Significance
Folding mechanisms of large proteins are often complicated by the existence of kinetic traps that impede progress toward the native conformation. We have tested the role of chain connectivity in creating such traps by permuting the sequence of a small α/β/α sandwich protein, the chemotaxis response regulator Y. An approach combining experimental and native-centric simulations reveals that chain entropy and aliphatic-rich sequences conspire to create frustrated species whose structures and stabilities vary with connectivity. The initial events in folding reflect not a random collapse driven by the hydrophobic effect but rather the accumulation of substructures favored by low-contact-order nonpolar interactions in the polypeptide. The conserved global free-energy minimum of the native conformation ultimately resolves these early frustrations in folding.
Keywords: CF-SAXS, Gō models, CheY permutants, protein-folding intermediates
Abstract
Folding of globular proteins can be envisioned as the contraction of a random coil unfolded state toward the native state on an energy surface rough with local minima trapping frustrated species. These substructures impede productive folding and can serve as nucleation sites for aggregation reactions. However, little is known about the relationship between frustration and its underlying sequence determinants. Chemotaxis response regulator Y (CheY), a 129-amino acid bacterial protein, has been shown previously to populate an off-pathway kinetic trap in the microsecond time range. The frustration has been ascribed to premature docking of the N- and C-terminal subdomains or, alternatively, to the formation of an unproductive local-in-sequence cluster of branched aliphatic side chains, isoleucine, leucine, and valine (ILV). The roles of the subdomains and ILV clusters in frustration were tested by altering the sequence connectivity using circular permutations. Surprisingly, the stability and buried surface area of the intermediate could be increased or decreased depending on the location of the termini. Comparison with the results of small-angle X-ray–scattering experiments and simulations points to the accelerated formation of a more compact, on-pathway species for the more stable intermediate. The effect of chain connectivity in modulating the structures and stabilities of the early kinetic traps in CheY is better understood in terms of the ILV cluster model. However, the subdomain model captures the requirement for an intact N-terminal domain to access the native conformation. Chain entropy and aliphatic-rich sequences play crucial roles in biasing the early events leading to frustration in the folding of CheY.
Highly denatured states of globular proteins resemble statistical random coils when examined with low-resolution techniques such as X-ray scattering (1) and hydrodynamic analyses (2). However, a higher-resolution view provided by experimental models (3–6) and simulations (7) shows that the conformational ensemble is biased toward low-contact-order (CO) structures, e.g., α-helices, β-turns, and β-hairpins, which form and melt in less than a few microseconds. During folding, these nascent structures presumably coalesce into higher-order assemblies of ever-increasing free energy until reaching the transition-state ensemble (TSE) that leads to the native conformation. From another perspective, this assembly process mediates a global collapse of the chain in an unfavorable solvent (8). Landscape theory (9) posits that, in the simplest scenario, native-like substructures appear and lead without pause to the TSE and the native conformation in an apparent two-state fashion. However, simulations have found that topological frustration, e.g., the premature formation of a substructure that impedes access to the productive TSE, can lead to the accumulation of intermediates (I) that must unfold to some extent to traverse the folding reaction coordinate successfully (8, 10, 11). Experimental and computational studies on the folding of the α-subunit of Trp synthase (12, 13), the chemotaxis response regulator Y (CheY) (10, 14), a pair of apo-flavodoxins (8, 15, 16), and tandem titan domains (17) revealed frustration in the form of off-pathway intermediates (IOFF). Thus, as-yet unexplored aspects of sequence and structure can add complexity to folding reactions.
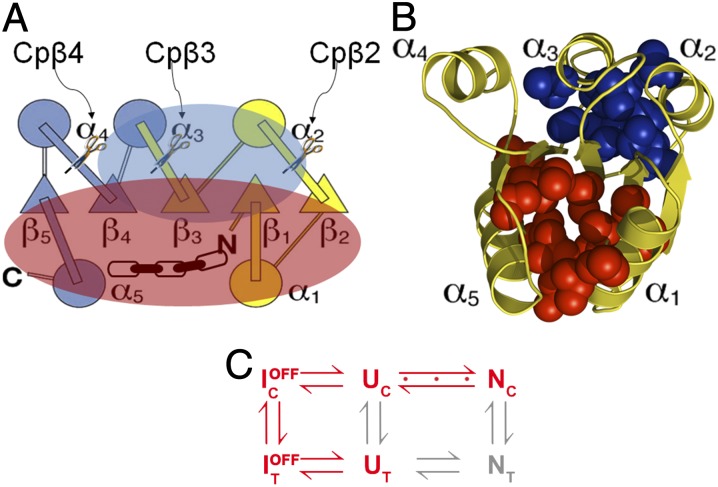
The observed inverse relationship between CO and folding rate constant (18) implies that elements of secondary structure that are near in sequence and near in space will associate preferentially over those that are distant in sequence. However, if such low-CO substructures are not involved in the productive TSE, they could serve as sources of frustration. A case in point is CheY, a member of the very common flavodoxin-fold family with its classic α/β/α sandwich architecture. The (β/α)5 motif displays the α1 and α5 helices on one face of the parallel β-sheet and the α2, α3, and α4 helices on the opposing face (Fig. 1 A and B). The proposed kinetic folding mechanism (Fig. 1C) (14) involves two parallel folding channels defined by the cis and trans isomers of the prolyl peptide bond between K109/P110. The unfolded proteins (U) in both the major trans (Ut) (90%) and minor cis (Uc) (10%) channels sample an off-pathway submillisecond intermediate (IBP), IBPt and IBPc, respectively, before the rate-limiting isomerization reaction in the IBPt→IBPc step. IBPc unfolds to the Uc state before accessing the productive TSE leading to the native conformation (N) in the Uc →Nc step. Further complicating the mechanism is an on-pathway intermediate, ION, between Uc and Nc that has been observed by equilibrium NMR measurements (19) and in Gō-model simulations (10) but not by circular dichroism (CD) or flow fluorescence (FL) experiments.
