Significance
Translation requires aminoacyl-tRNAs that are mainly formed by acylating tRNAs with the corresponding amino acids. Methanogenic archaea synthesize Cys-tRNA in an unusual indirect fashion. They attach a precursor amino acid, phosphoserine, to tRNACys, which is then converted to cysteine. This study shows that the indirect Cys-tRNA formation is carried out in a multienzyme complex assembled by a translation factor. Complex formation markedly promotes reaction efficiency. Because the indirect Cys-tRNA formation is the ancestral pathway of Cys biosynthesis in archaea, this complex may represent a remnant of a primordial machinery for Cys coding.
Keywords: methanogen, aminoacyl-tRNA, protein complex
Abstract
Methanogenic archaea lack cysteinyl-tRNA synthetase; they synthesize Cys-tRNA and cysteine in a tRNA-dependent manner. Two enzymes are required: Phosphoseryl-tRNA synthetase (SepRS) forms phosphoseryl-tRNACys (Sep-tRNACys), which is converted to Cys-tRNACys by Sep-tRNA:Cys-tRNA synthase (SepCysS). This represents the ancestral pathway of Cys biosynthesis and coding in archaea. Here we report a translation factor, SepCysE, essential for methanococcal Cys biosynthesis; its deletion in Methanococcus maripaludis causes Cys auxotrophy. SepCysE acts as a scaffold for SepRS and SepCysS to form a stable high-affinity complex for tRNACys causing a 14-fold increase in the initial rate of Cys-tRNACys formation. Based on our crystal structure (2.8-Å resolution) of a SepCysS⋅SepCysE complex, a SepRS⋅SepCysE⋅SepCysS structure model suggests that this ternary complex enables substrate channeling of Sep-tRNACys. A phylogenetic analysis suggests coevolution of SepCysE with SepRS and SepCysS in the last universal common ancestral state. Our findings suggest that the tRNA-dependent Cys biosynthesis proceeds in a multienzyme complex without release of the intermediate and this mechanism may have facilitated the addition of Cys to the genetic code.
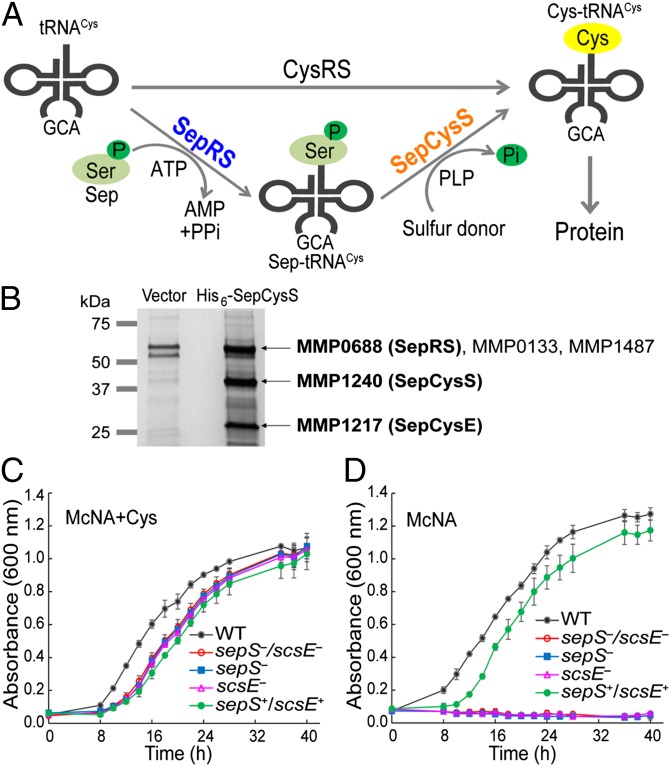
Translation requires a full set of aminoacyl-tRNAs (aa-tRNAs) with precisely matched amino acids and tRNAs (1). The aa-tRNAs are mainly synthesized via direct attachment of amino acids to their corresponding tRNAs by aa-tRNA synthetases (aaRSs), which specifically recognize the amino acid:tRNA pairs to maintain the fidelity of protein synthesis (2, 3). However, many methanogenic archaea lack the Cys-tRNA synthetase (CysRS) that acylates tRNACys with Cys; instead, they rely on an indirect pathway to produce Cys-tRNACys (4). In a two-step process, tRNACys is initially aminoacylated with O-phosphoserine (Sep) by O-phosphoseryl-tRNA synthetase (SepRS) to form Sep-tRNACys, which is then converted to Cys-tRNACys by the tRNA-dependent modifying enzyme Sep-tRNA:Cys-tRNA synthase (SepCysS) with a sulfur donor (Fig. 1A) (4).
Fig. 1.
SepCysE is an essential factor for tRNA-dependent Cys biosynthesis. (A) The direct and indirect pathways for Cys-tRNACys formation. The direct pathway acylates tRNACys with Cys by CysRS. The indirect pathway acylates tRNACys with Sep by SepRS (encoded by sepS), and then Sep is converted to Cys by SepCysS (encoded by pscS) with a sulfur donor. (B) Pull-down of His6-tagged SepCysS expressed in M. maripaludis. Proteins purified from 200 mL cells transformed with the empty vector (left lane) or with the vector for His6-SepCysS expression (right lane) were separated by SDS/PAGE, in-gel tryptic digested, and analyzed by liquid chromatography-tandem mass spectrometry analysis. The locus tag of identified proteins (>60% coverage) only present in the right lane are labeled in bold. (C) Growth of the M. maripaludis strains in the defined medium (McNA) with 1 mM l-Cys. (D) Growth of the M. maripaludis strains in McNA without Cys. scsE−, ΔsepS/ΔscsE mutant with SepRS expressed from a vector; sepS+/scsE+, ΔsepS/ΔscsE mutant with both SepRS and SepCysE expressed from a vector; sepS−, ΔsepS/ΔscsE mutant with SepCysE expressed from a vector; sepS−/scsE−, ΔsepS/ΔscsE double mutant. Data are mean ± SDs from three replicate cultures.
