MP in restraining aberrant stomatal patterning starts to be apparent.
Fig. 2.
MP functions to restrain aberrant stomatal patterning at an early seedling developmental stage. (A–J) Confocal images of pTMM::LUC-NLS-GFP cotyledons in WT (A–E) and arf5-1 (F–J) background, respectively. Epidermal cell periphery is highlighted by propidium iodide (red) staining. Brackets indicate clustered stomata or abnormal stomatal lineage cells. (Scale bars: 50 μm.)
MP Acts to Repress STOMAGEN Expression in Mesophyll to Inhibit Stomatal Development.
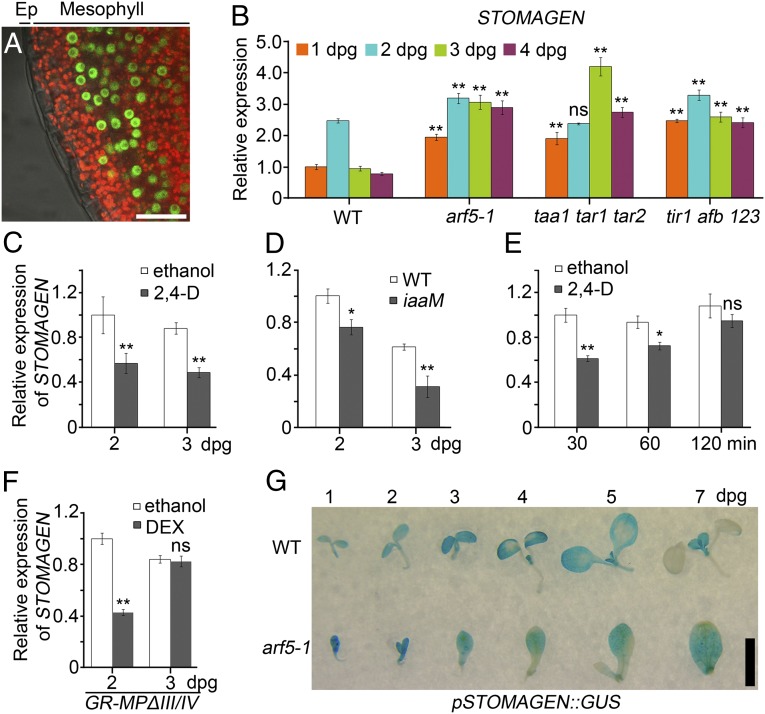
We investigated the expression pattern of MP in leaves of transgenic seedlings expressing MP fused to GFP under the control of the MP native promoter (pMP::MP-GFP) (2). Strong nucleus-localized GFP signals were observed in mesophyll cells rather than in epidermal cells at 3 dpg (Fig. 3A). A previous report showed that MP–GFP is strongly expressed in mesophyll but weakly in epidermis at 1 dpg, and restricted to mesophyll at 2 dpg (48). Combined with our observation, these results indicate that epidermal cells should express very low, if any, levels of MP after 2 dpg. The strong expression of MP in mesophyll suggests that MP might function non–cell-autonomously in the regulation of stomatal development. So far, the short peptide STOMAGEN is known to be a unique mobile factor that is expressed in mesophyll cells and then migrates to the epidermis to regulate stomatal development (18). Because MP acts as a transcription factor mediating auxin signaling (51), we entertained the possibility that MP-mediated auxin signaling negatively regulates stomatal development likely through repressing STOMAGEN expression. Indeed, STOMAGEN expression peaks at 2 dpg and drastically decreases at 3 dpg in WT seedlings as shown by quantitative RT-PCR (Fig. 3B). In contrast, STOMAGEN transcripts in taa1 tar1 tar2, tir1 afb1 afb2 afb3, and arf5-1 sustain significantly higher levels than those in WT at 1, 3, and 4 dpg, respectively, and hardly decline at 3 and 4 dpg (Fig. 3B), which might account for the phenotypic differences between arf5-1 and WT appearing at 3 dpg (Fig. 2 C and H). Because the regulation of STOMAGEN expression at 2 and 3 dpg appears to be critical for stomatal development, hereafter only STOMAGEN expression at 2 and 3 dpg will be analyzed.
Fig. 3.
MP-mediated auxin signaling represses STOMAGEN expression. (A) Confocal images of pMP::MP-GFP leaves at 3 dpg. The merged signals of bright field, Cy5, and GFP channels are shown. Cy5 shows chlorophyll autofluorescence (red) in mesophyll. The nuclear GFP signals (green) indicate MP-GFP. Ep, epidermis. (B–F) STOMAGEN qRT-PCR in the seedlings with indicated genotypes and/or treatments. Data are means ± SDs (n = 3) with Student t test (*P < 0.01; **P < 0.001; ns, no significant differences). Statistical analyses are made by comparing data from mutants (B) and 35S::iaaM (D) with those from WT at the same age, respectively. (C) Seeds were grown in medium with ethanol control or 150 nM 2,4-D for indicated time, and statistical analyses are made by comparing data from different treatments at the same age. (E) The 2-d-old seedlings were treated in liquid medium with ethanol control or 150 nM 2,4-D for indicated time, and statistical analyses are made by comparing data from different treatments at the same time point. (F) 35S::GR-MPΔIII/IV seeds were grown in medium with ethanol control or 5 μM DEX for indicated time, and statistical analyses are made by comparing data from different treatments at the same age. (G) Time-course GUS staining of WT-like (WT) and arf5-1 siblings segregated from pSTOMAGEN::GUS;arf5-1/+. (Scale bars: A, 25 μm; G, 2.5 mm.)