Fig. 1.
(A) Topology diagram of CheY. The N-terminal folding subdomain is highlighted in yellow, and the C-terminal folding subdomain is highlighted in blue. The effects of each permutation on the continuity of cluster 1 (blue), and cluster 2 (red) are shown. (B) Clusters of ILV residues are superimposed on the crystal structure of CheY (Protein Data Bank ID code: 3CHY). Cluster 1 (blue) has a lower CO and resides on the α2/α3/α4 side of the central β-sheet. The larger cluster 2 (red) contains high-CO contacts and resides on the α1/α5 side of the β-sheet. (C) The folding mechanism of WT CheY. The major pathway is highlighted in red. The Uc →Nc step, involving the on-pathway intermediate, is designated by the triple dots.
Mutational analysis (20, 21) has revealed a nucleation-condensation folding mechanism for CheY in which the N-terminal subdomain [residues 1–70, (βα)1–2β3] serves as the nucleus for the subsequent condensation of the C-terminal subdomain [residues 70–129, α3(βα)4–5] (Fig. 1A). However, native-centric simulations identified contacts between the N- and C-terminal subdomains, centered around (βα)3–4, early in folding that are incompatible with access to the productive TSE and that lead to frustration in the folding mechanism (10). Another perspective is provided by the branched chain aliphatic side chains (BASiC) hypothesis, which supposes that large clusters of isoleucine, leucine, and valine (ILV) side chains serve as cores of stability in folding intermediates (11, 14). Both these clusters have been shown to have a high contact density (22). CheY has two ILV clusters, each serving to fuse the surface helices to each other and to the central β-sheet (Fig. 1B). The smaller cluster (cluster 1) contains 10 side chains and primarily links α2(βα)3β4 on one face of the β-sheet; the larger cluster (cluster 2) contains 15 side chains and links the β-strands to α1 and α5. The sequence spanned by the smaller cluster agrees closely with the (βα)3–4 segment identified as the source of frustration in the simulations and, importantly, involves only low-CO contacts. If cluster 1 were to form early and, by sequestering β3, impede the development of the productive TSE in the N-terminal subdomain, (βα)1–2β3, the BASiC hypothesis would provide an alternative explanation for frustration in the folding of CheY.
Permutations in the sequence of CheY provide a means to compare the subdomain model and the ILV cluster model as explanations for the frustration in folding detected by simulations and experiments. By fusing the natural N and C termini with a short linker peptide (Gly-Ala-Gly) and inserting new termini in the loops after β2, β3, and β4, it is possible to cleave within the N-terminal subdomain (Cpβ2), between the subdomains (Cpβ3), and within the C-terminal subdomain (Cpβ4). Related to the ILV clusters, Cpβ2 cleaves cluster 1 and leaves cluster 2 largely intact, Cpβ3 cleaves both clusters, and Cpβ4 cleaves only cluster 2 (Fig. 1A). Our simulations and experiments on these permutants show that aspects of both models describe the relationships between sequence, structure, and frustration in the folding of CheY. The results also show that frustration can be modulated by sequence permutations that can bias the initial stages of folding toward the productive TSE and away from kinetic traps.
Results
Permutations Differentially Affect the Secondary Structures of the Folded States of the Permutants.
We introduced sequence permutations into the F14N variant of CheY, denoted “CheY*”, to increase the stability of the platform and its tolerance for the introduction of the linker and the new termini; the folding mechanism for CheY* is unchanged from the WT protein (20). The new N termini for Cpβ2, Cpβ3, and Cpβ4 become D38, D64, and E89, respectively. An additional glycine residue at position −1 is a remnant of the cleaved hexahistidine affinity tag (SI Methods). The far-UV CD spectrum of Cpβ2 is markedly different from CheY* (Fig. S1), with the relative intensities of the double minima at ∼210 and ∼222 nm reversed from those of the CheY* protein. Unfortunately, the substantial perturbation of the secondary structure precluded Gō-model simulations that rely on knowledge of the native structure. The CD spectra of Cpβ3 and Cpβ4 display the same relative double minima as CheY*; the spectrum of Cpβ4 is coincident with CheY*, and Cpβ3 is reduced in amplitude by ∼15% (Fig. S1). Although the secondary structure of Cpβ3 appears to fray to some extent, the basic β/α/β architecture is preserved. Therefore, both Cpβ3 and Cpβ4 were deemed to be good candidates for a combined experimental and computational analysis of their folding mechanisms.
Stability Analysis of the Permutants.