SepRS and SepCysS always coexist and are confined to certain archaea including most methanogens, methanotrophic archaea, and the Archaeoglobus and Ferroglobus species. Phylogenetic studies suggest that the SepRS/SepCysS pathway was the ancestral mechanism of Cys biosynthesis and coding in archaea (5). Later on, the bacterial Cys biosynthesis pathway and the class I CysRS were horizontally transferred to some archaeal lineages; this replaced the indirect pathway (5–7). The question of why some archaea preserve the SepRS/SepCysS pathway is unclear. Interestingly, the SepRS/SepCysS containing archaea have higher Cys content in their proteome (∼1.3% on average) than other archaea (∼0.7% on average), suggesting that the tRNA-dependent Cys biosynthesis correlates with high Cys content in archaea (8).
Both SepRS and SepCysS have been characterized. SepRS is a class II aaRS, and its structure consists of an α4-tetramer (9, 10) that binds two tRNACys molecules in the crystal structure (10) and in solution (11). SepCysS has a similar structure to the pyridoxal-5′-phosphate–dependent Cys desulfurases (12). It is a dimer with the active site located near the dimer interface (12). Three conserved Cys residues of the active site are required for sulfur transfer (13, 14). A binary complex of SepRS and SepCysS that may promote reaction efficiency and sequester Sep-tRNACys was proposed (15). However, formation of this complex caused both SepRS and SepCysS to dissociate from their oligomeric states into monomers (15), and thus this complex is unlikely to be physiologically active. Furthermore, addition of SepRS to the SepCysS reaction does not improve Cys-tRNACys production (16). Here we present a translation factor, SepCysE, essential for tRNA-dependent Cys biosynthesis in methanococci. It forms a ternary complex with SepRS and SepCysS that may enable substrate channeling of Sep-tRNACys.
Results
SepCysE Is Essential for tRNA-Dependent Cysteine Biosynthesis in Methanococcus maripaludis.
To study the physiological interactions of proteins involved in the tRNA-dependent Cys biosynthesis pathway, the His6-tagged SepCysS (MMP1240) was overexpressed in M. maripaludis for a pull-down experiment. Metal affinity chromatography followed by mass spectrometry analysis revealed a ∼25-kDa protein (MMP1217 identified with 72% coverage) associated with SepCysS (Fig. 1B). We named this protein “SepCysE” (SepRS/SepCysS pathway enhancer, encoded by scsE). The primary sequence of SepCysE is not related to any protein of known function. Furthermore, mass spectrometry analysis also confirmed the presence of SepRS (66% coverage) in a ∼60-kDa protein band (Fig. 1B), which additionally contains two contaminating proteins of similar size (MMP0133 and MMP1487). These results indicate that SepCysS physiologically interacts with SepRS and SepCysE.
To confirm the involvement of SepCysE in cysteine biosynthesis, mutagenesis studies were performed in M. maripaludis. This archaeon has both the direct and indirect pathways for Cys-tRNACys formation (Fig. 1A). However, deletion of sepS (encoding SepRS) causes Cys auxotrophy (4), indicating that the indirect pathway is the primary manner of de novo Cys biosynthesis. In this study we constructed a ΔsepS/ΔscsE double mutant. As expected, this mutant required Cys in the medium for growth (Fig. 1C); expression of both SepRS and SepCysE from a shuttle vector restored its growth without Cys (Fig. 1D). However, expression of either SepRS or SepCysE alone (confirmed by Western blot; Fig. S1) was unable to rescue Cys auxotrophy (Fig. 1D). These genetic results demonstrate that SepCysE is essential for the tRNA-dependent Cys biosynthesis in methanococci.
SepRS, SepCysS, and SepCysE Assemble into a Stable Ternary Complex with High Affinity for tRNACys.
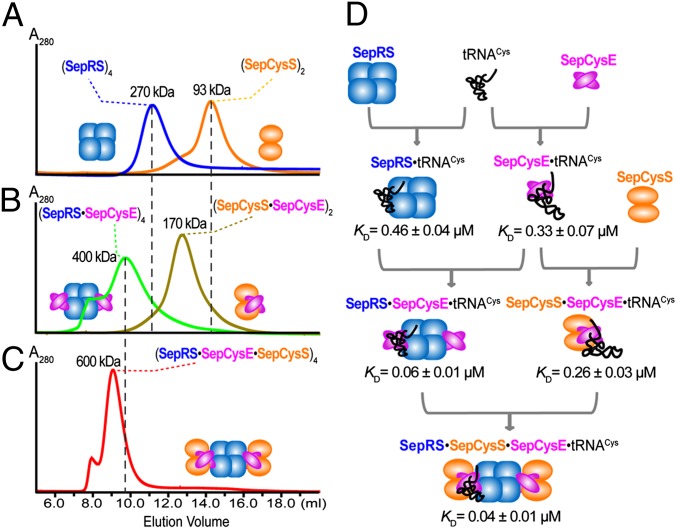
The interactions between recombinant Methanocaldococcus jannaschii SepRS, SepCysS, and SepCysE were studied by gel filtration chromatography using a Superdex 200 size-exclusion column (GE Healthcare). SepRS and SepCysS eluted with an apparent Mr of 270,000 and 93,000, respectively (Fig. 2A), consistent with a tetrameric SepRS structure (monomeric Mr = 66,000) (9, 10) and a dimeric SepCysS structure (monomeric Mr = 47,000) (12). They appeared as separate peaks when mixed together (at concentrations up to 30 µM) before loading, suggesting that SepRS and SepCysS cannot form a stable complex under the experimental condition. On the other hand, when SepRS and SepCysE were mixed together (1:1 molar ratio), gel filtration showed a single peak corresponding to an apparent Mr of ∼400,000 (Fig. 2B). This pattern suggests that both proteins fully associated with each other in a complex composed of four SepRS and four SepCysE (monomeric Mr = 26,000) molecules. Similarly, the mixture of SepCysS and SepCysE (1:1 molar ratio) eluted with an apparent Mr of 170,000, matching a complex composed of two SepCysS and two SepCysE (Fig. 2B). The mixture of SepRS, SepCysS, and SepCysE (1:1:1 molar ratio) also yielded a single peak with an apparent Mr of ∼600,000 (Fig. 2C), matching a ternary complex composed of four SepRS, four SepCysE, and four SepCysS molecules.