Our findings that both 2,4-D treatment and iaaM overexpression significantly reduce SMI (Fig. 1 A–D, Q, and R) prompt us to analyze STOMAGEN expression in these conditions. Consistent with the phenotypes, 2,4-D treatment results in a significant decrease in STOMAGEN expression at 2 and 3 dpg compared with mock treatment (Fig. 3C), and the expression of STOMAGEN in iaaM overexpressors is significantly reduced compared with that in WT (Fig. 3D), respectively, indicating that auxin inhibits stomatal development likely through repressing STOMAGEN expression. To explore whether STOMAGEN expression is sensitive to transiently applied auxin, we monitored a time-course response of STOMAGEN expression to auxin treatment and found that STOMAGEN expression is significantly reduced on 30 min auxin treatment, and then progressively elevated to the untreated level at 120 min (Fig. 3E). The rapid response of STOMAGEN expression to auxin treatment indicates that STOMAGEN should be one of the primary auxin-responsive genes. The progressive recovery of STOMAGEN expression after 30 min of auxin treatment might be a consequence of negative feedback conferred by rapidly up-regulated primary auxin-responsive genes, including genes of AUX/IAA and GH3 families, which inhibit ARF activity and reduce active auxin level (52, 53), respectively.
To confirm the MP repression of STOMAGEN expression, we analyzed the effects of GR-MPΔIII/IV on STOMAGEN transcription. Compared with mock-treated GR-MPΔIII/IV, DEX-treated GR-MPΔIII/IV seedlings express significantly reduced STOMAGEN transcripts at 2 but not at 3 dpg (Fig. 3F), indicating that the down-regulation of STOMAGEN expression at 2 dpg might be sufficient to reduce SMI. It is important to note that DEX-induced GR-MPΔIII/IV has no such dramatic effects as 150 nM of 2,4-D on stomatal development (Fig. 1 Q and T), which might be ascribed to their differential influences on STOMAGEN expression at 3 dpg (Fig. 3 C and F). The reasons for our failure to observe the reduction of STOMAGEN expression in DEX-induced GR-MPΔIII/IV seedlings at 3 dpg remains to be determined. Moreover, LUC activity assay was performed in the populations including WT-like seedlings (WT) and the arf5-1 homozygous siblings (arf5-1/−) segregated from the transgenic pSTOMAGEN::LUC;arf5-1/+ plants. In 10 independent transgenic lines, the LUC activities in arf5-1/− are consistently much higher than those in WT (Fig. S2). For visual inspection, histochemical staining of GUS activity with pSTOMAGEN::GUS;arf5-1/+ transgenic seedlings was performed. Consistent with a previous report (18), GUS activity is only detected in the mesophyll rather than in the epidermis (Fig. S3A), and broad at 1 dpg in WT cotyledons, gradually confined to the tips of cotyledons (4 and 5 dpg) until eventually almost invisible (7 dpg) with the development of cotyledons (Fig. 3G). In contrast, GUS activity in arf5-1/− always broadly expands throughout the cotyledons even at as late as 7 dpg (Fig. 3G), indicating that MP regulates the spatiotemporal expression pattern of STOMAGEN. Interestingly, the GUS activity appears to expand to the epidermis in arf5-1/− (Fig. S3B), indicating that trace amounts of MP and/or other ARFs might efficiently restrict STOMAGEN expression at an undetectable level in WT epidermis, whereas the repression of STOMAGEN by MP in mesophyll is likely much more elastic, making mesophyll a critical place where MP controls stomatal development by regulating STOMAGEN expression.
MP Regulates Stomatal Development Through Repressing STOMAGEN Expression.
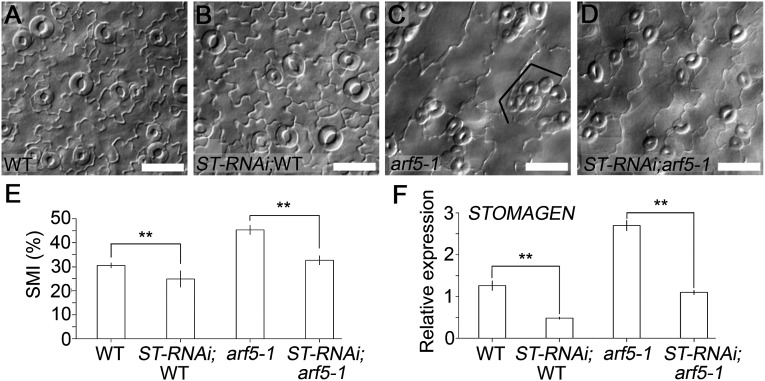
To examine the genetic interaction between MP and STOMAGEN, we silenced STOMAGEN in arf5-1/+ through artificial miRNA (18). The results demonstrate that STOMAGEN expression is significantly reduced in both WT and arf5-1/– seedlings segregated from the same STOMAGEN-RNAi;arf5-1/+ transgenic line (Fig. 4F), and that the down-regulation of STOMAGEN leads to a prominent decrease in SMIs in WT and arf5-1/– cotyledons, respectively (Fig. 4 A–E). It is important to note that the large stomatal clusters consisting of over four stomata, otherwise obviously present in arf5-1/– mutant, are absent in the STOMAGEN-RNAi;arf5-1/– transgenic seedlings (Fig. 4 C and D). The intermediate stomatal phenotype observed for STOMAGEN-RNAi;arf5-1/– may indicate that there exist more genes other than STOMAGEN that are modulated by MP to regulate stomatal development. These results indicate that MP regulates stomatal development, at least in part, through repressing STOMAGEN expression.