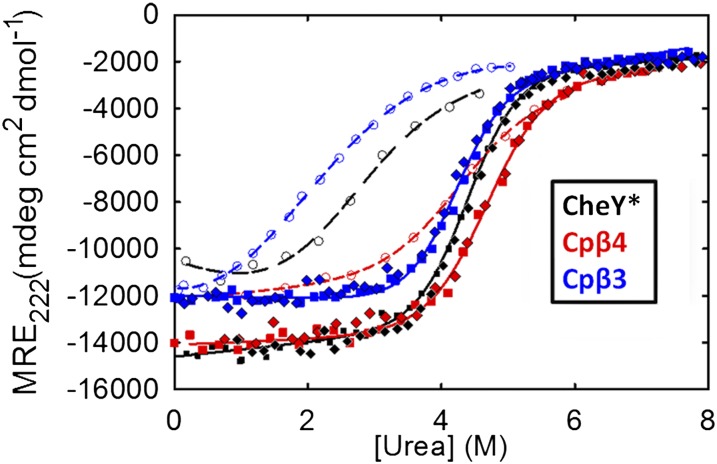
The concerted disruption of secondary and tertiary structures with increasing concentrations of urea revealed an apparent two-state process, N⇌U, for CheY* and Cpβ2 (Fig. 2 and Table S1). Fits of the data to a linear dependence of free energy of folding on the denaturant concentration (23) showed that the stabilities varied from 2.11 kcal·mol−1 for Cpβ2 to 8.0 kcal·mol−1 for CheY* protein (Table S1). The denaturant dependence of the free energy of folding, the m-value [a measure of the change in buried surface area (24)], varied from 0.77 kcal·mol−1⋅Murea−1 for Cpβ2 to 1.99 kcal·mol−1⋅Murea−1 for CheY*. Notably, Cpβ3 displayed a complex equilibrium unfolding reaction, with noncoincident CD and FL denaturation transitions (Fig. S2C). The lower midpoint of the FL unfolding transition most likely reflects the introduction of the new N terminus only a few residues downstream from the single tryptophan, W58. The stability estimated for the global unfolding reaction, indicated by the CD transition, is 6.79 kcal·mol−1. Although the titration data for Cpβ4 could be fit to a two-state model, kinetic analysis (see below) revealed the presence of a stable intermediate and dictated a three-state model. The melting temperatures estimated from the temperature dependence of heat capacities calculated by the simulations (Fig. S3) are in the same rank order as the midpoint points in the urea titrations (Fig. 2): Cpβ3 < CheY* < Cpβ4. Experimental thermal melts by both DSC and CD were irreversible, and a reliable experimental measurement of the melting temperatures could not be obtained. Further experiments on Cpβ2 were not pursued.
Fig. 2.
Analysis of N and IBP stability. Filled symbols display the urea melts derived from the ellipticity at 222 nm for CheY* and each of the permuted variants; the denaturant-induced unfolding reactions are fully reversible. The open symbols display the urea dependence of the ellipticity at 222 nm after 5 ms of refolding. With the exception of Cpβ4, the solid and dashed lines show the fits of these data to two-state equilibrium models. The data for Cpβ4 are fit to a three-state model.
Kinetic Analysis of Permutant Folding.
We monitored the dynamic responses of the permutants to rapid changes in the denaturant concentration in the microseconds-to-hundreds of seconds time range with a combination of continuous-flow (CF), stopped-flow (SF), and manual-mixing (MM) techniques interfaced with FL, circular dichroism (CD), and small-angle X-ray–scattering (SAXS) detection. For CheY*, a large-amplitude FL phase occurs within the 25-μs dead time of CF refolding, followed by a small-amplitude phase lasting several hundred microseconds. The subsequent formation of the native state occurs in hundreds of seconds and has been attributed to the trans → cis isomerization of the K109–P110 peptide bond (14) (Fig. S4). Unfortunately, refolding along the cis channel for the permutants could not be resolved because of its small amplitude in direct refolding experiments and interrupted unfolding experiments. A pair of unfolding reactions were observed in the seconds-to-hundreds of seconds time range; the interconversion of the native cisP110 conformer to its trans counterpart, Nc →Nt, controls unfolding in the transition zone and the direct unfolding of the native cisP110 to the unfolded cisP110, Nc →Uc, controls unfolding at high denaturant concentrations. Similar overall responses were observed for Cpβ3 and Cpβ4, with the exception that the direct unfolding of the native cisP110 conformer was accelerated for Cpβ4.
Stability and Secondary Structure of Submillisecond Intermediate States.
The orders of magnitude in time (from microseconds to hundreds of seconds) separating the folding reactions for all three proteins enabled us to measure the stability of the product of the microsecond reaction, IBP, and its CD spectrum. By plotting the ellipticity at 222 nm after 5 ms of refolding in varying final denaturant concentrations, the stability can be estimated by fitting the resulting titration curve to a two-state model (Fig. 2). The IBP species for Cpβ3 is significantly less stable than for CheY*, 0.84 kcal·mol−1 vs. 2.02 kcal·mol−1, and the m-value is also decreased (Table S1). Very surprisingly, IBP stability is much greater for Cpβ4 (4.31 kcal·mol−1) than for CheY*, and the m-value is increased (Table S1). Comparison of the denaturation curves for folded Cpβ4 and its IBP species (Fig. 2) shows that the two curves overlap between 3 and 5 M urea. By fixing the thermodynamic parameters for the IBP⇌U reaction to those extracted from the burst-phase titration data, the stability and m-value for the N⇌U reaction could be estimated by fitting the equilibrium titration data for Cpβ4 to a three-state model. The difference in free energy between its native and unfolded forms is 8.19 kcal·mol−1, and the m-value is 1.80 kcal·mol−1·M−1, comparable to that of CheY*.
We obtained the CD spectra of the IBP species by refolding jumps to the same final urea concentration in the folded baseline and varying the detection wavelengths in the far-UV range. The IBP species for CheY*, Cpβ3, and Cpβ4 recover ∼85%, ∼80%, and ∼90% of their native ellipticities at 222 nm within 5 ms (Fig. 2). The subtle but significant differences previously observed between the IBP and native states of CheY* (14) (Fig. S5) indicate that the aromatic side chains have not yet attained their native packing. In contrast, the very similar shapes of the spectra for the IBP and native states of Cpβ4 and Cpβ3 show that an exciton coupling, likely between the side chains in a cluster of phenylalanines on the α1/α5 side of the β-sheet (Fig. S5 C and D), is present in the IBP state for both permutants.