Fig. 2.
The M. jannaschii SepRS, SepCysS, and SepCysE form a complex with high affinity for tRNACys. (A) Gel filtration of SepRS (blue) and SepCysS (orange). (B) Gel filtration of a SepRS and SepCysE mixture (green) and a SepCysS and SepsCysE mixture (brown). (C) Gel filtration of a SepRS, SepCysS, and SepCysE mixture (red). Each protein was present at 30 µM. The shoulders of the major SepRS⋅SepCysE and SepRS⋅SepCysE⋅SepCysS peaks were probably due to protein aggregation under the experimental conditions. (D) Affinity of the M. jannaschii proteins for tRNACys. KD of proteins for tRNACys binding were determined by protein titration in a filter-binding assay that quantifies free tRNA and protein⋅tRNA complex. Data are mean ± SDs (n = 3).
The effect of complex formation on tRNACys binding was measured with the M. jannaschii proteins by a filter-binding assay (17). SepRS and SepCysE showed similar affinities for tRNACys, exhibiting KD values of 0.46 ± 0.04 and 0.33 ± 0.04 µM, respectively (Fig. 2D and Fig. S2). This demonstrates that SepCysE by itself binds tRNACys. On the other hand, SepCysS (at concentrations up to 10 µM) did not show measurable binding to tRNACys even at a stoichiometry of 2,000 molecules per tRNA. The formation of binary complexes with SepCysE significantly increased the affinities of both SepRS and SepCysS for tRNACys (Fig. 2D). Furthermore, the SepRS⋅SepCysE⋅SepCysS ternary complex exhibited the strongest binding to tRNACys, with a KD value ∼17-fold lower than that of SepRS alone (Fig. 2D). This result demonstrates that formation of the SepRS⋅SepCysE⋅SepCysS complex greatly increases the protein affinity for tRNACys.
Complex Formation Accelerates Reaction Rate.
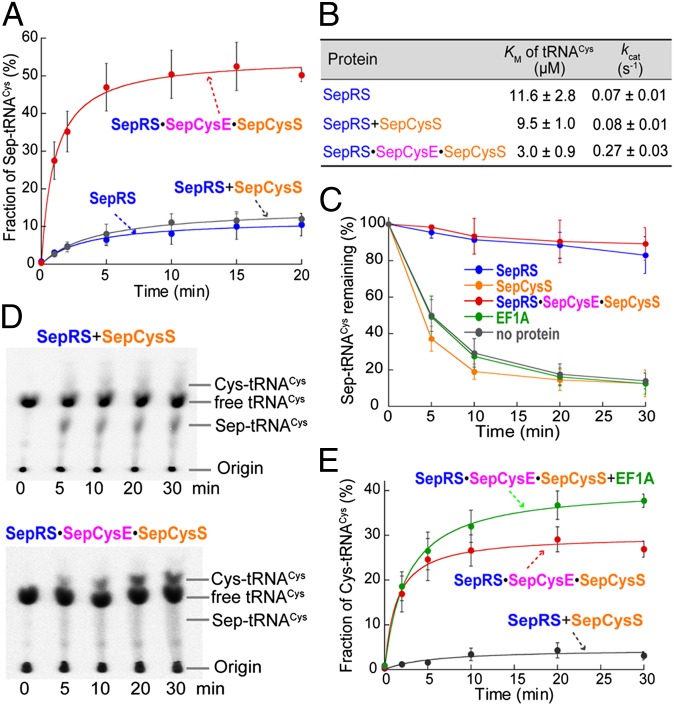
The benefit of complex formation on enzymatic reactions was investigated by three assays. First, the Sep acylation assay showed that the M. jannaschii SepRS only charged tRNACys to a plateau level of ∼10% (Fig. 3A). Steady-state kinetics showed that the KM (of tRNACys) and kcat of this reaction (Fig. 3B) were comparable to the values determined for the M. maripaludis SepRS (18). The addition of SepCysS to the SepRS reaction did not change the activity significantly, whereas the addition of SepCysE together with SepCysS markedly improved the activity (Fig. 3A). The SepRS⋅SepCysE⋅SepCysS ternary complex raised the plateau level of Sep-tRNACys production to 50%, decreased the KM of tRNACys by about fourfold, and increased the catalytic efficiency (kcat/KM) by ∼18-fold compared with SepRS alone (Fig. 3 A and B). These results indicate that complex formation improves the substrate binding and turnover of SepRS.
Fig. 3.
Complex formation accelerates reaction rate and protects Sep-tRNACys from deacylation. (A) Time course of Sep-tRNACys formation by SepRS (blue), SepRS+SepCysS (gray), or the SepRS⋅SepCysE⋅SepCysS complex (red) determined in the Sep acylation assay. (B) Table summarizing steady-state kinetics of Sep-tRNACys formation. (C) Protection of Sep-tRNACys from deacylation. The 32P-labeled Sep-tRNACys (10 µM) was incubated at pH 7.5 with 10 µM each protein at 60 °C. (D) Representative phosphorimages of time-dependent Cys-tRNACys formation by SepRS+SepCysS (Upper) and the SepRS⋅SepCysE⋅SepCysS complex (Lower). (E) Time course of Cys-tRNACys formation by SepRS+SepCysS (gray), SepRS⋅SepCysE⋅SepCysS (red), and SepRS⋅SepCysE⋅SepCysS+EF1A (green). All enzymes are recombinant M. jannaschii proteins expressed and purified from E. coli. Data are mean ± SDs (n = 3).