Fig. 4.
STOMAGEN acts genetically downstream of MP to regulate stomatal development. (A–D) Differential interference contrast microscopy images of abaxial cotyledon epidermis of 8-dpg seedlings with indicated genotypes. (Scale bars: 50 μm.) ST-RNAi;WT (B) and ST-RNAi;arf5-1 (D) is segregated from the same ST-RNAi;arf5-1/+ line. Bracket indicates clustered stomata. (E) SMIs calculated from A–D. Data are means ± SDs (n = 9). (F) STOMAGEN qRT-PCR in the seedlings with indicated genotypes at 4 dpg. Data are means ± SDs (n = 3). Significant differences were determined with Student t test; **P < 0.001. ST, STOMAGEN.
MP Directly Binds to the STOMAGEN Promoter.
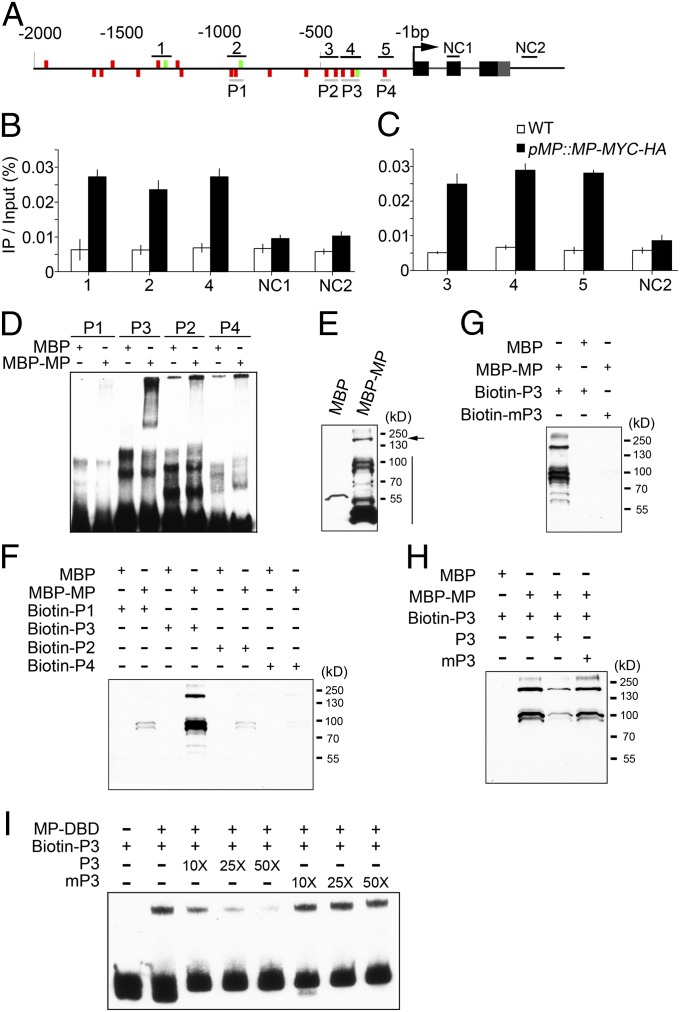
Based on the demonstration that ARFs mediate auxin signaling through binding to AuxREs (54), and our finding that STOMAGEN expression is regulated by MP (Fig. 3 B, F, and G and Fig. S2), we examined whether these elements exist in the STOMAGEN promoter. Indeed, we identified 17 putative (TGTC) and three canonical (TGTCTC) AuxREs in the STOMAGEN promoter (Fig. 5A). To determine whether MP binds to the STOMAGEN promoter in vivo, we generated transgenic Arabidopsis plants expressing MP fused to a fragment encoding MYC-HA epitope under the control of the MP native promoter in the arf5-1 mutant background (pMP::MP-MYC-HA;arf5-1), and performed ChIP-quantitative PCR (qPCR) assay. The results demonstrate that the specific MP-occupied sites are spread throughout the 1,500 base pairs upstream of the translational start codon (−1,500 bp; Fig. 5B). Consistent with the finding that the AuxRE-rich region is mainly located within −500 bp, multiple sites within this region were found to be bound by MP (Fig. 5C).
Fig. 5.
