Significance
Legionella pneumophila, the Legionnaires’ disease-causing bacterial pathogen, translocates a myriad of bacterial proteins, called effectors, into host cells. These proteins exploit normal host cellular functions to facilitate bacterial intracellular growth. To identify the functions of these bacterial effectors has been a major challenge. Here, we determined the structure of one such effector, substrate of Icm/Dot transporter (SidC), which was previously thought to be a vesicle-tethering factor for recruiting endoplasmic reticulum (ER) vesicles to the bacterial vacuoles. Surprisingly, our data uncovered a canonical catalytic triad resembling that of cysteine proteases. We further demonstrated that SidC possesses ubiquitin ligase activity, which is required for the enrichment of ER proteins and ubiquitin conjugates to bacterial vacuoles. Collectively, our data on SidC define a unique family of ubiquitin ligases.
Keywords: pathogen infection, membrane trafficking, ubiquitination, ER vesicle, type IV secretion system
Abstract
The activity of proteins delivered into host cells by the Dot/Icm injection apparatus allows Legionella pneumophila to establish a niche called the Legionella-containing vacuole (LCV), which is permissive for intracellular bacterial propagation. Among these proteins, substrate of Icm/Dot transporter (SidC) anchors to the cytoplasmic surface of the LCV and is important for the recruitment of host endoplasmic reticulum (ER) proteins to this organelle. However, the biochemical function underlying this activity is unknown. Here, we determined the structure of the N-terminal domain of SidC, which has no structural homology to any protein. Sequence homology analysis revealed a potential canonical catalytic triad formed by Cys46, His444, and Asp446 on the surface of SidC. Unexpectedly, we found that SidC is an E3 ubiquitin ligase that uses the C–H–D triad to catalyze the formation of high–molecular-weight polyubiquitin chains through multiple ubiquitin lysine residues. A C46A mutation completely abolished the E3 ligase activity and the ability of the protein to recruit host ER proteins as well as polyubiquitin conjugates to the LCV. Thus, SidC represents a unique E3 ubiquitin ligase family important for phagosomal membrane remodeling by L. pneumophila.
The ubiquitous bacterium, Legionella pneumophila, is found in aquatic environments where it infects freshwater protozoa. Development of the potentially fatal Legionnaires’ disease occurs when susceptible individuals inhale contaminated aerosols (1). After engulfment by phagocytes, L. pneumophila uses its Dot/Icm type IV secretion system to deliver a large number of effector proteins that modulate host cellular processes, leading to the creation of a specialized Legionella-containing vacuole (LCV) that provides the environment for robust intracellular bacterial growth (2). The mature LCV is characterized by an enrichment of a particular phosphoinositide lipid, PI(4)P (3–5), and by the accumulation of endoplasmic reticulum (ER) proteins, presumably captured by intercepting vesicles derived from the ER (6, 7). Another feature of the LCV is enrichment of polyubiquitin conjugates around the vacuolar membrane (8). This sophisticated membrane remodeling process is achieved by the coordinated activity of effector proteins delivered into the host through the Dot/Icm apparatus (9, 10). However, the mechanism underlying these processes is still not well established. Recently, the Legionella effector proteins substrate of Icm/Dot transporter (SidC) (11) and its paralog SdcA were proposed to function as vesicle fusion tethering factors. Both proteins were shown to recruit ER vesicles to the LCV while anchored on the LCV via a specific C-terminal phosphatidylinositol-4-phosphate [PI(4)P]-binding domain (3, 11, 12). Bacterial vacuoles harboring the ΔsidC-sdcA mutant bacteria recruit ER-derived vesicles less efficiently, and in vitro experiments further showed that the N-terminal 70-kDa fragment of SidC binds to ER vesicles in Dictyostelium and macrophage lysates (12). However, the biochemical mechanism for SidC-mediated ER recruitment remains unclear.
Posttranslational modification by ubiquitin (Ub) regulates a myriad of cellular pathways, including protein homeostasis (13), cell signaling (14), and membrane trafficking (15, 16). Protein ubiquitination requires the sequential activities of a cascade of enzymes known as ubiquitin activating enzyme E1, ubiquitin conjugating enzyme E2, and ubiquitin ligase E3 (17). Ubiquitin E3s belong to two major groups: the really interesting new gene (RING)-domain and the homologous to the E6AP carboxyl terminus (HECT)-domain families (18, 19). E3s in the RING family facilitate the transfer of Ub from the E2 catalytic cysteine directly to a substrate (18). However, HECT-domain E3s use their own active cysteine to form a thioester bond-linked E3∼Ub intermediate before transferring the Ub to specific substrates.