Second, the stability of Sep-tRNACys was measured in a hydrolysis protection assay. Although its half-life at pH 7.5 was ∼30 min at 37 °C (Fig. S3), it was only ∼5 min at 60 °C (Fig. 3C). This suggests that Sep-tRNACys is unstable in hyperthermophiles, e.g., M. jannaschii with a growth optimum at 85 °C. When SepRS or the SepRS⋅SepCysE⋅SepCysS complex was present, no significant hydrolysis was observed for over 30 min (Fig. 3C). In contrast, the hydrolysis was irrespective to the presence of SepCysE, SepCysS or elongation factor 1A (EF1A) (Fig. 3C and Fig. S3). These results suggest that SepRS in the SepRS⋅SepCysE⋅SepCysS complex can protect Sep-tRNACys from deacylation before its transfer to SepCysS.
Third, the overall activity of Cys-tRNACys production was determined with sodium sulfide as a sulfur donor. When SepRS and SepCysS were incubated with Sep, ATP, Na2S, and tRNACys, the final product Cys-tRNACys was only formed up to 4% of the total tRNACys (Fig. 3 D and E). The intermediate, Sep-tRNACys, was produced as a major product (Fig. 3D), suggesting that Sep→Cys conversion was a rate-limiting step. However, when the SepRS⋅SepCysE⋅SepCysS ternary complex was incubated with the substrates, Sep-tRNACys did not accumulate (Fig. 3D), suggesting that Sep is quickly converted to Cys in the complex. About 28% of the total tRNACys was converted to Cys-tRNACys, with a 14-fold faster initial velocity (0.84 ± 0.18 molCys-tRNA/min⋅molSepCysS−1) than that without SepCysE (0.06 ± 0.02 molCys-tRNA/min⋅molSepCysS−1) (Fig. 3E). These results suggest that complex formation markedly improves the overall production of Cys-tRNACys. Furthermore, the presence of the M. jannaschii translation EF1A raised the plateau level of Cys-tRNACys production to 38% (Fig. 3E), suggesting that cognate Cys-tRNACys is protected by EF1A against deacylation.
The N-Terminal Domain of SepCysE Is Important for Complex Formation.
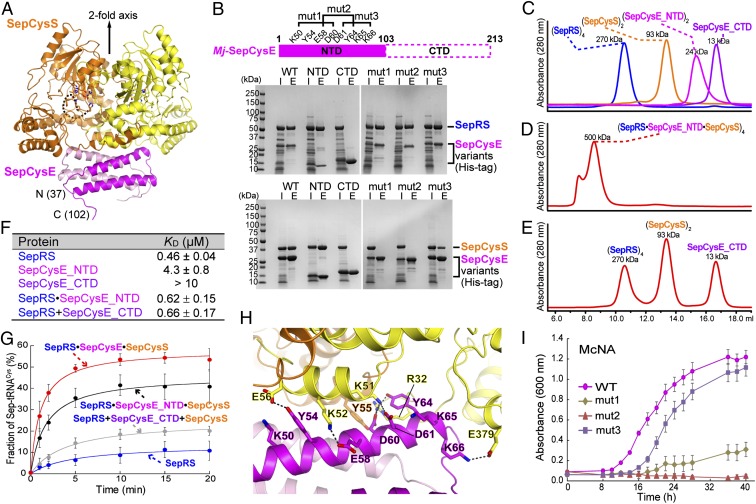
To reveal the structural basis of protein–protein interactions, an X-ray crystal structure of the M. jannaschii SepCysS⋅SepCysE complex was determined at 2.8-Å resolution (Table S1). In this structure, SepCysS and SepCysE are arranged in an α2β2 complex (Fig. 4A), consistent with the stoichiometry determined by gel filtration. The complex exhibits a twofold axis of symmetry through the dimer interface of each protein (Fig. 4A). The SepCysS structure in this complex is quite similar to that of the Archaeoglobus fulgidus SepCysS dimer (12) with an rmsd of 1.1 Å between 688 corresponding Cα atoms (Fig. S4A). This suggests that SepCysS did not undergo large conformational changes upon association with SepCysE. Only the C-terminal part of SepCysS (residues 295–396) moved slightly toward SepCysE (Fig. S4A). This region possibly interacts with the tRNACys acceptor stem (12, 13); thus, its movement may affect tRNA binding.
Fig. 4.
Crystal structure and characterization of the M. jannaschii SepCysS⋅SepCysE complex. (A) Overall structure of a M. jannaschii (Mj) SepCysS⋅SepCysE complex. It consists of a SepCysS dimer (orange and yellow) and a SepCysE dimer (magenta and pink) arranged in an α2β2 organization with twofold axis symmetry. The dashed line indicates a disordered loop at the SepCysS active site. (B) Pull-down analysis of WT and mutant Mj-SepCysE. CTD, Mj-SepCysE C-terminal domain (residues 104–213); E, elution from the nickel chromatography purification; I, input of the crude extracts after heat treatment at 70 °C; mut1, the SepCysE Lys50Ala/Tyr54Ala/Glu58Ala/Asp60Ala variant; mut2, the SepCysE Glu58Ala/Asp60Ala/Asp61Ala/Tyr64Ala variant; mut3, the SepCysE Asp61Ala/Tyr64Ala/Lys65Ala/Lys66Ala variant; NTD, Mj-SepCysE N-terminal domain (residues 1–103). (C) Gel filtration of SepRS (blue), SepCysS (orange), SepCysE_NTD (magenta), and SepCysE_CTD (purple). (D) Gel filtration of a SepRS, SepCysS, and SepCysE_NTD mixture. (E) Gel filtration of a SepRS, SepCysS, and SepCysE_CTD mixture. Each protein was present at 30 µM. (F) Table summarizing the affinity of the M. jannaschii proteins for tRNACys. KD of proteins for tRNACys binding were determined in a filter-binding assay. Data are mean ± SDs (n = 3). (G) Time courses of Sep-tRNACys formation by M. jannaschii SepRS (blue), SepRS⋅SepCysE⋅SepCysS (red), SepRS⋅SepCysE_NTD⋅SepCysS (black), and SepRS+SepCysE_CTD+SepCysS (gray) determined in the Sep acylation assay. Data are mean ± SDs (n = 3). (H) Interactions of SepCysS and SepCysE via hydrogen bonds. (I) Growth of the M. maripaludis ΔsepRS/ΔscsE strain expressing SepRS together with the SepCysE variants (WT, mut1, mut2, or mut3) in the McNA (Cys-free) medium. Data are mean ± SDs from three replicate cultures.