MP directly associates with the AuxREs in a STOMAGEN promoter. (A) STOMAGEN locus comprising the 2-kb promoter region and transcribed region. AuxREs including TGTCTC (green bar) and TGTC (red bar) on the plus (Upper) and minus strand (Lower) of a 2-kb upstream STOMAGEN gene are indicated. Black lines denote fragments amplified in ChIP-qPCR (B and C). Negative controls 1 and 2 (NC1 and NC2) for ChIP-qPCR are located in the second exon (black box) and the region downstream of 3′ UTR (gray box), respectively. Gray lines indicate probes used in EMSA (D and I) and DNA–protein pull-down assays (F–H). (B and C) qPCR of fragments (as in A) from ChIP of pMP::MP-MYC-HA seedlings and WT with anti-HA antibody. Data are means ± SDs (n = 3). (D) EMSA assay using indicated proteins and probes. Various probes as indicated in A were used for EMSA assay (D). (E–H) Western blot analyses of the indicated input (E) and pulled-down proteins in DNA–protein pull-down assay (F–H) with anti-MBP monoclonal antibody. Arrow and vertical line indicate the full-length and degradation fragments of MBP–MP, respectively. Biotin-labeled probes (as in A) were used for DNA–protein pull-down assay with MBP and MBP–MP being as prey (F). Biotin-labeled wild-type (biotin-P3) or mutated (biotin-mP3) AuxRE-containing probes were used in G. Biotin-P3 was used to pull down MBP–MP with unlabeled P3 and mP3 as competitors, respectively (H). (I) EMSA assay using indicated proteins and probes. MP–DBD and biotin-P3 are protein and probe, respectively. Unlabeled P3 and mP3 were used as cold competitors, respectively. 10×, 25×, and 50× indicate the amount of cold competitors relative to that of labeled probe.
We then performed EMSA to confirm the specific interactions of MP with these sites of STOMAGEN promoter in vitro, and found a site (P3) within the −500-bp AuxRE-rich region that strongly interacts with recombinant maltose binding protein tagged MP (MBP-MP) (Fig. 5D). In the EMSA assay, smear-like signals or no clear shifted bands are obvious (Fig. 5D). To circumvent this problem, we performed a DNA–protein pull-down assay. The results show that the truncated fragments of MBP-MP with various molecular weights exist (Fig. 5E), and some of the fragments (comparing pull-down with input) together with the full-length of MBP-MP are strongly pulled down by P3 and weakly by P1, P2, and P4, whereas MBP is hardly pulled down by these probes (Fig. 5 E and F), consistent with the EMSA results (Fig. 5D). Furthermore, DNA–protein pull-down assay demonstrates that biotin-labeled mutated P3 fails to pull down MBP-MP (Fig. 5G) and that nonlabeled intact but not mutated P3 reduces the association of MBP-MP with biotin-labeled P3 (Fig. 5H), indicating that the interaction of MBP-MP with P3 is specific and depends on AuxREs. Moreover, His-tag MP-DNA binding domain (DBD, residues 120–274) (55), which was shown to be able to homodimerize rather than multimerize because of lacking C-terminal domain III/IV (56), was expressed in bacteria and purified. The purified MP-DBD and biotin-labeled P3 probe were used for EMSA assay. The results show that a single shifted band is obvious when MP-DBD is added, and the band intensity is reduced by the competition from intact rather than mutated cold competitor (Fig. 5I), indicating that the MP-DBD-P3 interaction is specific and depends on AuxREs, consistent with the results from DNA–protein pull-down assay (Fig. 5 F–H). The lack of higher-order shifted bands in the MP-DBD EMSA assay indicates the possible contribution of higher-order MP-DNA complexes due to multimerization to smear-like signals in full-length of MP EMSA assay (Fig. 5D). Taken together, these results indicate that MP directly binds to STOMAGEN promoter in an AuxRE-dependent manner.
MP Regulates STOMAGEN Expression in an AuxRE-Dependent Manner in Vivo.
To identify which AuxREs within the STOMAGEN promoter might be mainly responsible for responding to MP in planta, we performed dual-LUC assay, which has been extensively used to analyze gene expression regulation (57, 58). Surprisingly, the reporter genes driven by the STOMAGEN promoter are activated, but not suppressed, by MP in tobacco (Fig. S4 A–C), which is seemingly contradictory to the negative regulation of STOMAGEN by MP in Arabidopsis. To further address this question, we generated another reporter gene driven by the ARR15 promoter, which is shown to be repressed by MP in Arabidopsis (3), and found that ARR15 promoter is also significantly activated by MP in tobacco (Fig. S4D). These results indicate two possibilities: one is that the activation of STOMAGEN and ARR15 by MP in tobacco is nonspecific (i.e., MP could constitutively activate the reporters regardless of upstream elements in tobacco), and the other is that the dual-LUC results could reflect the specific regulation of STOMAGEN and ARR15 promoters by MP in tobacco, regardless of activation or repression. To distinguish these two possibilities, we first made deletion fragments of STOMAGEN promoter and examined their responses to MP in tobacco using dual-LUC assay. The results show that the deletion of −500-bp AuxRE-rich region of the STOMAGEN promoter leads to a complete loss of response to MP (Fig. S4 A–C), consistent with the ChIP-qPCR, DNA–protein pull-down, and EMSA results (Fig. 5), demonstrating that MP cannot constitutively activate the reporter in tobacco. Next, we made a series of multiple mutations of AuxREs within −500-bp region of the STOMAGEN promoter, and demonstrated that at least three AuxREs (Fig. S4E, i–iii), which are shown to be bound by MP in ChIP-qPCR, DNA–protein pull-down, and EMSA assays (Fig. 5), are mainly responsible for the reporter to respond to MP (Fig. S4F), and that the STOMAGEN reporters bearing quadruple to sextuple AuxRE mutations (designated MIV, MV, and MVI; Fig. S4E) fail to respond to MP (Fig. S4F), indicating that MP specifically regulates STOMAGEN promoter in an AuxRE-dependent manner in tobacco.
