Significance
The enzymes required for the incorporation of host fatty acids into the membrane phospholipids of Gram-positive bacterial pathogens are unknown. Fatty acid kinase (Fak) is a new enzyme in lipid metabolism that requires two proteins to form acyl-phosphate: an ATP binding-domain protein (FakA) that interacts with a fatty acid binding protein (FakB). The analysis of Staphylococcus aureus mutants reveals that Fak is essential for phospholipid synthesis from extracellular fatty acids and also impacts the transcription of numerous virulence factors. This study reveals the function for two large bacterial protein families and their essential role in host fatty acid metabolism by pathogens, and connects Fak to the regulation of virulence factor transcription in S. aureus.
Abstract
Extracellular fatty acid incorporation into the phospholipids of Staphylococcus aureus occurs via fatty acid phosphorylation. We show that fatty acid kinase (Fak) is composed of two dissociable protein subunits encoded by separate genes. FakA provides the ATP binding domain and interacts with two distinct FakB proteins to produce acyl-phosphate. The FakBs are fatty acid binding proteins that exchange bound fatty acid/acyl-phosphate with fatty acid/acyl-phosphate presented in detergent micelles or liposomes. The ΔfakA and ΔfakB1 ΔfakB2 strains were unable to incorporate extracellular fatty acids into phospholipid. FakB1 selectively bound saturated fatty acids whereas FakB2 preferred unsaturated fatty acids. Affymetrix array showed a global perturbation in the expression of virulence genes in the ΔfakA strain. The severe deficiency in α-hemolysin protein secretion in ΔfakA and ΔfakB1 ΔfakB2 mutants coupled with quantitative mRNA measurements showed that fatty acid kinase activity was required to support virulence factor transcription. These data reveal the function of two conserved gene families, their essential role in the incorporation of host fatty acids by Gram-positive pathogens, and connects fatty acid kinase to the regulation of virulence factor transcription in S. aureus.
The pathway for the uptake and incorporation of host fatty acids (FA) by bacterial pathogens is important to understanding their physiology and determining if type II fatty acid synthesis (FASII) inhibitors will be useful antibacterial therapeutics. FASII is an energy-intensive process, and the advantage of being able to use host FA for membrane assembly obviously allows ATP to be diverted to the synthesis of other macromolecules. Gram-positive pathogens are capable of incorporating extracellular FA into their phospholipids. In species related to Streptococcus agalactiae, extracellular FA can completely replace endogenously synthesized FA, rendering the FASII inhibitors ineffective growth inhibitors if sufficient FA are present (1, 2). In contrast, FASII inhibitors exemplified by the enoyl-acyl carrier protein (ACP) reductase therapeutic AFN-1252 are effective against Staphylococcus aureus, even when extracellular FA are abundant (2, 3). A major impediment to our understanding of this diversity is that the pathway and enzymes responsible for the incorporation of extracellular FA is not established in the Firmicutes. A recent analysis of a ΔplsX knockout strain of S. aureus ruled out a role for either acyl-CoAs or acyl-ACP as intermediates in host FA metabolism and instead provided evidence for the existence of a new enzyme that phosphorylates FA, called FA kinase (Fak) (4) (Fig. 1A). Host FA are phosphorylated by Fak, and the acyl-PO4 formed is either used by the PlsY glycerol-3-phosphate acyltransferase or converted to acyl-ACP by PlsX. The acyl-ACP may be either elongated by FASII or used by PlsC. Despite the strong evidence for their existence, the genes/proteins that encode this novel enzyme in FA metabolism were previously unidentified. This study shows that Fak activity requires two separate proteins of previously unknown function. A kinase domain protein (called FakA) interacts with a fatty acid binding protein (called FakB) to form acyl-PO4. The two FakBs of S. aureus have different FA binding specificities. The inactivation of Fak severely attenuates virulence factor production in S. aureus. Fak represents a new pathway for the incorporation of host FA into phospholipids by Gram-positive pathogens, and the analysis of the deletion mutants illustrates that Fak activity also positively regulates the expression of virulence factors in S. aureus.
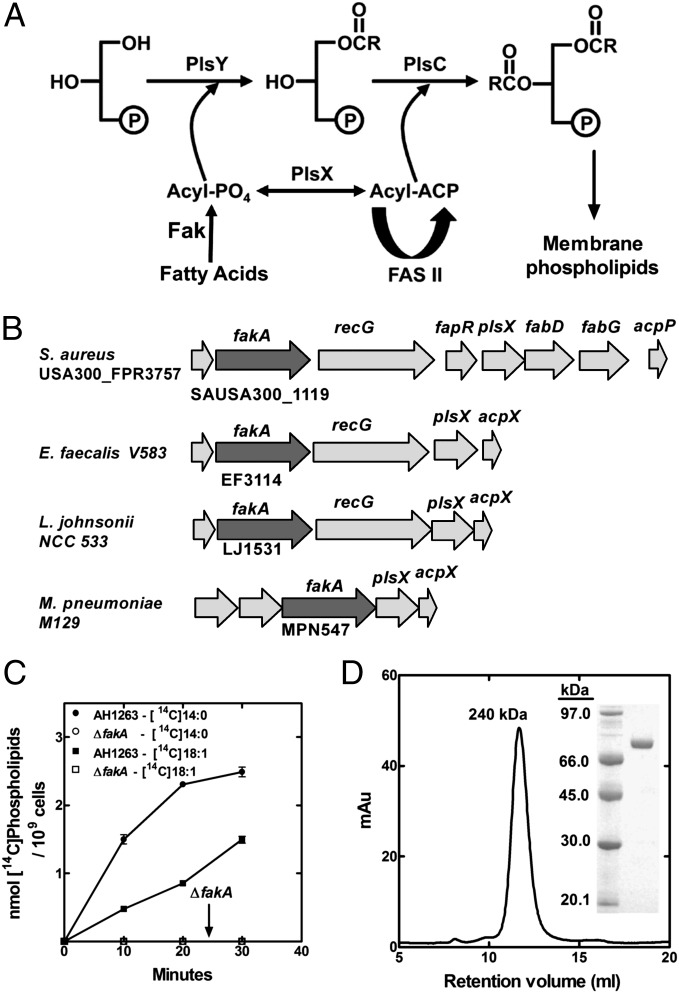
Fig. 1.