Made up of 213 aa, SepCysE belongs to the Pfam family DUF2100. There is no structural homolog in the Protein Data Bank; thus it represents a previously unidentified structure. Its N-terminal domain (SepCysE_NTD, residues 37–103) is composed of antiparallel helix bundles in direct contact with SepCysS, and its C-terminal domain (SepCysE_CTD, residues 104–213) is disordered in our structure. We then determined the structure of the SepCysS⋅SepCysE_NTD complex at 3.0-Å resolution (Table S1). Deletion of SepCysE_CTD did not cause substantial conformational changes in either SepCysS or SepCysE_NTD (Fig. S4B). These results suggest that SepCysE_NTD is sufficient to form a complex with SepCysS, and that SepCysE_CTD is flexible and may work independently of SepCysE_NTD.
Two experiments confirmed the essential role of SepCysE_NTD in complex formation. First, the His-tagged N- and, C-terminal domains of SepCysE were separately expressed in Escherichia coli together with SepRS or SepCysS for pull-down experiments. SepCysE_NTD brought down both SepRS and SepCysS, whereas SepCysE_CTD failed to do so (Fig. 4B). Second, the complex formation with SepCysE_NTD was analyzed by gel filtration chromatography. SepCysE_NTD and SepCysE_CTD eluted as dimer and monomer, respectively (Fig. 4C). When SepCysE_NTD was mixed with both SepRS and SepCysS at a 1:1:1 molar ratio, a single peak eluted with an apparent Mr of 500,000 matching a tetramer of SepRS⋅SepCysE_NTD⋅SepCysS (Fig. 4D). On the other hand, SepCysE_CTD failed to form a complex with either SepRS or SepCysS (Fig. 4E).
Although SepCysE_NTD is sufficient to link SepRS and SepCysS into a ternary complex, two lines of evidence suggest that SepCysE_CTD is still required for other functions. First, the NTD or CTD alone did not improve the binding affinity of SepRS for tRNACys (Fig. 4F). This suggests that full-length SepCysE is required for improved tRNA binding. Second, the addition of either NTD or CTD moderately increased the Sep acylation activity, but to a lower level than full-length SepCysE (Fig. 4G). This suggests that the NTD and CTD are both required for maximal enhancement of aminoacylation activity.
Conserved Residues Coordinate Interactions of SepCysS and SepCysE.
The SepCysS⋅SepCysE protein–protein interface revealed a basic region of SepCysS to interact with an acidic region of SepCysE (Fig. S4C), forming a hydrogen-bond network (Fig. 4H). Chiefly, the negatively charged, highly conserved Glu58, Asp60, and Asp61 residues in SepCysE form H bonds with the conserved SepCysS residues Arg32, Lys51, and Lys52 (Fig. S5). Two experiments confirmed the importance of these residues for complex formation. First, three His-tagged SepCysE variants (mut1:Lys50Ala/Tyr54Ala/Glu58Ala/Asp60Ala, mut2:Glu58Ala/Asp60Ala/Asp61Ala/Tyr64Ala, and mut3:Asp61Ala/Tyr64Ala/Lys65Ala/Lys66Ala) were expressed in E. coli, and their ability to form a complex with SepRS or SepCysS was examined by pull-down experiments (Fig. 4B). Although all three variants associated with SepRS, SepCysE mut1, and SepCysE mut2 did not bring down SepCysS, this indicates that SepCysE residues Glu58, Asp60, and Asp61 are important for interaction with SepCysS but dispensable for interaction with SepRS. Second, expression of SepCysE mut1 or SepCysE mut2 together with SepRS in the M. maripaludis ΔsepRS/ΔscsE mutant strain did not rescue Cys auxotrophy, whereas coexpression of SepCysE mut3 with SepRS exhibited near WT growth (Fig. 4I); this indicates that Glu58, Asp60, and Asp61 of SepCysE are necessary for Cys-tRNACys production by the indirect pathway.
Discussion
SepCysE Is an Essential Translation Factor for tRNA-Dependent Cys-tRNACys Formation.
Taken together, our data demonstrate that SepCysE is an essential component of the tRNA-dependent Cys-tRNACys biosynthetic pathway that links Cys biosynthesis with translation in methanococci. SepCysE acts as a scaffold that facilitates formation of a ∼600-kDa tetrameric SepRS⋅SepCysE⋅SepCysS complex. This leads to (i) an increase in affinity of SepRS and SepCysS for tRNACys, (ii) enhanced aminoacylation rate, (iii) protection of the labile Sep-tRNACys especially at elevated temperatures, and (iv) improved overall efficiency of Cys-tRNACys production.