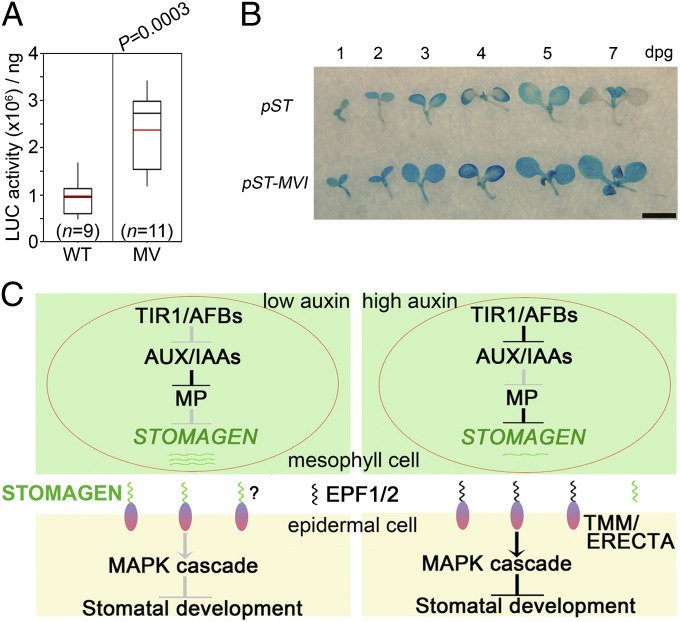
To further determine whether the AuxRE-mediated association of STOMAGEN promoter with MP identified in EMSA, DNA–protein pull-down, and dual-LUC assays is meaningful in Arabidopsis, we made four constructs expressing LUC or GUS driven by wild-type (pSTOMAGEN::LUC or pSTOMAGEN::GUS) and mutant STOMAGEN promoters bearing quintuple or sextuple AuxREs mutations (pSTOMAGEN-MV::LUC or pSTOMAGEN-MVI::GUS), respectively, and introduced them into Arabidopsis. Analysis of luciferase activity shows that pSTOMAGEN-MV::LUC seedlings express considerably more LUC proteins than pSTOMAGEN::LUC seedlings (Fig. 6A). Time-course GUS staining demonstrates that the GUS activity is elevated, spatially expanded, and temporally extended in the cotyledons of pSTOMAGEN-MVI::GUS seedlings, compared with that in the cotyledons of pSTOMAGEN::GUS seedlings (Fig. 6B). In an attempt to test the binding capacity of MP to STOMAGEN promoter harboring sextuple AuxRE mutations in pSTOMAGEN-MVI::GUS seedlings, we used an anti-MP antibody that we generated to perform the ChIP assay. Western blot analysis indicates that this antibody is able to detect the induced MP in transgenic plants (pXVE::MP-Myc and pXVE::MP), but not the endogenous MP (Fig. S5 A–C), probably due to its low abundance in the wild type. We tried to use this antibody to perform a ChIP assay with wild-type seedlings, but found no significant binding of MP to STOMAGEN promoter (Fig. S5D), indicating that the antibody is not suitable for ChIP assay in wild-type seedlings. Although we do not have the necessary materials to detect the requirement of AuxREs within the STOMAGEN promoter for MP binding to the STOMAGEN promoter in Arabidopsis seedlings by ChIP assay, the results from the LUC assay with pSTOMAGEN-MV::LUC seedlings (Fig. 6A) and GUS staining with pSTOMAGEN-MVI::GUS seedlings (Fig. 6B) strongly indicate that the AuxRE-mediated regulation of STOMAGEN by MP obtained in EMSA, DNA–protein pull-down, and dual-LUC assays is meaningful in planta. Therefore, we conclude that AuxREs within the −500-bp region are mainly responsible for the MP repression of STOMAGEN expression in Arabidopsis.
Fig. 6.