Due to the important role of protein ubiquitination in eukaryotes, it is not surprising that a number of bacterial pathogens and symbionts exploit the host ubiquitin pathway (20, 21). For example, the Pseudomonas syringae protein AvrPtoB contains a RING-like domain and functions as a ubiquitin ligase to inhibit host programmed cell death (22). The effector SopA from Salmonella enterica is a HECT-like ubiquitin ligase with a catalytic domain organized in a bilobed architecture similar to the conventional HECT E3s (23). The Shigella effector IpaH has a C-terminal domain that carries ubiquitin ligase activity but bears no sequence and structural resemblance to other ubiquitin ligases (24).
In L. pneumophila, a number of Dot/Icm effectors are predicted to contain regions with sequence similarity to the F-box or U-box domain (25, 26). Several of such predicted F-box containing effectors, including LegAU13/AnkB (27–29), LegU1, and LicA (27) interact with components of the Skp–Cullin–F-box (SCF) ubiquitin ligase complex. In vitro assays have further validated the ubiquitin E3 ligase activity for LegU1, LegAU13/AnkB (27), and LubX (30). Using yeast two-hybrid or other biochemical methods, several specific host and bacterial proteins that are targeted for ubiquitination by Legionella E3 ligases have been identified. The U-box type E3 ligase LubX polyubiquitinates the host kinase Clk1 (30) and the Legionella effector SidH (31). The F-box protein LegU1 specifically directs the ubiquitination of the host chaperone protein BAT3 (27). The cooption of the host ubiquitin pathway by L. pneumophila is further supported by a seminal discovery that the bacterium recruits polyubiquitinated species around the bacterial phagosome shortly after bacterial uptake (8). Despite these findings, much remains to be discovered regarding effectors and their targets in the exploitation of the host ubiquitin pathway by L. pneumophila.
Here we report the crystal structure of the N-terminal portion of SidC, which revealed a canonical catalytic triad containing a cysteine, a histidine, and an aspartate residue. We found that ectopic expression of this domain altered the intracellular ubiquitination pattern. In vitro experiments demonstrated the formation of high–molecular-weight ubiquitinated conjugates in a manner that is dependent on the catalytic residue C46. We further showed that the SidC paralog SdcA has ubiquitin ligase activity but with a preference for a different E2. Finally, wild-type SidC, but not the C46A mutant, can fully complement the defect of ΔsidC-sdcA mutant in the recruitment of ER markers during infection. Our results demonstrate that the Legionella effector SidC defines a unique family of ubiquitin ligases, the activity of which facilitates the maturation of LCV by remodeling its protein composition.
Results
Crystal Structure of the N-Terminal Domain of SidC Reveals a Canonical Catalytic Triad.
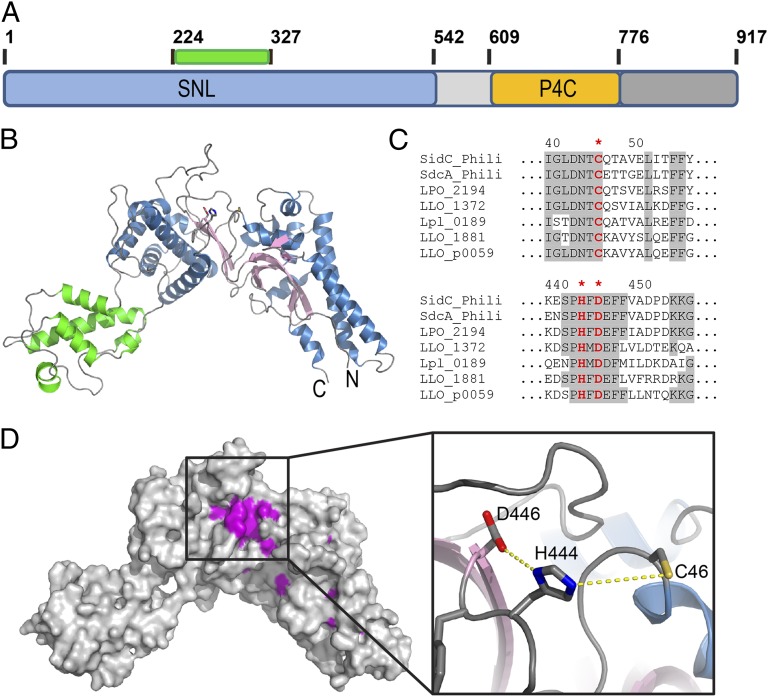
SidC is a large protein containing 917 amino acid residues with a conserved N-terminal domain and a C-terminal PI(4)P binding domain, P4C (Fig. 1A). The N-terminal portion of SidC has sequence homology to other Legionella proteins lacking this PI(4)P binding domain, indicating the presence of a functional independent module (amino acids 1–542) within the N terminus (SI Appendix, Figs. S1 and S2). We named this conserved domain as SidC N-terminal ubiquitin ligase (SNL) domain.
Fig. 1.