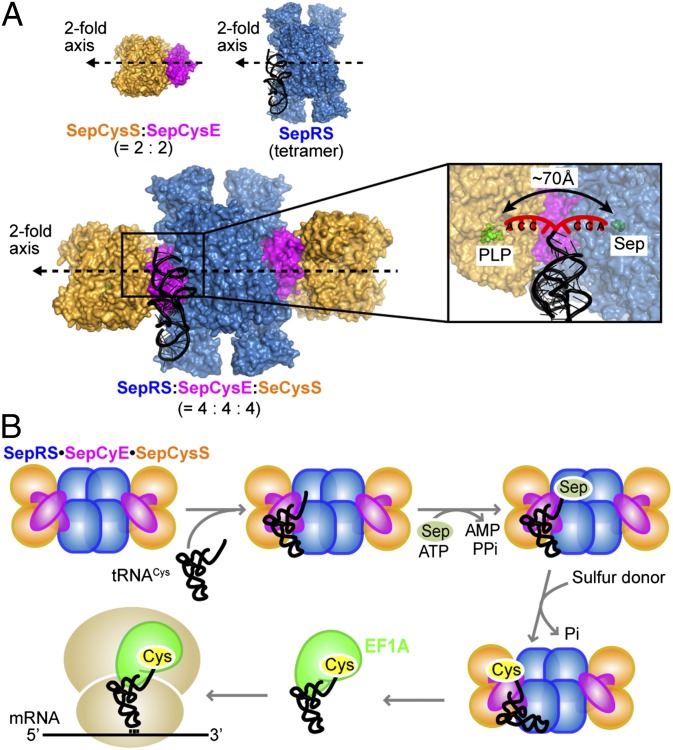
These advantages of complex formation are supported by a SepRS⋅SepCysE⋅SepCysS model obtained by docking the SepCysS⋅SepCysE complex onto the A. fulgidus SepRS⋅tRNACys structure (10) (Fig. 5A). In this model, SepRS and SepCysS bind to opposite sides of tRNACys; this allows the movement of tRNA between the two enzymes without dissociation from the complex. The active sites of SepRS and SepCysS are about 70 Å apart, a distance close enough for the phosphoserylated 3′ CCA terminus of tRNACys to flip between these two active sites. Such a mechanism is used by the mischarged 3′ tRNA termini to move from the aminoacylation site to the editing domain in certain aaRSs (19, 20). Thus, this structural model suggests that the SepRS⋅SepCysE⋅SepCysS ternary complex facilitates substrate channeling (21–23) of the unstable Sep-tRNACys species between the different enzymes in the complex.
Fig. 5.
Proposed scheme of tRNACys substrate channeling. (A) Docking model of SepRS⋅SepCysE⋅SepCysS in complex with tRNACys. The SepCysS⋅SepCysE complex (PDB ID code 3WKR) is docked onto the A. fulgidus SepRS⋅tRNACys complex (PDB ID code 2DU3) (10) by overlapping the symmetric axis of these two complexes. SepRS is in blue, SepCysS is in orange, SepCysE is in magenta, and tRNACys is in black with the discriminator base and the 3′ CCA terminus highlighted in red (Inset). (B) Proposed scheme of Cys-tRNACys formation for translation in methanococci.
Accordingly, we propose the following scheme of tRNA-dependent Cys biosynthesis (Fig. 5B). First, SepCysE assembles SepRS and SepCysS into a ternary complex that recognizes tRNACys. SepRS mainly interacts with the anticodon loop and acceptor stem of tRNACys (10), whereas SepCysE may interact with other parts of tRNACys. Next, the 3′ CCA terminus of tRNACys is phosphoserylated at the active site of SepRS and sequentially transferred to the active site of SepCysS. Finally, after conversion of Sep to Cys on tRNACys with a sulfur donor, the complex releases Cys-tRNACys that is immediately captured by EF1A and delivered to the ribosome for protein synthesis.
Complex Formation Is a Common Feature of tRNA-Dependent Amino Acid Modification.
Similar to the indirect Cys-tRNACys biosynthesis in methanogens, many bacteria and archaea lack asparaginyl-tRNA synthetase (AsnRS) and/or glutaminyl-tRNA synthetase (GlnRS) and rely on indirect pathways for asparagine and glutamine biosynthesis and aa-tRNA formation. In these processes, the nondiscriminating aaRSs (ND-aaRSs) charge tRNAAsn and tRNAGln with Asp (24) and Glu (25), respectively; then they are converted to Asn-tRNAAsn (24, 26) and Gln-tRNAGln (27), respectively, by amidotransferases (AdTs). The ND-aaRSs and AdTs can form complexes called transamidosomes (28), which could allow channeling of the mischarged aa-tRNAs from the ND-aaRSs to the AdTs without dissociation from the complexes (29, 30). Several different types of transamidosome and regulatory features have been discovered (31). (i) The Asn transamidosome in Thermus thermophilus requires tRNAAsn as a scaffold for formation and is stable over the sequential reactions (28, 29). It enhances the aminoacylation activity, protects the mischarged Asp-tRNAAsn from deacylation, and increases the overall production of Asn-tRNAAsn (28). (ii) The Gln and Asn transamidosomes in Helicobacter pylori are transiently formed in the presence of tRNAGln and tRNAAsn, respectively (32, 33). A protein component Hp0100 can facilitate the formation of a stable Asn transamidosome in H. pylori (34). (iii) The Gln transamidosome in Methanothermobacter thermautotrophicus is tRNA independent and unstable through Gln-tRNAGln formation (35). It has no major effect on the kinetics of aminoacylation or transamidation (35, 36). These varied transamidosomes are probably adaptations to different organisms’ metabolism. Presumably, complex formation in indirect aa-tRNA biosynthetic pathways is selected to (i) improve reaction efficiency via substrate channeling and (ii) safeguard the genetic code against mistranslation by sequestering the mischarged aa-tRNAs. Hence, it will be interesting to investigate whether such a mechanism also applies to the tRNA-dependent Sec-tRNASec formation.
SepRS, SepCysS, and SepCysE Coevolved.