Compaction of CheY* and Cpβ4 by CF-SAXS.
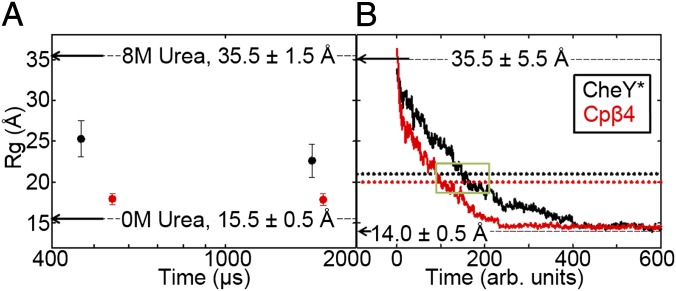
The very surprising increases in the stability and the apparent compaction for the IBP species for Cpβ4, the latter implied by the increased m-value for its urea melt, motivated us to measure its radius of gyration (Rg) in the ∼100-μs–to–1-ms time range by CF-SAXS. The urea-denatured states of CheY* and Cpβ4 display Rgs of ∼35 Å, slightly smaller than predicted for space-filling random coils of 129 amino acids, 38 Å (25). CheY* collapses to an apparent Rg of ∼25 Å within the ∼100-μs dead time, experiences a further compaction to ∼23 Å by 1 ms, and ultimately contracts to an Rg of 15 Å in the native conformation (Fig. 3A). In distinct contrast, Cpβ4 collapses to a near-native Rg, ∼18 Å within ∼100 μs and remains unchanged after 2.4 ms before contracting to the 15.5 Å Rg of the native state (Fig. 3A). Although the change in connectivity does not have a discernible effect on the size of the unfolded ensemble, the cleavage of the chain after β4 and the fusion of the natural N and C termini cause Cpβ4 to collapse more rapidly to a near-native Rg.
Fig. 3.
Dimensional analysis of CheY* and Cpβ4 during folding by SAXS and simulations. The radius of gyration for CheY* (black) and Cpβ4 (red) from CF-SAXS (A) and the average Rg from Gō-model simulations (CheY*: n = 46; Cpβ4: n = 32) in which the intermediate was observed (B) as a function of folding time. Statistical analysis of the simulations finds the intermediate to be highly populated within the average time values of the first and last occurrences (green box; see Table S2 for details).The unweighted Rg values of ION and IOFF species from simulations are shown as dotted lines. Arrows indicate the Rg values and their estimated uncertainties under equilibrium conditions for the folded and the unfolded states (A). Ninety-three points were collected within the mixer channel from 142–2,400 μs and averaged over 20 scans. After low-quality data points were removed, the remaining data were binned into two parts, 142–959 μs and 1,055–2,400 μs. CheY* Rg = 25.3 ± 2.2 Å (n = 11, 142–791 μs) and 22.6 ± 2.0 Å (n = 15 1,223–1,944 μs). Cpβ4 Rg = 18.0 ± 0.7 Å (n = 21, 142–959 μs) and 17.8 ± 0.7 Å (n = 33 1,055–2,304 μs).
Topological Frustration by Simulations.
The significant differences in the stabilities of the IBP species of these proteins are surprising, given the similarity of the kinetic responses observed. Unfortunately, the small amplitude of the refolding reaction along the cis channel precluded the use of global analysis to resolve the folding mechanism of the permutant proteins. Therefore we used Gō-model simulations to resolve the underlying structural basis of the differences in the IBP stability and infer the kinetic model that is most consistent with the experimental observables. Previous experimental work has concluded that the IOFF is not a consequence of the proline isomerization reaction (14, 21). Likewise, in computational work where the trans geometry was enforced via harmonic restraints, CheY still was able to access the folded state from the unfolded configurations. Although, the folded state is destabilized by 2.1 kcal·mol−1 relative to flexible Pro110 (26), the relative energy landscapes of the cis and trans channels are similar in the native and intermediate states (10, 11).