Crystal structure of the N-terminal SNL domain of SidC (amino acids 1–542). (A) Schematic diagram of the domain structure of SidC. SidC contains an N-terminal SNL domain (blue) and a C-terminal PI(4)P binding domain, P4C (yellow). The green bar indicates the position of an inserted subdomain within the SNL domain. (B) Ribbon diagram of the overall structure of the SNL domain. The SNL domain has two subdomains. The main subdomain contains a two layered β-sheet (pink) flanked by two clusters of α-helices (blue). The all α-helical insertion subdomain is shown in green. (C) Multiple sequence alignment reveals two clusters of conserved residues. The first cluster contains an invariable cysteine C46, and the second cluster contains invariable H444 and D446. (D) Mapping of the invariable residues across all SNL domains to the structure. Identical residues in the two conserved residues clusters shown in C form a patch on the surface. (Inset) A zoom-in view of this conserved residue patch unveils the presence and the structural arrangement of a canonical catalytic triad formed by C46, H444, and D446.
The structure of the SNL domain was determined by single isomorphous replacement with anomalous signal (SIRAS) using crystals soaked with a mercury containing compound (SI Appendix, Fig. S3 and Table S1). The structure reveals that the SNL domain has a crescent-like overall shape consisting of two subdomains (Fig. 1 A and B). A small all-alpha helical domain (in green) is inserted into the rest of the protein between residues 224 and 327 (SI Appendix, Figs. S2 and S4). The main domain contains a core of two layered β-sheets, which is sandwiched between two clusters of α helices (Fig. 1B and SI Appendix, Fig. S4). Structural homology search with the Dali server (32) did not yield any significant hits, which restricts the functional assignment based on structural homology. Because many Legionella effector proteins are enzymes, we hypothesized that SidC may be an enzyme. In this case, the catalytic residues responsible for the presumed catalytic function should be conserved across all SidC homologs. To test this hypothesis, we performed multiple sequence alignment analyses. The alignment revealed two clusters of conserved residues within the SNL domain (Fig. 1C and SI Appendix, Fig. S2). Strikingly, when all of the identical residues across the SNL-domain family members were mapped to the crystal structure, some of these residues emerging on the surface of the structure formed a continuous patch (Fig. 1D and Movie S1). Within this patch, three completely identical residues, C46, H444, and D446, are arranged in a way that is reminiscent of the classical catalytic triad found in cysteine-based proteases (33), deubiquitinases (DUB) (34), and other cysteine-based enzymes. Interestingly, the area encompassing this conserved residue patch shows a concentrated negative electrostatic potential (SI Appendix, Fig. S5), suggesting a potential site for protein-protein or protein-substrate interactions. Taken together, the sequence and structure analyses of the SNL domain raised the possibility that this domain is an enzyme containing a conserved Cys-His-Asp catalytic triad.
Ectopic Expression of SidC Alters the Pattern of Intracellular Ubiquitinated Species.
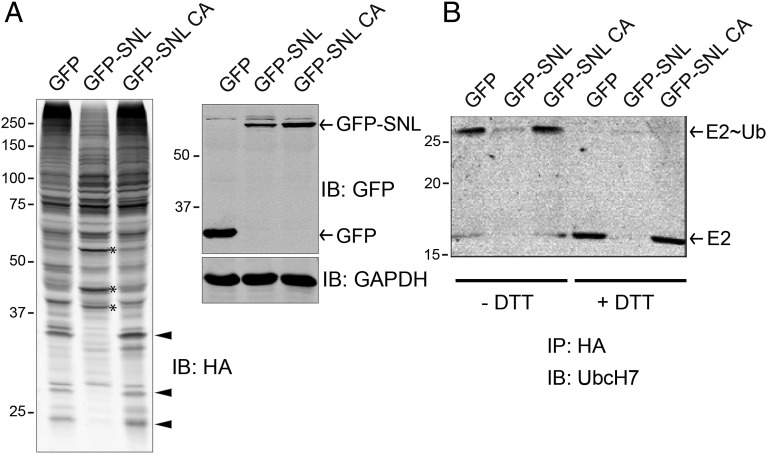
The enzymatic activity of SidC was tested using commercially available cysteine protease kits or polyubiquitin chains as potential DUB substrates. However, we did not detect either kind of activity (SI Appendix, Fig. S6). To interrogate the potential cysteine-related enzymatic activities of SidC, we coexpressed the GFP-tagged wild-type SNL domain of SidC or its C46A mutant with human HA-tagged ubiquitin (HA-Ub) in 293T cells. Whole cell lysates were separated in SDS/PAGE gels and probed for HA. The pattern of ubiquitinated species was significantly different in cells expressing the wild-type SNL domain from that in cells expressing the SNL C46A mutant or GFP control (Fig. 2A). Some ubiquitinated species were more prominent (indicated by an asterisk), whereas others were absent (indicated by arrowheads) in transfected cells (Fig. 2A). This result suggests that SidC affects the host ubiquitin pathway. To identify potential proteins with an altered ubiquitination state, we used the stable isotope labeling by amino acids in cell culture (SILAC) method (35). HEK293T cells were first cotransfected with HA-Ub and GFP-tagged SNL domain (either the wild type or its C46A mutant). After labeling cells with heavy or light medium, cell lysates were prepared and ubiquitinated species were immunoprecipitated with anti-HA antibody coated beads. The samples were further analyzed by mass spectrometry (SI Appendix, Fig. S7 and Tables S2 and S3). Strikingly, SILAC analysis showed that five of the nine top hits with reduced ubiquitination in cells expressing the wild-type SNL domain belong to the ubiquitin-conjugating enzyme E2 family (SI Appendix, Table S2).