The recruitment of the SepRS⋅SepCysE⋅SepCysS ternary complex for tRNA-dependent Cys biosynthesis raises the question of whether these three proteins coevolved. SepRS and SepCysS are conserved in all methanogens except the Methanobrevibacter and Methanosphaera species. They are ancient enzymes that were possibly already present at the time of the last universal common ancestral state (5, 37). On the other hand, SepCysE is conserved in class I methanogens (the Methanobacteriales, Methanococcales, and Methanopyrales orders) but missing in class II (the Methanomicrobiales order) and class III (the Methanosarcinales order) methanogens (Fig. S6). Two lines of evidence support that SepCysE is as ancient as SepRS and SepCysS. First, the sequence-based phylogenetic pattern of SepCysE homologs is consistent with that of the ribosomal RNA and methanogenesis genes (Fig. S7A). Therefore, SepCysE is at least as old as the class I methanogen lineage, which evolved before other methanogens and may have appeared as the ancestral Euryarchaeota (38). Second, the ratios of evolutionary distances (RED) analysis suggests that SepRS, SepCysS, and SepCysE share a common evolutionary history. In RED analysis, the evolutionary distances (Ed) of an experimental gene are plotted against a set of control genes, and a linear plot would indicate the same evolutionary history of the experimental gene and the control genes (6, 39). The SepCysE Ed values correlate with those of SepRS and SepCysS, and the Ed of all three of these genes correlate with the average Ed of the control genes (the ribosomal protein L2P, leucyl-tRNA synthetase, and protein translocase SecY) (Fig. S7B), which are believed to have primarily undergone vertical evolution (6). This suggests that SepRS, SepCysS, and SepCysE were all vertically inherited and coevolved with the organismal lineages. A recent structure-based phylogenetic analysis suggests that SepRS/SepCysS predated the tRNA-independent pathway and arose as the first Cys biosynthetic route (40). In this scenario, the SepRS⋅SepCysE⋅SepCysS complex probably represents a remnant of an ancient translation apparatus that facilitated the addition Cys to the genetic code.
If SepCysE coevolved with SepRS and SepCysS, and is essential for efficient tRNA-dependent Cys biosynthesis, one may ask why only class I methanogens preserved it. We propose two thoughts: (i) Class II and III methanogens acquired the bacterial Cys biosynthetic pathway as well as CysRS through horizontal gene transfer (5–7). Biochemical studies have shown that the bacterial Cys biosynthesis enzymes are active in some Methanosarcina species (41), suggesting that they have redundant pathways for Cys biosynthesis. On the other hand, class I methanogens lack a recognizable bacterial pathway for Cys biosynthesis (Fig. S6), and therefore need to maintain a highly efficient tRNA-dependent Cys biosynthesis pathway. (ii) Class I methanogens have many extreme thermophiles and hyperthermophiles, and the mesophily of some methanococci is a recent adaptation (42), whereas class II and III methanogens are mostly mesophiles. Because Sep-tRNACys is unstable at high temperatures and the formation of a SepRS⋅SepCysE⋅SepCysS complex affords protection from hydrolysis, class I methanogens—which include thermophiles—may have higher selective pressure to retain SepCysE for complex assembly.
Materials and Methods
Measurement of Protein Binding Affinity to tRNACys.
The affinity of proteins for tRNACys was measured by a filter-binding assay (17) with modification (SI Materials and Methods).
Measurement of Sep-tRNACys Formation.
The activity of Sep-tRNACys formation was measured by Wolfson assay (43) with modification (SI Materials and Methods). Kinetic constants were derived from plotting the initial velocity versus [tRNA]. The plots were fitted to the Michaelis–Menten curve using KaleidaGraph 4.0 (Synergy Software).
Measurement of Cys-tRNACys Formation.
All steps of the Cys-tRNACys formation were performed in an anaerobic chamber with an atmosphere of 95% (vol/vol) N2 and 5% (vol/vol) H2. The assay used 1 mM Na2S as the sulfur donor and was carried out as described (16) with modification (SI Materials and Methods).
Crystallization and Structure Determination.
The crystals of both the SepCysS⋅SepCysE and SepCysS⋅SepCysE_NTD complexes were obtained by sitting-drop vapor-diffusion method. Both structures were solved by molecular replacement. Additional details are in SI Materials and Methods. Atomic coordinates have been deposited in the Protein Data Bank (PDB) under the PDB ID codes 3WKR (SepCysS⋅SepCysE) and 3WKS (SepCysS⋅SepCysE_NTD).
Supplementary Material
Acknowledgments
We thank Drs. William B. Whitman, Jiqiang Ling, Xiao-Long Zhou, and Patrick O’Donoghue for insightful discussions. For assistance with data collection, we thank the Photon Factory beam line staff. This work was supported by National Institute of General Medical Sciences Grant GM22854 (to D.S.); Department of Energy Office of Basic Energy Sciences Grant DE-FG02-98ER20311 (to D.S., for the genetic aspects of this study); and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan (I.T.). A.N. was a Japan Society for the Promotion of Science Postdoctoral Fellow for Research Abroad.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3WKR (SepCysS⋅SepCysE) and 3WKS (SepCysS⋅SepCysE_NTD)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411267111/-/DCSupplemental.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Wang E. Transfer RNA: A dancer between charging and mis-charging for protein biosynthesis. Sci China Life Sci. 2013;56(10):921–932. doi: 10.1007/s11427-013-4542-9. [DOI] [PubMed] [Google Scholar]
- 4.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307(5717):1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 5.O’Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. The evolutionary history of Cys-tRNACys formation. Proc Natl Acad Sci USA. 2005;102(52):19003–19008. doi: 10.1073/pnas.0509617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farahi K, Pusch GD, Overbeek R, Whitman WB. Detection of lateral gene transfer events in the prokaryotic tRNA synthetases by the ratios of evolutionary distances method. J Mol Evol. 2004;58(5):615–631. doi: 10.1007/s00239-004-2582-2. [DOI] [PubMed] [Google Scholar]