AuxREs within the −500-bp region are critical for repression of STOMAGEN expression in vivo. (A) Box plot showing luciferase activities in pSTOMAGEN::LUC and pSTOMAGEN-MV::LUC transgenic lines. WT and MV indicate WT and quintuple AuxRE-mutated STOMAGEN promoters as described in Fig. S4 E and F, respectively. Box comprises values within median 50%. Error bars indicate highest or lowest value. Black and red lines represent median and average, respectively. n represents the number of individual lines. P value (two-tailed unpaired Student t test) is shown above. (B) Time-course examination of GUS activities in pSTOMAGEN::GUS and pSTOMAGEN-MVI::GUS transgenic lines. WT and MVI indicate WT and sextuple AuxRE-mutated STOMAGEN promoters as described in Fig. S4 E and F, respectively. (Scale bar: 2.5 mm.) (C) A model for auxin regulation of stomatal development. When auxin levels are low (Left), AUX/IAA proteins are stabilized and repress MP, leading to high STOMAGEN expression in mesophyll. The accumulated STOMAGEN peptides, which migrate to the epidermis, might compete for TMM or ERECTA with EPF1/2 and inhibit the TMM/ERECTA-MAPK signaling cascade, thus promoting stomatal development. Auxin signaling through TIR1/AFBs activates MP (Right) in mesophyll cells, resulting in repression of STOMAGEN and inhibition of stomatal development by probable activation of EPF1/2-TMM/ERECTA-MAPK signaling. Arrows and T-bars indicate attenuated (gray) or enhanced (black) activation and inhibition, respectively.
Discussion
The present work shows that auxin inhibits stomatal development (Fig. 1), extending our knowledge about the physiological roles of auxin. Furthermore, we revealed the molecular mechanism underlying auxin control of stomatal development by linking auxin signaling with a core stomatal development pathway. When auxin level in mesophyll is low, AUX/IAA proteins are stabilized and accumulated, thus inhibiting MP activity and resulting in high STOMAGEN expression in mesophyll. The resulting STOMAGEN peptide then migrates from mesophyll to epidermis to promote stomatal production probably by competitively inhibiting the EPF1/2-TMM/ERECTA-MAPK pathway (Fig. 6C, Left). When auxin is at high levels in mesophyll, AUX/IAA proteins undergo nuclear auxin receptors TIR1/AFB-mediated degradation by 26S proteasome, thus releasing MP and leading to repression of STOMAGEN transcription. The low production of STOMAGEN peptide probably makes the EPF1/2-TMM/ERECTA-MAPK pathway highly activated in epidermal cells, thus causing attenuated stomatal production (Fig. 6C, Right).
During the revision of our paper, a study reports that altered auxin transport, made either by mutation of auxin efflux transporters or by inhibition of auxin transport with inhibitors, affects stomatal patterning (59). However, the precise physiological role of auxin in the regulation of stomatal development, the regulatory signaling pathway, as well as the underlying molecular mechanisms have basically not been revealed. In the present study, we demonstrate that either altered auxin level or modified auxin signaling could significantly influence stomatal development. Moreover, we established a molecular model for auxin inhibition of stomatal development by showing that MP-mediated nuclear auxin signaling acts non–cell-autonomously to repress a peptide gene STOMAGEN expression in mesophyll. This work advances our knowledge about auxin’s role in stomatal development and the underlying molecular mechanism. To note, it is shown in this recent work that exogenously applied auxin fails to lead to pronounced stomatal development phenotype, based on the production of stomatal clusters but not SMI (59). In the present study, exogenous 2,4-D application results in a significant decrease in SMI (Fig. 1 A, B, and Q). Because WT rarely produces clustered stomata according to the one-cell spacing rule, and auxin acts to inhibit but not promote stomatal production (This study), statistical analysis of SMI rather than stomatal cluster number should be informative for elucidating the role of exogenously applied auxin in stomatal development.
It is reported that MP is expressed in embryonic cells and acts non–cell-autonomously to specify an extraembryonic cell (hypophysis) to become the primary root meristem founder cell during embryogenesis by facilitating auxin transport into hypophysis, and directly transactivating TMO7, a gene encoding a mobile transcription factor that moves from embryonic cell to hypophysis to specify embryonic root initiation (2). In this study, we found that MP inhibits stomatal development mainly through directly repressing STOMAGEN expression in mesophyll, although we could not rule out the epidermal role, if any, of MP in stomatal development, providing new insights into the non–cell-autonomous action of MP in the regulation of postembryonic development. The mutants or genetically modified plants with abnormalities (enhancement or attenuation) in either auxin biosynthesis or signaling usually display phenotypic alterations in the whole plant, including altered plant height, branching, and floral organ development, or extreme phenotypes, including rootless seedlings, single cotyledon, and infertility (1, 4, 12, 40, 60–62). The regulation of STOMAGEN expression and stomatal development by MP-mediated auxin signaling occurs in the mesophyll where photosynthesis mainly takes place (Fig. 6C), providing the possibility that coordinating stomatal development with photosynthesis can be achieved by manipulating auxin signaling specifically in the mesophyll without disturbing the whole body plan.