Fig. 2.
Ectopic expression of SidC altered the intracellular ubiquitination pattern. (A) HEK293T cells were cotransfected with HA-ubiquitin and GFP control, GFP-tagged wild-type SidC SNL domain, or its C46A mutant. Whole cell lysates were prepared and analyzed by Western blot with anti-HA (Left), anti-GFP, and anti-GAPDH as a loading control (Right). Arrowheads highlight several bands that are positive in GFP and GFP-SNL C46A controls but are diminished in the presence of wild-type SNL domain. Asterisk denotes bands that are more prominent in the sample expressing wild-type SNL domain. (B) HEK293T cells were cotransfected with HA-ubiquitin and other indicated plasmids. Cells lysates were prepared and incubated with anti-HA beads to immunoprecipitate HA-Ub tagged species. The precipitated samples were prepared in SDS loading buffer without (three lanes on the left) or with (three lanes on the right) DTT and subjected to Western blot analysis. In the presence of DTT, the E2∼Ub complex was fully reduced to E2, indicating a thioester linkage between E2 and ubiquitin.
To further validate the mass spectrometry results, a similar experiment was performed and the immunoprecipitated proteins were solubilized with SDS with or without DTT and analyzed by Western blot using specific antibody against the E2 enzyme UBE2L3/UbcH7. Indeed, the amount of ubiquitinated UbcH7 was significantly reduced in the presence of the wild-type SNL domain compared with the GFP control or the SNL C46A mutant (Fig. 2B). In the presence of DTT, the E2∼Ub bands were shifted down relative to their corresponding E2 band (Fig. 2B, last three lanes). This observation implies that the linkage between the C-terminal carboxyl group of the Ub molecule and E2 is not an isopeptide bond, which would be resistant to DTT treatment. Instead, the Ub of the immunoprecipitated E2∼Ub appears to be linked via a thioester bond to the catalytic cysteine of the E2, which is readily reduced by DTT (Fig. 2B, last three lanes). These data further suggest that SidC is not a DUB, which cleaves the isopeptide linkage between Ub and target proteins or within Ub chains. Given the fact that the ectopically expressed SNL domain can discharge the activated Ub (linked to the E2 through a thioester bond) from the E2, we hypothesize that SidC functions as a ubiquitin ligase that efficiently hijacks the charged Ub from E2s and then attaches the Ub moiety to other SidC-specific targets. In agreement with this hypothesis, ectopic expression of the SNL domain did not change the total levels of UbcH7; instead, it only exhausted the pool of E2 charged with ubiquitin (SI Appendix, Fig. S8). This hypothesis also explains the observation shown in Fig. 2A that some ubiquitinated species were diminished (presumably due to competition with endogenous E3s for charged E2s), whereas others appeared more prominent in the presence of the SNL domain (presumably due to the ubiquitin ligase activity of SidC).
N-Terminal Domain of SidC/SdcA Is a Ubiquitin E3 Ligase.