- 7.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64(1):202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klipcan L, Frenkel-Morgenstern M, Safro MG. Presence of tRNA-dependent pathways correlates with high cysteine content in methanogenic Archaea. Trends Genet. 2008;24(2):59–63. doi: 10.1016/j.tig.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Kamtekar S, et al. Toward understanding phosphoseryl-tRNACys formation: The crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase. Proc Natl Acad Sci USA. 2007;104(8):2620–2625. doi: 10.1073/pnas.0611504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukunaga R, Yokoyama S. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat Struct Mol Biol. 2007;14(4):272–279. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 11.Hauenstein SI, Hou YM, Perona JJ. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem. 2008;283(32):21997–22006. doi: 10.1074/jbc.M801838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukunaga R, Yokoyama S. Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: Crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus. J Mol Biol. 2007;370(1):128–141. doi: 10.1016/j.jmb.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 13.Helgadóttir S, Sinapah S, Söll D, Ling J. Mutational analysis of Sep-tRNA:Cys-tRNA synthase reveals critical residues for tRNA-dependent cysteine formation. FEBS Lett. 2012;586(1):60–63. doi: 10.1016/j.febslet.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Catalytic mechanism of Sep-tRNA:Cys-tRNA synthase: Sulfur transfer is mediated by disulfide and persulfide. J Biol Chem. 2012;287(8):5426–5433. doi: 10.1074/jbc.M111.313700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys) Nat Struct Mol Biol. 2008;15(5):507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauenstein SI, Perona JJ. Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J Biol Chem. 2008;283(32):22007–22017. doi: 10.1074/jbc.M801839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90(12):5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohn MJ, Park HS, O’Donoghue P, Schnitzbauer M, Söll D. Emergence of the universal genetic code imprinted in an RNA record. Proc Natl Acad Sci USA. 2006;103(48):18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285(5430):1074–1077. [PubMed] [Google Scholar]
- 20.Palencia A, et al. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat Struct Mol Biol. 2012;19(7):677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava DK, Bernhard SA. Metabolite transfer via enzyme-enzyme complexes. Science. 1986;234(4780):1081–1086. doi: 10.1126/science.3775377. [DOI] [PubMed] [Google Scholar]
- 22.Hausmann CD, Praetorius-Ibba M, Ibba M. An aminoacyl-tRNA synthetase:elongation factor complex for substrate channeling in archaeal translation. Nucleic Acids Res. 2007;35(18):6094–6102. doi: 10.1093/nar/gkm534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29(4):419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker HD, Kern D. Thermus thermophilus: A link in evolution of the tRNA-dependent amino acid amidation pathways. Proc Natl Acad Sci USA. 1998;95(22):12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J Bacteriol. 1986;165(1):88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382(6592):589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 27.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331(6152):187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 28.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: A dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell. 2007;28(2):228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Blaise M, et al. Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 2010;29(18):3118–3129. doi: 10.1038/emboj.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010;467(7315):612–616. doi: 10.1038/nature09411. [DOI] [PubMed] [Google Scholar]
- 31.Saad NY, et al. Two-codon T-box riboswitch binding two tRNAs. Proc Natl Acad Sci USA. 2013;110(31):12756–12761. doi: 10.1073/pnas.1304307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer F, et al. The asparagine-transamidosome from Helicobacter pylori: A dual-kinetic mode in non-discriminating aspartyl-tRNA synthetase safeguards the genetic code. Nucleic Acids Res. 2012;40(11):4965–4976. doi: 10.1093/nar/gks167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huot JL, et al. Gln-tRNAGln synthesis in a dynamic transamidosome from Helicobacter pylori, where GluRS2 hydrolyzes excess Glu-tRNAGln. Nucleic Acids Res. 2011;39(21):9306–9315. doi: 10.1093/nar/gkr619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva GN, et al. A tRNA-independent mechanism for transamidosome assembly promotes aminoacyl-tRNA transamidation. J Biol Chem. 2013;288(6):3816–3822. doi: 10.1074/jbc.M112.441394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rampias T, Sheppard K, Söll D. The archaeal transamidosome for RNA-dependent glutamine biosynthesis. Nucleic Acids Res. 2010;38(17):5774–5783. doi: 10.1093/nar/gkq336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhaskaran H, Perona JJ. Two-step aminoacylation of tRNA without channeling in Archaea. J Mol Biol. 2011;411(4):854–869. doi: 10.1016/j.jmb.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavran JM, et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA. 2007;104(27):11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blank CE. Not so old Archaea - the antiquity of biogeochemical processes in the archaeal domain of life. Geobiology. 2009;7(5):495–514. doi: 10.1111/j.1472-4669.2009.00219.x. [DOI] [PubMed] [Google Scholar]
- 39.Farahi K, Whitman WB, Kraemer ET. RED-T: Utilizing the Ratios of Evolutionary Distances for determination of alternative phylogenetic events. Bioinformatics. 2003;19(16):2152–2154. doi: 10.1093/bioinformatics/btg284. [DOI] [PubMed] [Google Scholar]
- 40.Zhang HY, Qin T, Jiang YY, Caetano-Anollés G. Structural phylogenomics uncovers the early and concurrent origins of cysteine biosynthesis and iron-sulfur proteins. J Biomol Struct Dyn. 2012;30(5):542–545. doi: 10.1080/07391102.2012.687520. [DOI] [PubMed] [Google Scholar]
- 41.Borup B, Ferry JG. Cysteine biosynthesis in the Archaea: Methanosarcina thermophila utilizes O-acetylserine sulfhydrylase. FEMS Microbiol Lett. 2000;189(2):205–210. doi: 10.1111/j.1574-6968.2000.tb09231.x. [DOI] [PubMed] [Google Scholar]
- 42.Keswani J, et al. Phylogeny and taxonomy of mesophilic Methanococcus spp. and comparison of rRNA, DNA hybridization, and phenotypic methods. Int J Syst Bacteriol. 1996;46(3):727–735. doi: 10.1099/00207713-46-3-727. [DOI] [PubMed] [Google Scholar]
- 43.Wolfson AD, Pleiss JA, Uhlenbeck OC. A new assay for tRNA aminoacylation kinetics. RNA. 1998;4(8):1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.