In this study, we performed dual-LUC assay in tobacco to analyze MP regulation of STOMAGEN expression and found regulatory effects opposite to those obtained in Arabidopsis. Of 23 ARFs in Arabidopsis, five Q-rich ARFs, including ARF5/MP, ARF6, ARF7, ARF8, and ARF19, are thought to activate gene expression according to transient expression assay with DR5 or P3(4×) reporter (63–65). However, it is known that MP not only activates (2, 64) but also represses (3) gene expression in different developmental contexts, indicating that activation or repression might be determined by the spatiotemporal availabilities of MP-interacting partners, coactivator or corepressor (65). According to this prediction, MP might constitutively exhibit activation or repression effects on a AuxRE-containing promoter when transiently expressed in a given context (such as specific tissue or cell type) where either coactivator or corepressor is predominant. To test this possibility, we used the ARR15 promoter, which is shown to be repressed by MP in an AuxRE-dependent manner in Arabidopsis (3), in dual-LUC assay. The results show that the ARR15 promoter is also significantly activated by MP in tobacco (Fig. S4D), indicating that the MP-interacting coactivator might be predominant over the corepressor in tobacco leaves. To verify the specificity, we made deletion and mutation in the STOMAGEN promoter, respectively, and found that both deletion of the −500-bp AuxRE-rich region and mutations of AuxREs in the −500-bp region lead to complete loss of response of STOMAGEN promoter to MP (Fig. S4 C and F), indicating that the regulation of STOMAGEN promoter by MP is specific and depends on AuxREs in the −500-bp region. These results are further confirmed by results from LUC and GUS activity analyses in Arabidopsis (Fig. 6 A and B). Taken together, these results demonstrate that, regardless of activation or repression, the AuxREs responsible for response to MP could be reliably identified through transient dual-LUC assay in tobacco. Importantly, our combined results from analyses of STOMAGEN gene expression in mp mutant (Fig. 3B) and transgenic plants overexpressing GR-MPΔIII/IV (Fig. 3F), STOMAGEN promoter activity in mp mutant (Fig. 3G and Fig. S2), and genetic interaction between MP and STOMAGEN (Fig. 4 A–E) strongly demonstrate that STOMAGEN is repressed, but not activated, by MP in Arabidopsis. Nevertheless, the mechanisms that render the opposite regulation of STOMAGEN by MP in tobacco and Arabidopsis remain to be explored.
MP is a versatile regulator of plant growth and development. The physiological processes involving MP include embryonic root initiation (2), SAM function (3), floral primordial initiation (4), and vascular development (1). It is not surprising that MP is implicated in other developmental processes, including stomatal development. Although MP regulates vascular development, the expression of MP is not restricted to vascular tissue. In fact, MP is strongly expressed in mesophyll at the early seedling developmental stage, as shown by our and others’ results (Fig. 3A) (48). The expression pattern of MP makes it possible that MP regulates mesophyll-derived signals. Stomata, the epidermal pores facilitating gas and water exchange, play important roles in optimizing photosynthetic efficiency and adaptability. The regulation of stomatal development by MP indicates that MP should play a role in photosynthesis. The vascular tissue of plants functions to transport water, salt, and phytohormones as well as organic compounds, including photosynthetic products, thus connecting all plant tissues. The efficient coordination of photosynthesis and the transportation system is critical for optimizing plant growth and development, which could be easily achieved by placing them under control of the same regulator; this might be the biological significance for MP regulation of stomatal development and vascular development. That the regulation of STOMAGEN by MP occurs in mesophyll makes such regulation sense the changes of both photosynthesis and vascular cargoes efficiently and rapidly, thus timely adjusting STOMAGEN expression and stomatal development to coordinating photosynthesis and the transportation system.
Previous studies reported that GLV/RGF/CLE-like peptides interplay with auxin to regulate root gravitropism by regulating auxin transport (66, 67). Another report showed that ARF7-mediated induction of peptide gene IDA by auxin is involved in the regulation of lateral root initiation (67, 68). However, the molecular mechanism underlying auxin-induced IDA expression is unknown. In this study, we provide another example of auxin–peptide cross-talk using stomatal development as a model system and reveal the underlying mechanism by showing that MP directly binds to the STOMAGEN promoter and represses its expression, which advances our understanding of auxin–peptide cross-talk in the regulation of plant development. Stomatal development is under complex regulation of an endogenous program, developmental cues, including phytohormones (e.g., BR and auxin), and environmental cues, including light and carbon dioxide. How these signals are integrated to optimize stomatal development and plant growth awaits further investigation.
Materials and Methods
Plant materials, growth conditions, and treatments, quantitative RT-PCR, GUS staining, ChIP, EMSA, DNA–protein pull-down, and luciferase assays as well as all primers used (Table S1) are in SI Materials and Methods.
Phenotypic Analyses.
Cotyledons of 8-d-postgermination seedlings were transiently preserved in 5% (wt/vol) NaOH solution at 100 °C for ∼10 s, washed with distilled water, and placed in the clear solution (glycerol/chloral hydrate/water, 1: 8: 1, vol/wt/vol) overnight or for a few days. Two images at 200× magnification (0.2 mm2) were captured per cotyledon using a Leica DM2500 microscope with Nomarski optics. A Leica TCS SP5II confocal laser scanning microscope was used to capture Cy5 channel signal and GFP fluorescent images.
ChIP and EMSA Assays.
ChIP was performed with seedlings of pMP::MP-MYC-HA transgenic lines and WT. IP/input (%) was calculated by comparing the Ct values of immunoprecipitate and input from each genotype. For the EMSA assay, a kit was used with 200 ng of recombinant MBP or MBP-MP and 20 fmol biotin-labeled probes, according to manufacturer’s instructions (Thermo Scientific).