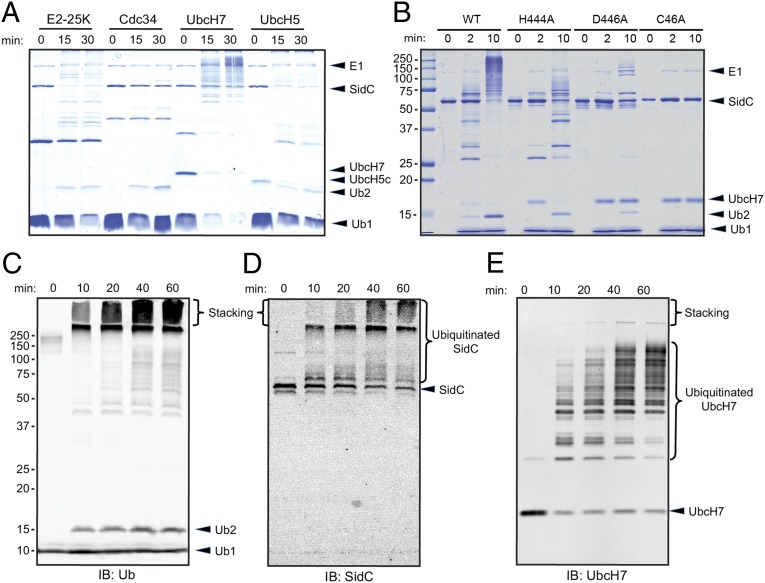
To test whether SidC is a ubiquitin E3 ligase, we performed in vitro ubiquitination assays using purified recombinant proteins (Materials and Methods). Assays were performed with the SNL domain of SidC (amino acids 1–542), E1, and a set of four representative E2s (bovine E2-25K, human Cdc34/UBE2R1, UbcH7/UBE2L3, and UbcH5c/UBE2D3). As expected, in the absence of SidC, di-ubiquitin (di-Ub) chains were formed in the presence of E2-25K or Cdc34, demonstrating that the purified ubiquitin E1 and E2 enzymes are functional (SI Appendix, Fig. S9). By contrast, characteristic ubiquitin ladders were observed when the wild-type SNL domain was incubated with E2-25K, UbcH7, and UbcH5c but were barely visible with Cdc34 (Fig. 3A). Because SidC appeared most active in the presence of UbcH7 (Fig. 3A), which was also among the top SILAC hits (SI Appendix, Table S2), we used UbcH7 in all our following ubiquitin E3 ligase assays. Next, we examined the ubiquitin ligase activity of SidC mutants in which the proposed catalytic triad was disrupted. Compared with the wild-type SNL domain protein, both the H444A and D446A showed reduced activity, whereas the activity was completely abolished in the C46A mutant (Fig. 3B).
Fig. 3.
The SNL domain of SidC has E3 ubiquitin ligase activity. (A) In vitro ubiquitination assays with the SNL domain of SidC (amino acids 1–542) and 4 representative E2s: E2-25K, Cdc34, UbcH7 and UbcH5. (B) E3 activity assay of SidC H444A, D446A, and C46A mutants. The activity of the C46A mutant was completely abolished, whereas the activities of the H444A and D446A mutants were significantly reduced. (C) Time-dependent ubiquitination assay. The reactions were performed with E1, UbcH7, SNL domain, and wild-type ubiquitin. The reaction mixtures were analyzed by Western blot with anti-ubiquitin antibody. Within the stacking gel, an increased amount of heavily polyubiquitinated species was observed during the time course. (D) Western blot of the same materials as in C with anti-SidC antibody. The high–molecular-weight ubiquitin species were positive for SidC. (E) Western blot of the same samples as in C with anti-UbcH7. UbcH7 was also ubiquitinated during the in vitro reaction.
To analyze the nature of the polyubiquitinated species, time-dependent reactions with physiologically relevant amounts of enzymes were performed. The time-course experiments showed the gradual appearance of high–molecular-weight polyubiquitinated species and a small amount of di-Ub chains. The majority of these high–molecular-weight ubiquitin species were retained in the stacking gel (Fig. 3C). Probing the blot with a SidC-specific antibody revealed that the ubiquitin species retained in the stacking gel and at the top part of the separation gel were products of SidC autoubiquitination (Fig. 3D). Western blot with antiserum against UbcH7 also revealed poly- or multiple monoubiquitination of this E2 (Fig. 3E). These ubiquitinated E2s contributed to the ubiquitin signals derived from ubiquitin species in the middle range of molecular weights observed in Fig. 3C. E3 ligases often display autoubiquitination in in vitro assays, and in most instances autoubiquitination is considered as an activity readout (36). In the case of SidC, autoubiquitination was not detected under infection conditions (SI Appendix, Fig. S10).
We further compared the ubiquitin ligase activity of the SNL domain (1–542) with the full-length SidC and the full-length SidC paralog SdcA. Both the SNL domain and the full-length SidC protein showed comparable activity (SI Appendix, Fig. S11). Intriguingly, SdcA exhibited a ubiquitin ligase activity, but with a different E2 preference. Unlike SidC, SdcA efficiently catalyzed ubiquitin polymerization in the presence of UbcH5 (SI Appendix, Fig. S12). This observation suggests that L. pneumophila encodes two seemingly redundant genes to maximize its ability to hijack the host ubiquitin system. Together, these data demonstrate that SidC and its paralog SdcA are bona fide ubiquitin ligases that have a broad and nonoverlapping specificity for ubiquitin-conjugating E2 enzymes.