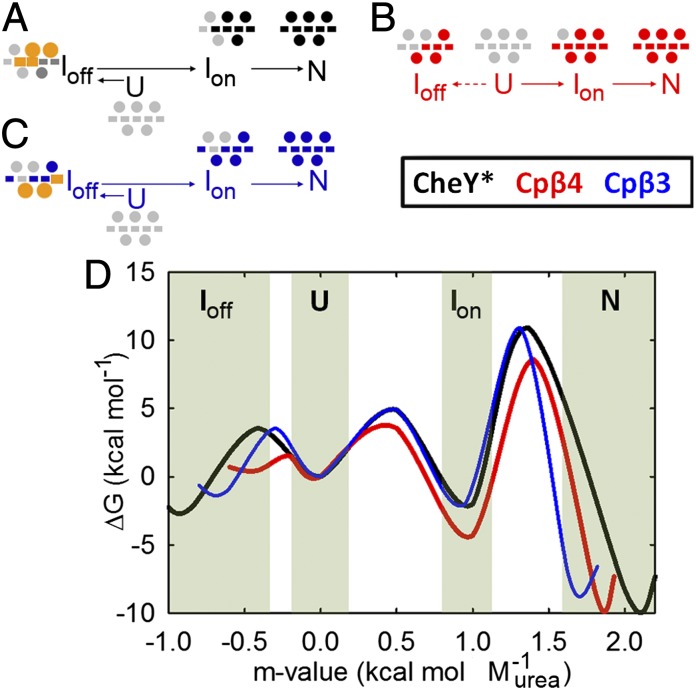
Despite using a model in which native interactions are predominantly favored, the model can capture frustration arising from the formation of native interactions in an incorrect order (27). Fig. 4 shows the influence of chain connectivity on the topological frustration as deduced from folding simulations of CheY* and its circular permutants. Our results are consistent with those reported earlier (10) and show that the folding of CheY* proceeds with significant frustration that arises from the competition of interactions between N-terminal, C-terminal, and interfacial native contacts. At the fraction of total native contacts formed (Qtotal) = 0.4, local unfolding or backtracking of interfacial contacts (negative slope) between the N and C termini coincides with the sudden increase in the contacts of the N-terminal subdomain (Fig. 4 A and D). These prematurely formed contacts in the C subdomain partially unfold before folding proceeds to the native conformation. Similar results are observed for Cpβ4; however, the interfacial frustration is markedly reduced (Fig. 4 B and E). In Cpβ3 this interfacial frustration is absent (Fig. 4 C and F) because the new termini disrupt the frustrated region. These results are consistent with the ILV cluster model for folding, in that the WT connectivity is driven to fold to the off-pathway IBP species by the premature formation of cluster 1 spanning the interfacial contacts (14). The local connectivity of the larger cluster 2 in Cpβ4 enables it to outcompete the formation of cluster 1.
Fig. 4.
Frustration observed in Gō-model simulations. (A–C) Ensemble averaged fractional contacts of the N-terminal subdomain (red), C-terminal subdomain (blue), and subdomain interface (green) are plotted as a function of fractional total native contacts for CheY* (A), Cpβ4 (B), and Cpβ3 (C). (D–F) The interfacial region is dissected in D–F where β3–β4 contacts are shown in magenta, α2–α3 contacts are shown in black, and α5–C-terminal contacts are shown in green. The C-terminal subdomain is dissected into fragments of β4–β5 contacts (gold) and α3–α4 contacts (blue) for CheY* (D), Cpβ4 (E), and Cpβ3 (F).
Notably, a minor restructuring event in the N-terminal subdomain is observed late in folding at Qtotal = 0.6 in all connectivities. This second event corresponds to the loosening of structure that is observed routinely in the folding of α-helices before final maturation of the tertiary structure and is not comparable to early frustration (27).
Kinetic Simulations.
More detailed structural insights into the folding mechanisms are gleaned through simulations from the time evolution of Rg and the corresponding time courses of the mean fraction of secondary structure contacts formed for the representative folding trajectories of CheY* and permutants. Cpβ4 collapses faster than CheY* (Fig. 3B) before both approach a common Rg of ∼14 Å in their respective native conformations. Examination of individual trajectories for CheY* and Cpβ4 (Fig. S6) revealed pauses, reflecting the transient occupancy of partially folded states with discrete Rg values. Of 100 kinetic trajectories, only about half pass through this intermediate and persist long enough to be observable. Therefore, the intermediate can be regarded as a nonobligate ION. Statistical analysis suggests that the intermediate for Cpβ4 is slightly more compact (20.2 vs. 21.3 Å), appears earlier (86 vs. 97 time units), and disappears sooner (104 vs. 151 time units) than its CheY* counterpart (Fig. 3B and Table S2). The Rgs for these intermediates are in remarkably good agreement with those observed by SAXS after 1 ms of folding, ∼23 Å for CheY* and ∼18 Å for Cpβ4 (Fig. 3A). The differences in the folding kinetics of CheY* and the permutants may reflect the extent of frustration that arises during the folding of each system.
To characterize the intermediates structurally, we extracted structures sampled during kinetic folding simulations that fall within 20 Å < Rg < 22 Å and measured the probability of forming native contacts in this ensemble. The results for CheY* are consistent with previous work (10) in which the Nheptad was identified as the structured region encompassing (βα)1–3β4 (Fig. S7A). Further, a subsection of the Nheptad with the highest probability of contact formation is apparent at the subdomain interface, β3–β4, a region previously described as an area where topological frustration is present (5). Through a similar analysis of Cpβ4 (Fig. S7B) and Cpβ3 (Fig. S7C), the differences in chain connectivity were found to have structural repercussions on the early-folding intermediates. The probability of forming contacts in the frustrated region is diminished as the Nheptad is lengthened to include α5/β5.
Discussion
CheY is a member of the flavodoxin fold family of proteins whose α/β/α sandwich architecture represents one of the more common motifs in biology. Unlike the flavodoxins, CheY has a conserved cis proline that controls the access to the native conformation (14, 20). Like CheY, however, a pair of homologous flavodoxins sample a kinetic trap before successfully traversing the productive TSE (8, 16). Elucidating the molecular basis for the frustration in folding for CheY has implications for an entire motif.
CheY*: The results presented here on the F14N variant of CheY are consistent with previous experimental and computational work on the WT protein (10, 14). By Gō-model simulations, topological frustration arises at the subdomain interface before partially unfolding to resume folding from the N→C terminus. This result is consistent with experimental data that show nonnative Phe packing in the IBP species (Fig. S5). The nonnative packing of IBP along with the backtracking observed by simulations (Fig. 4 A and D; see also ref. 14) and the negative m-value observed through global analysis of experimental data (14) argue that CheY populates an off-pathway kinetic trap, IOFF. Mechanistic details gleaned from the simulations suggest that low-CO ILV contacts in cluster 1 drive early folding events and lead to the premature formation of the subdomain interface.