Supplementary Material
Acknowledgments
We thank Dolf Weijers for kindly providing the pMP::MP-GFP line; Zhong Zhao, Hong-Tao Liu, Jia-Wei Wang, Jie Xu, Chang-Song Yin, Wan-Qi Liang, and Da-Bing Zhang for technical assistance on the ChIP and paraffin section; Yali He for assistance in statistic analysis; and the Arabidopsis Biological Resource Center at Ohio State University for mutants. This work was supported by National Natural Science Foundation of China Grants 90917014, 91217307, and 30830012 (to H.-Q.Y.), the China Innovative Research Team, Ministry of Education, and 111 Project B14016.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400542111/-/DCSupplemental.
References
- 1.Berleth T, Jürgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- 2.Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464(7290):913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Z, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465(7301):1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi N, et al. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell. 2013;24(3):271–282. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Calderón Villalobos LI, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8(5):477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 7.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 8.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 9.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 10.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 11.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann DC, Sack FD. Stomatal development. Annu Rev Plant Biol. 2007;58:163–181. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- 14.Torii KU. Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012;17(12):711–719. doi: 10.1016/j.tplants.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Hetherington AM, Woodward FI. The role of stomata in sensing and driving environmental change. Nature. 2003;424(6951):901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- 16.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21(14):1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol. 2009;19(10):864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 18.Sugano SS, et al. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463(7278):241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 19.Lee JS, et al. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012;26(2):126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara K, et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50(6):1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 21.Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309(5732):290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- 22.Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296(5573):1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 23.Yang M, Sack FD. The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell. 1995;7(12):2227–2239. doi: 10.1105/tpc.7.12.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304(5676):1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19(1):63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampard GR, Lukowitz W, Ellis BE, Bergmann DC. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell. 2009;21(11):3506–3517. doi: 10.1105/tpc.109.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18(10):2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanaoka MM, et al. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20(7):1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445(7127):501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 30.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445(7127):537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 31.Lai LB, et al. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell. 2005;17(10):2754–2767. doi: 10.1105/tpc.105.034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohki S, Takeuchi M, Mori M. The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat Commun. 2011;2:512. doi: 10.1038/ncomms1520. [DOI] [PubMed] [Google Scholar]
- 33.Katsir L, Davies KA, Bergmann DC, Laux T. Peptide signaling in plant development. Curr Biol. 2011;21(9):R356–R364. doi: 10.1016/j.cub.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudesblat GE, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14(5):548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 35.Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell. 2009;21(9):2624–2641. doi: 10.1105/tpc.109.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482(7385):419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano CP, Robson PR, Smith H, Estelle M, Klee H. Transgene-mediated auxin overproduction in Arabidopsis: Hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol. 1995;27(6):1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- 38.Mashiguchi K, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(45):18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Won C, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(45):18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133(1):177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 41.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133(1):164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romano CP, Hein MB, Klee HJ. Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev. 1991;5(3):438–446. doi: 10.1101/gad.5.3.438. [DOI] [PubMed] [Google Scholar]
- 43.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Lukowitz W, Roeder A, Parmenter D, Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;116(1):109–119. doi: 10.1016/s0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- 45.Rademacher EH, et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell. 2012;22(1):211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 46.Cole M, et al. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136(10):1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- 47.Lau S, De Smet I, Kolb M, Meinhardt H, Jürgens G. Auxin triggers a genetic switch. Nat Cell Biol. 2011;13(5):611–615. doi: 10.1038/ncb2212. [DOI] [PubMed] [Google Scholar]
- 48.Krogan NT, Ckurshumova W, Marcos D, Caragea AE, Berleth T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012;194(2):391–401. doi: 10.1111/j.1469-8137.2012.04064.x. [DOI] [PubMed] [Google Scholar]
- 49.Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16(13):1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuo J, Niu QW, Frugis G, Chua NH. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002;30(3):349–359. doi: 10.1046/j.1365-313x.2002.01289.x. [DOI] [PubMed] [Google Scholar]
- 51.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17(5):1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staswick PE, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17(2):616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276(5320):1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 55.Besnard F, et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature. 2014;505(7483):417–421. doi: 10.1038/nature12791. [DOI] [PubMed] [Google Scholar]
- 56.Boer DR, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156(3):577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 57.Meng Y, Li H, Wang Q, Liu B, Lin C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell. 2013;25(11):4405–4420. doi: 10.1105/tpc.113.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi H, et al. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell. 2013;25(10):3770–3784. doi: 10.1105/tpc.113.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le J, et al. Auxin transport and activity regulate stomatal patterning and development. Nat Commun. 2014;5:3090. doi: 10.1038/ncomms4090. [DOI] [PubMed] [Google Scholar]
- 60.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291(5502):306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 62.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20(13):1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19(3):309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 64.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96(10):5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitford R, et al. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell. 2012;22(3):678–685. doi: 10.1016/j.devcel.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Delay C, Imin N, Djordjevic MA. Regulation of Arabidopsis root development by small signaling peptides. Front Plant Sci. 2013;4:352. doi: 10.3389/fpls.2013.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumpf RP, et al. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA. 2013;110(13):5235–5240. doi: 10.1073/pnas.1210835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.