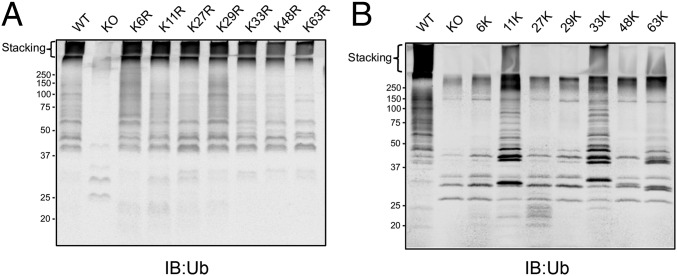
The N-Terminal Domain of SidC Catalyzes Multiple Types of Ubiquitin Chain Linkage.
We next investigated the linkages of the heavily polyubiquitinated species made by SidC. The linkage within Ub chains is formed between the C-terminal carboxyl group of a distal Ub and the ε-amino group on a lysine or the N-terminal amino group of the proximal Ub. Ub has seven lysine residues in addition to the N-terminal methionine, implying eight possible linkages in poly-Ub chains. We first tested the activity of SidC with seven ubiquitins, each carrying a single lysine to arginine mutation. Similar to wild-type ubiquitin, all single K to R ubiquitin mutants produced high–molecular-weight polyubiquitin species that were retained within the stacking gel. By contrast, a Ub mutant carrying all seven K to R mutation (K0) did not show evidence of poly-Ub formation (Fig. 4A). These data suggest that SidC is capable of catalyzing the formation of poly-Ub chain through multiple lysine residues, but not via the N-terminal methionine. To further dissect the linkage preference of SidC, we assayed the activity of SidC with ubiquitin mutants containing six K-to-R mutations with only one native lysine remaining. Although less efficient compared with wild-type ubiquitin, high–molecular-weight polyubiquitinated species did form in the presence of Ub-11K and Ub-33K (Ub mutant with only one native lysine at positions 11 and 33, respectively). Much weaker poly-Ub signals were observed with mutant Ub-63K and Ub-48K (Fig. 4B). These results indicate that SidC is able to use multiple lysine residues for ubiquitin polymerization with a preference for K11 and K33-linked chains.
Fig. 4.
The ubiquitin linkage preference by the SNL domain of SidC. (A) In vitro ubiquitination assays using E1, UbcH7, SNL domain, and a set of ubiquitin mutants with a single K-to-R mutation. K0 represents the ubiquitin mutant carrying all seven K-to-R to mutations. (B) In vitro ubiquitination assays using E1, UbcH7, SNL domain, and a set of ubiquitin containing a single native lysine with six K-to-R mutations.
Ubiquitin Ligase Activity of SidC Is Required for the Recruitment of ER Proteins and Ubiquitin to the LCV.
SidC and its paralog SdcA have been shown to recruit ER vesicles to the LCV (12). We thus investigated the relevance of the E3 ligase activity of SidC in Legionella infection. A Dictyostelium discoideum strain stably expressing GFP-HDEL (37) (a GFP fusion with the ER retention marker HDEL) was infected with relevant Legionella strains and the bacteria were immunostained with an anti-Legionella antibody. In D. discoideum infected with wild-type L. pneumophila, more than 50% of the LCVs were positive for GFP-HDEL 2 h after uptake (Fig. 5 A and B). In contrast, such recruitment did not occur in infections with a Dot/Icm deficient strain. The association of GFP-HDEL with the LCVs created by the ΔsidC-sdcA mutant was significantly reduced. Importantly, this defect was almost fully restored by a plasmid expressing wild-type SidC but not by its catalytically inactive C46A mutant (Fig. 5 A and B). The C46A mutation did not have detectable effects on the expression or the translocation of the protein by the bacteria (SI Appendix, Fig. S13). These results demonstrate that the ubiquitin ligase activity of SidC is critical for its role in the recruitment of ER components to the bacterial phagosome.
Fig. 5.
The E3 ubiquitin ligase activity is required for the ER marker recruitment to the bacterial phagosome. (A) Images show the recruitment of the ER marker GFP-HDEL (green) to the LCVs in D. discoideum cells infected with the indicated Legionella strains (red). (Scale bars, 2 μm.) WT: L. pneumophila Philadelphia-1 wild-type strain Lp02; dotA: the type IV secretion system defective strain Lp03; ΔsidC-sdcA: the SidC and SdcA double deletion mutant of the Lp02 strain, ΔsidC-sdcA(pSidC); and ΔsidC-sdcA(pSidC C46A): ΔsidC-sdcA strain complemented with a plasmid expressing wild-type or C46A mutant SidC. (B) Percent of cells containing GFP-HDEL positive LCVs counted from three independent assays under the conditions infected with the indicated Legionella strains. *P < 0.01 compared with the infection with wild-type Legionella strain.