Cpβ2: By introducing new termini between β2 and α2, the Cpβ2 permutant cleaves the N-terminal subdomain while leaving cluster 1 essentially intact and cluster 2 discontinuous. The observation that Cpβ2 is incapable of adopting the CheY* fold demonstrates an essential role for the intact N-terminal subdomain because cluster 2 is discontinuous in CheY* and in all three permutants. This conclusion is consistent with the results of a previous mutational analysis, which found the N-terminal subdomain to be a central feature of the productive TSE (21).
Cpβ3: The introduction of new termini between β3 and α3 leaves the two subdomains intact but cleaves both ILV clusters and the Nheptad. Notably, the FL and CD titrations are noncoincident, suggesting that multiple species are present before the global unfolding reaction. However, because the kinetic response is similar to that of CheY* under strongly unfolding conditions, Cpβ3 transverses the same barriers as CheY* (Fig. S4). The additional faster phase in unfolding may reflect a small fraction of the protein moving through a parallel channel in a limited range of unfolding conditions.
Although the amplitude of the CD spectrum of the IBP species for Cpβ3 is decreased by only ∼15% from its CheY* counterpart, the stability is reduced markedly, from 2.02 kcal·mol−1 for IBPt in CheY* to 0.84 kcal·mol−1 in Cpβ3 (Table S1), and the m-value is reduced from 0.83 to 0.59 kcal·mol−1·M−1. We attribute the decreased stability of IBPt to the cleavage of cluster 1, postulated to be a key stabilizing component of the IBPt species for WT CheY (14). Interestingly, the loss in stability is accompanied by native-like packing of the Phe cluster on the α1/α5 face of the β-sheet (Fig. 2D).
Simulations show the elimination of the interfacial frustration of the subdomains for Cpβ3, which would be expected if β3 and β4 are segregated to opposite ends of the chain. The absence of early frustration in the Cpβ3 simulations may reflect the marginal stability of the IBP species, as has been observed previously for a CheY homolog, NT-NtrC (14). In contrast to CheY*, frustration in Cpβ3 arises late in folding around the β1α1/β5α5 interface on the opposite face of the β-sheet (Fig. S8). The high Q values where this frustration occurs are not consistent with the small m-value for the IBP species for Cpβ3 and likely reflect annealing reactions often seen in the late stages of folding in Gō-model simulations when helix repacking often occurs.
The structural basis for the altered folding properties in Cpβ3 also can be visualized in 2D contact probability maps derived from the simulations (Fig. S7C). For its IOFF species, CheY* has a high probability of contacts in the α2(βα)3β4 region, but Cpβ3 does not. Indeed, the region of high probability of native contacts in Cpβ3 shifts to the β1α1 and β5α5 segments that are covalently linked by permutation of the sequence.
Cpβ4: The introduction of new termini between β4 and α4 in Cpβ4 cleaves the C-terminal subdomain while leaving cluster 1 intact and cluster 2 discontinuous. The coincidence of the far-UV CD spectra of CheY* and Cpβ4 (Fig. S1) shows that an intact C-terminal subdomain is not essential for proper folding and is in agreement with the view that the C-terminal subdomain forms after the TSE (21). The resultant IBP species folds more rapidly, is both more stable and more compact than CheY*, and has native-like packing of its phenylalanine cluster. The increased stability of IBP provides a logical explanation for the accelerated unfolding reaction, via the Hammond effect (Fig. 5) and argues for its assignment as an on-pathway intermediate. These surprising experimental results are in very good agreement with the predictions of decreased frustration from an off-pathway intermediate and a more compact on-pathway intermediate including β1, α1, β5, and α5 in the simulations.
Fig. 5.
Structures of intermediates and the simplified folding free-energy surfaces. The sequence of events in folding is indicated by the arrows. The proline isomerization step, occurring between the cis and trans IBP species, is not shown. Structured components of each species as determined by Gō-model simulations. Elements in gray are not yet formed; colored elements [A: black, CheY*; B: red, Cpβ4; C: blue, Cpβ3] are significantly structured; elements implicated in topological frustration are orange. (D) Reaction coordinate diagrams for CheY* (black), Cpβ3 (blue), and Cpβ4 (red). The barrier heights were estimated using the Kramer’s formalism with a prefactor of 1 μs, and m-values were calculated from equilibrium and kinetic experiments, when available. Each permutant would have a unique unfolded ensemble, but the free energies have been aligned for direct comparison.
The 2D contact probability map of the Cpβ4 folding intermediate reveals an intact Nheptad and a high probability for contacts between the covalently connected β1α1 and β5α5 sequences. The linkage of the natural termini leads to the preferential formation and stabilization of a species that corresponds to the ION for CheY*. The decreased frustration for Cpβ4 likely reflects both the destabilization of the C-terminal subdomain via cleavage and the increased competition from the more rapidly forming and stable extended Nheptad, including the β1α1/β5α5 complex.
Early Folding Events by CF-SAXS, Simulations, and CF-FL.
The faster collapse of unfolded Cpβ4 observed by CF-SAXS (Fig. 3A) and simulations (Fig. 3B) is not reflected in the CF-FL data, which found essentially identical relaxation times for Cpβ4 and CheY* (Fig. S4). The discrepancy can be traced to the small m-value for the 300-μs phase and the implied small change in buried surface area accompanying this reaction. The commonality of the relaxation time of this phase for Cpβ3, Cpβ4, and CheY* strongly suggests a local folding event at the single Trp residue that does not reflect the global collapse monitored by CF-SAXS and simulations.