It has been reported that L. pneumophila recruits polyubiquitin conjugates around the bacterial phagosomes shortly after infection (8). This recruitment is nearly abolished in the infection with the ΔsidC-sdcA mutant strain (38). We tested whether the E3 ligase activity of SidC/SdcA is responsible for the recruitment of ubiquitin conjugates. U937 cells were infected with relevant Legionella strains, and the bacteria were immunostained with an anti-Legionella antibody and the specific polyubiquitin antibody FK1. The recruitment of polyubiquitin species was observed in cells infected with wild-type bacteria but not with the ΔdotA or the ΔsidC-sdcA mutants. This defect was reversed by expression of wild type but not of the catalytically inactive C46A mutant in a plasmid. Previous studies showed that in strain AA100/130b, the F-box protein AnkB appeared to be essential for the recruitment of ubiquitin materials to LCVs (28). However, here we showed that in strain Lp02 (39), the enrichment of polyubiquitin species was not impaired by the deletion of ankB (SI Appendix, Fig. S14). Thus, the E3 ligase activity of SidC/SdcA, but not AnkB, is required for the enrichment of ubiquitinated species to the LCV at least in strain Lp02.
Discussion
Through the Dot/Icm transporter, L. pneumophila delivers ∼300 experimentally verified substrates into the host (9, 10). However, it has been a vast challenge to assign them an exact function in infection due to the scarcity of conserved functional motifs within these effector proteins. The Legionella effector SidC was such an example. SidC and its paralog SdcA were first assigned as vesicle tethering factors for promoting the recruitment and fusion of ER-derived vesicles to the bacterial phagosome (12). Recently, crystal structures of the N-terminal portion of SidC were reported (38, 40). In both publications, the N-terminal portion of SidC was concluded to function as an ER vesicle tethering factor. Here, we determined the crystal structure of the N-terminal portion of SidC (1–542), which we named SNL domain. The structure of the SNL domain is very similar to recently reported structures with an rmsd of less than 1.2 Å to both structures. However, by careful structural and biochemical analysis, we discovered that the N-terminal SNL domain of SidC possesses a cysteine-based ubiquitin ligase activity.
Compared with other cysteine based ubiquitin ligases, the SNL-domain family has some unique features. First, the SNL domain has no detectable primary and tertiary structural similarity to any known protein, suggesting a unique fold of this domain, as also suggested in the two previous publications (38, 40). Second, the SNL domain has a conserved catalytic C–H–D triad. The aspartic and histidine residues within the triad likely render the catalytic cysteine a stronger nucleophile than just a single catalytic cysteine found in other ubiquitin ligases. This feature may enhance the kinetics of Ub transfer from E2s to SidC and may also be advantageous during the competition with host Ub ligases for the pool of activated E2∼Ub. Third, in HECT E3s, the catalytic cysteine is located at the C-terminal small lobe, which can move a large distance toward the N-terminal lobe through a hinge-like motion. This conformational flexibility is believed to allow the transfer of Ub from E2 to the E3 catalytic cysteine (41–43). However, the catalytic cysteine of the SNL domain is localized at the center of a surface of its main subdomain (Fig. 1 B and D). This structural organization suggests that the Ub transfer by the SNL domain may not involve large movements of the catalytic cysteine, which further indicates that the binding site for E2∼Ub is in close proximity to the catalytic cysteine. To address this hypothesis, future structural and biochemical experiments are warranted to identify the E2 binding site on the SNL domain. Nevertheless, given these unique features, we conclude that the SNL domain of SidC defines a distinctive family of ubiquitin E3 ligases.
Although SidC and SdcA share 72% sequence identity, they exhibit differential preference for ubiquitin E2 conjugating enzymes. This discrimination of E2s suggests that these two proteins have evolved to maximally exploit components in the host ubiquitin pathway. This idea could explain why L. pneumophila has maintained two such highly similar proteins in evolution.
Modulation of multiple host cellular processes is essential for the intracellular life cycle of L. pneumophila (44). Given the fact that ubiquitination regulates a myriad of cellular functions, including cell cycle, cell death, trafficking, and immune responses (13), it is not surprising that L. pneumophila codes for at least six proteins containing F-box domains or U-box domains, which are hallmarks of multicomponent E3 ligase complexes (20). Among these, LubX and LegU1 are bona fide E3 ligases that target SidH, a Dot/Icm substrate involved in cell death, and BAT3, a protein involved in the cell cycle, respectively (27, 30). Our observation that the E3 ligase activity of SidC is essential for its role in the recruitment of ER proteins to the bacterial phagosome indicates its role in modulating vesicle trafficking, which is distinct from that of LubX and LegU1. However, the intriguing question of how the E3 ubiquitin activity of SidC connects with ER vesicle trafficking remains to be explored. It is likely that SidC/SdcA rewires the functions of specific host proteins by ubiquitination. Recent data have shown that the small GTPase Rab1, which regulates the vesicular trafficking step from the ER to the cis-Golgi, is monoubiquitinated when macrophage cells are infected with wild-type L. pneumophila, but not with a ΔsidC-sdcA mutant strain (38). However, SidC apparently does not ubiquitinate Rab1 directly, because cotransfection of SidC and Rab1 fails to cause monoubiquitination of Rab1 (38). In agreement with this observation, we also did not detect direct ubiquitination of Rab1 by SidC. When wild-type SidC was cotransfected with either GFP-tagged wild-type Rab1, the dominant negative Rab1(S22N), or the constitutive active Rab1(Q70L), no ubiquitinated Rab1 was detected (SI Appendix, Fig. S15A). Furthermore, in vitro ubiquitination experiments using recombinant GST-tagged Rab1 failed to detect direct ubiquitination of Rab1 (SI Appendix, Fig. S15B). These data suggest that the monoubiquitination of Rab1 is mediated through an indirect unknown mechanism as proposed by the previous report (38).