Modulation of the Folding Landscape by Permutations.
Both experiments on and simulations of CheY*, Cpβ3, and Cpβ4 reveal that the initial events in the folding are dictated by the connectivity of the chain. In another case, Cpβ2, altering the chain connectivity leads to a distinctly different but well-defined thermodynamic state. The combined results for those sequences that can attain the wild-type native conformation can be displayed on a reaction coordinate diagram shown in Fig. 5D; the proposed structured elements for the various species are shown in Fig. 5 A–C.
The path from the unfolded state to the respective intermediates for CheY*, Cpβ3, and Cpβ4 is controlled by preferred interactions between low-CO elements of secondary structure. The varying structures, stabilities, and buried surface areas for these partially folded states can be understood in terms of the thermodynamic compulsion to minimize the chain entropy penalty and maximize the participation of their resident aliphatic side chains in one of two ILV clusters located on either face of the central β-sheet. For CheY*, cluster 1 forms early and stabilizes IOFF. For Cpβ3, cluster 1 is cleaved, and a fraction of cluster 2 drives the formation of a poorly folded fragile IOFF. For Cpβ4, the C-terminal elements of cluster 2 reinforce the Nheptad, resulting in a remarkably stable ION. Thus, the folding free-energy surface of CheY and its attendant frustration in folding can be modulated either by the destabilization of the off-pathway intermediate, Cpβ3, or by the stabilization of an on-pathway intermediate, Cpβ4. Although the initial sources of frustration for these permuted sequences are quite different, all can achieve essentially the same native conformation.
Subdomain vs. ILV Cluster Model for the Folding of CheY.
The totality of the results suggests that the ILV cluster model provides the more parsimonious and complete description of the early events in folding but that the subdomain model better captures the crucial TSE required to access the proper native fold. In other words, low-CO clusters of ILV residues can strongly influence the early stages of folding before subdomain and global cooperativity engage expanding portions of the sequence to reach the native conformation.
Perspective.
Chain entropy plays a crucial role in defining the energies and structures of partially folded states on the folding free-energy surface of CheY. Thus, frustration can be modulated and productive folding favored by altering the sequence connectivity and, thereby, the local chain entropy. The local-in-sequence local-in-space topology of βα-repeat proteins, including the Rossmann-fold, triosephosphate isomerase barrels, and the flavodoxin/CheY folds, make them prime candidates for frustration in the early stages of folding. The associated partially folded states not only may impede the folding reaction but also may serve to nucleate aggregation reactions in pathological sequence variants. Recognition of the early events in folding and the partially folded structures that they produce provides a rational basis for the design of small molecules that might inhibit aggregation by binding at the interfaces of these nascent kernels of structure.
Methods
Thermodynamic and Kinetic Experiments and Analysis.
Details regarding protein expression, purification, and thermodynamic and kinetic characterization have been described previously (14). For details see SI Methods.
Equilibrium SAXS.
Equilibrium measurements were collected as previously described (28). The protein concentration was 1.5 mg·mL−1 in 10 mM potassium phosphate buffer at pH 7.0 and 25 °C.
CF-SAXS.
CF-SAXS measurements were made as previously described (29). The total flow rate was 20 mL·min−1 using a 1:10 dilution of the unfolded protein for a final protein concentration of 1.5 mg·mL−1 in 10 mM potassium phosphate, 8 M urea at pH 7.0 and refolding with 0 M urea buffer.
Gō-Model Simulations.
System preparation and model.
Cpβ4 and Cpβ3 were modeled on the crystal structure of WT CheY from Escherichia coli (Protein Data Bank ID code: 3CHY) (30). Models of both permutants were constructed by joining together the N and C termini with a Gly-Ala-Gly peptide and cleaving the bond between residues 63 and 64 and between residues 88 and 89 for the Cpβ3 and Cpβ4 permutants, respectively. The protein-folding simulations were performed with an unrestrained prolyl-bond geometry using a coarse-grained model developed by Karanicolas and Brooks (31). See SI Methods for further details.
Molecular dynamics protocol.
Molecular dynamics simulations were performed using the CHARMM macromolecular mechanics package (32). All models were evolved through Langevin dynamics, by using a friction coefficient of 1.36 ps−1 and a molecular dynamics time step of 22 fs. The virtual bond lengths were kept fixed using the SHAKE algorithm. For each permutant, 100 independent folding simulations were each performed for 2 × 108 dynamics steps at 0.87 Tf, where Tf is the folding transition temperature estimated as a temperature corresponding to the peak in the specific heat curve, Cv (T) (Fig. S3). See SI Methods for details.
Supplementary Material
Acknowledgments
We thank Jill Zitzewitz and Noah Cohen for helpful discussions, Ornella Bisceglia for helping with protein preparation, and Ronald Hills for help in revising the paper. This work was supported by the National Institutes of Health (NIH) through the Center for Multi-Scale Modeling Tools for Structural Biology Grant RR012255, the National Science Foundation through the Center for Theoretical Biological Physics Grant PHY0216576, the Division of Molecular and Cellular Biosciences Grant MCB1121942, National Center for Research Resources Grant 2P41RR008630-17, and NIH/National Institute of General Medical Sciences Grant 9 P41 GM103622-17. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract No. DE-AC02-06CH11357. This project was supported by Grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324230111/-/DCSupplemental.
References
- 1.Yoo TY, et al. Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J Mol Biol. 2012;418(3-4):226–236. doi: 10.1016/j.jmb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nozaki Y, Schechter NM, Reynolds JA, Tanford C. Use of gel chromatography for the determination of the Stokes radii of proteins in the presence and absence of detergents. A reexamination. Biochemistry. 1976;15(17):3884–3890. doi: 10.1021/bi00662a036. [DOI] [PubMed] [Google Scholar]
- 3.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14(1):76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Roder H, Maki K, Cheng H. Early events in protein folding explored by rapid mixing methods. Chem Rev. 2006;106(5):1836–1861. doi: 10.1021/cr040430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton WA, Thompson PA, Chan CK, Hage SJ, Hofrichter J. Fast events in protein folding. Structure. 1996;4(10):1133–1139. doi: 10.1016/s0969-2126(96)00121-9. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann H, et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(40):16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindorff-Larsen K, Trbovic N, Maragakis P, Piana S, Shaw DE. Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J Am Chem Soc. 2012;134(8):3787–3791. doi: 10.1021/ja209931w. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Recio J, Genzor CG, Sancho J. Apoflavodoxin folding mechanism: An alpha/beta protein with an essentially off-pathway intermediate. Biochemistry. 2001;40(50):15234–15245. doi: 10.1021/bi010216t. [DOI] [PubMed] [Google Scholar]
- 9.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14(1):70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Hills RD, Jr, Brooks CL., 3rd Subdomain competition, cooperativity, and topological frustration in the folding of CheY. J Mol Biol. 2008;382(2):485–495. doi: 10.1016/j.jmb.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hills RD, Jr, et al. Topological frustration in beta alpha-repeat proteins: Sequence diversity modulates the conserved folding mechanisms of alpha/beta/alpha sandwich proteins. J Mol Biol. 2010;398(2):332–350. doi: 10.1016/j.jmb.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Kondrashkina E, Kayatekin C, Matthews CR, Bilsel O. Microsecond acquisition of heterogeneous structure in the folding of a TIM barrel protein. Proc Natl Acad Sci USA. 2008;105(36):13367–13372. doi: 10.1073/pnas.0802788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finke JM, Onuchic JN. Equilibrium and kinetic folding pathways of a TIM barrel with a funneled energy landscape. Biophys J. 2005;89(1):488–505. doi: 10.1529/biophysj.105.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kathuria SV, Day IJ, Wallace LA, Matthews CR. Kinetic traps in the folding of beta alpha-repeat proteins: CheY initially misfolds before accessing the native conformation. J Mol Biol. 2008;382(2):467–484. doi: 10.1016/j.jmb.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 15.Nabuurs SM, Westphal AH, van Mierlo CPM. Extensive formation of off-pathway species during folding of an alpha-beta parallel protein is due to docking of (non)native structure elements in unfolded molecules. J Am Chem Soc. 2008;130(50):16914–16920. doi: 10.1021/ja803841n. [DOI] [PubMed] [Google Scholar]
- 16.Bollen YJM, Kamphuis MB, van Mierlo CPM. The folding energy landscape of apoflavodoxin is rugged: Hydrogen exchange reveals nonproductive misfolded intermediates. Proc Natl Acad Sci USA. 2006;103(11):4095–4100. doi: 10.1073/pnas.0509133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borgia MB, et al. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474(7353):662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plaxco KW, Simons KT, Ruczinski I, Baker D. Topology, stability, sequence, and length: defining the determinants of two-state protein folding kinetics. Biochemistry. 2000;39(37):11177–11183. doi: 10.1021/bi000200n. [DOI] [PubMed] [Google Scholar]
- 19.Garcia P, Serrano L, Rico M, Bruix M. An NMR view of the folding process of a CheY mutant at the residue level. Structure. 2002;10(9):1173–1185. doi: 10.1016/s0969-2126(02)00804-3. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz V, Lopez EM, Jager M, Serrano L. Kinetic characterization of the chemotactic protein from Escherichia coli, CheY. Kinetic analysis of the inverse hydrophobic effect. Biochemistry. 1994;33(19):5858–5866. doi: 10.1021/bi00185a025. [DOI] [PubMed] [Google Scholar]
- 21.López-Hernández E, Serrano L. Structure of the transition state for folding of the 129 aa protein CheY resembles that of a smaller protein, CI-2. Fold Des. 1996;1(1):43–55. [PubMed] [Google Scholar]
- 22.Hills RD, Jr, Brooks CL., 3rd Insights from coarse-grained Gō models for protein folding and dynamics. Int J Mol Sci. 2009;10(3):889–905. doi: 10.3390/ijms10030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 24.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4(10):2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohn JE, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101(34):12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hills RD., Jr . In: Protein Dynamics: Methods and Protocols, Methods in Molecular Biology, Methods in Molecular Biology. Livesay DR, editor. Totowa, NJ: Humana; 2014. pp. 123–140. [Google Scholar]
- 27.Clementi C, Jennings PA, Onuchic JN. Prediction of folding mechanism for circular-permuted proteins. J Mol Biol. 2001;311(4):879–890. doi: 10.1006/jmbi.2001.4871. [DOI] [PubMed] [Google Scholar]
- 28.Kathuria SV, et al. Microsecond barrier-limited chain collapse observed by time-resolved FRET and SAXS. J Mol Biol. 2014;426(9):1980–1994. doi: 10.1016/j.jmb.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graceffa R, et al. Sub-millisecond time-resolved SAXS using a continuous-flow mixer and X-ray micro-beam. J Synchrotron Radiat. 2013;20:1–6. doi: 10.1107/S0909049513021833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266(23):15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 31.Karanicolas J, Brooks CL., 3rd The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11(10):2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.