An exciting and challenging future direction is to identify specific host factors that are ubiquitinated by SidC/SdcA. We expect that proteins involved in the host ER-related membrane trafficking events are the potential targets of SidC/SdcA. The identification of these unknown factors would not only help explain the molecular mechanisms of pathogen–host interactions, but also would enhance our knowledge of basic cellular processes, in particular, membrane trafficking.
Materials and Methods
The materials and methods are described at length in SI Appendix, SI Materials and Methods. This includes cloning site-directed mutagenesis, expression and purification of SidC proteins crystallization and structural determination, in vitro ubiquitination assays, SILAC experiments, and Legionella infection analyses. References for the SI Appendix, SI Materials and Methods are also included.
Supplementary Material
Acknowledgments
We thank Dr. S. E. Ealick (Cornell University) for heavy atom containing reagents; Dr. S. Qian (Cornell University) for ubiquitin constructs; and Dr. S. D. Emr and Dr. V. Vogt (Cornell University) for critical reading of the manuscript. This work was supported by the Harry Samuel Mann Award (to F.H.); National Institutes of Health (NIH) Grants R01-GM094347 (to Y.M.), K02AI085403 (to Z.-Q.L.), R56AI103168 (to Z.-Q.L.), R01-GM102529 (to J.J.), and R01-GM097272 (to M.B.S.); the Welch Foundation Grant AU-1711 (to J.J.); and American Cancer Society RSG-11-146-01-DMC (to M.B.S.). The X-ray data were collected at MacChess beamline A1 and National Synchrotron Light Source beamline X4C. Cornell High-Energy Synchrotron Source is supported by the National Science Foundation (NSF) and NIH/National Institute of General Medical Sciences via NSF award DMR-0225180, and the MacCHESS resource is supported by NIH/National Center for Research Resources Award RR-01646.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4TRH (native SidC) and 4TRG (Hg-bound form)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402605111/-/DCSupplemental.
References
- 1.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15(3):506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006;2(5):e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu F, et al. Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc Natl Acad Sci USA. 2012;109(34):13567–13572. doi: 10.1073/pnas.1207903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toulabi L, Wu X, Cheng Y, Mao Y. Identification and structural characterization of a Legionella phosphoinositide phosphatase. J Biol Chem. 2013;288(34):24518–24527. doi: 10.1074/jbc.M113.474239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4(12):945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 7.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: Implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114(Pt 24):4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- 8.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: Exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2(4):e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6(3):e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lifshitz Z, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci USA. 2013;110(8):E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101(3):841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragaz C, et al. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;10(12):2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 13.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Hurley JH, Stenmark H. Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys. 2011;40:119–142. doi: 10.1146/annurev-biophys-042910-155404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125(Pt 2):265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 17.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 18.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 19.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10(6):398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 20.Hubber A, Kubori T, Nagai H. Modulation of the ubiquitination machinery by Legionella. Curr Top Microbiol Immunol. 2013;376:227–247. doi: 10.1007/82_2013_343. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2012;12(1):35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311(5758):222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 23.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15(1):65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 24.Singer AU, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15(12):1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Felipe KS, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187(22):7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36(11):1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 27.Ensminger AW, Isberg RR. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun. 2010;78(9):3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price CT, et al. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5(12):e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomma M, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12(9):1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 30.Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67(6):1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 31.Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6(12):e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Rosenstrom P. Dali server: Sonservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109(5):575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 34.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 1999;96(12):6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75(2):592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horenkamp FA, et al. Legionella pneumophila subversion of host vesicular transport by SidC effector proteins. Traffic. 2014;15(5):488–499. doi: 10.1111/tra.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 40.Gazdag EM, Schobel S, Shkumatov AV, Goody RS, Itzen A. The structure of the N-terminal domain of the Legionella protein SidC. J Struct Biol. 2014;186(1):188–194. doi: 10.1016/j.jsb.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Huang L, et al. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286(5443):1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 42.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36(6):1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verdecia MA, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11(1):249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 44.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.