The role of FakA in the incorporation of FA into S. aureus phospholipids. (A) Pathway for the incorporation of extracellular FA into phospholipids in S. aureus. Host FA are flipped to the interior of the cell and phosphorylated by Fak. The acyl-PO4 is either ligated to the 1-position of sn-glycerol-3-phosphate by PlsY or converted to acyl-ACP by PlsX. The acyl-ACP can either be elongated by the FASII enzymes or ligated into the 2-position of lysophosphatidic acid by PlsC. The phosphatidic acid product is the precursor to all membrane phospholipids. (B) The location of the fakA gene in relation to the plsX and acpP genes is illustrated in two Gram-positive pathogens (S. aureus and Enterococcus faecalis) and two fatty acid auxotrophs (L. johnsonii and M. pneumoniae) that lack FASII and must acquire FA from the host for survival. (C) Metabolic labeling of wild-type S. aureus strains AH1263 and its ΔfakA derivative. Cells were labeled with either [14C]oleic acid (18:1) or [14C]myristic acid (14:0) and incorporation of fatty acids into phospholipids measured. (D) Analytical size exclusion chromatography of FakA shows a single species with a peak corresponding to 240 kDa. (Inset) Coomassie-stained electrophoretic gel analysis of purified FakA.
Results
Identification of FakA.
Although Fak activity was detected in cell extracts (4), attempts to identify the responsible gene and protein by expression cloning or protein purification proved unsuccessful. Bacteria that lack FASII (e.g., Mycoplasma pneumoniae) rely exclusively on host FA and use the PlsX/Y/C acyltransferase system. There is a conserved gene encoding a putative kinase located adjacent to the plsX (acyl–acyl carrier protein:phosphate transacylase) and acpP (acyl carrier protein) genes in M. pneumoniae that was also associated with plsX in Gram-positive bacteria in general (Fig. 1B). We have named this gene fakA to reflect its biochemical function as a Fak uncovered in this research. This locus (first known as vfrB) most often exists in an operon with a gene encoding a protein with similarity to acid shock protein Asp23 (vfrA) (5). Genetic analysis shows vrfB (fakA) is a potent modifier of α-hemolysin production, but vfrA does not impact this phenotype (5). FakA belongs to a family of Gram-positive bacterial proteins of unknown function (COG1461) that contain an N-terminal domain similar to the di-Mg2+ ATP binding domain (Dak2) of dihydroxyacetone (Dha) kinase of Citrobacter freundii (6) (Fig. S1). We tested whether the fakA gene encoded the Fak by disrupting the gene and examining the uptake of exogenous FA in the knockout strain. The wild-type S. aureus strain AH1263 incorporated both myristic acid (14:0) and oleic acid (18:1) FA into phospholipid at a robust rate (Fig. 1C). However, the incorporation of both FAs was completely absent in the ΔfakA strain. Complementation of the ΔfakA stain with a plasmid expressing FakA restored FA uptake (Fig. S2). We also inactivated the leading gene in the operon, vfrA. Extracellular FA incorporation into phospholipid remained robust in the ΔvfrA strain (Fig. S2), ruling out an essential role for VfrA in FA uptake. FakA was expressed and purified from Escherichia coli (Fig. 1D, Inset). Purified FakA had an apparent size of 240 kDa by gel-filtration chromatography calibrated with typical globular proteins (Fig. 1D); however, analytical ultracentrifugation showed that FakA was a dimer (Table S1). The elongated shape (f/f0 = 1.64) of FakA accounted for its abnormal elution position in size-exclusion chromatography. FakA was completely inactive as a Fak in vitro. Soluble extracts of the ΔfakA strain lacked Fak activity, but acyl-PO4 formation was restored by the addition of purified FakA. Ultrafiltration and heat denaturation experiments indicated that there was a second protein in the ΔfakA cell extract that worked in concert with FakA to reconstitute Fak activity.
Identification of FakB.
Gram-positive bacteria express multiple representatives of the DegV protein family (COG1307) that have been structurally characterized as FA binding proteins (7, 8). The type of FA modeled into the structures deposited in the PDB depends on the source of the DegV homolog. A global screen for interacting proteins in Streptococcus pneumoniae (9) reported that the FakA homolog interacted with a DegV homolog and PlsX in this organism, suggesting that these proteins function together. S. aureus expresses two of these FA binding proteins (DegV homologs), which we have named FakB1 and FakB2 (Fig. 2A). Blocks of conserved residues mark this protein family (Fig. 2A), but the two FakB proteins in S. aureus are only 34% identical. Single- and double-knockout strains of the two fakB genes were constructed. The incorporation of [14C]18:1 was not affected in the ΔfakB1 strain, was significantly attenuated in the ΔfakB2 strain, and was eliminated entirely in the ΔfakB1 ΔfakB2 double-knockout strain (Fig. 2B). Neither of the single fakB knockouts were significantly attenuated in [14C]14:0 incorporation; however, the ΔfakB1 ΔfakB2 double knockout was unable to incorporate [14C]14:0 into phospholipid (Fig. 2C). These data showed that a FakB protein was required in addition to FakA for FA incorporation into phospholipid of S. aureus.
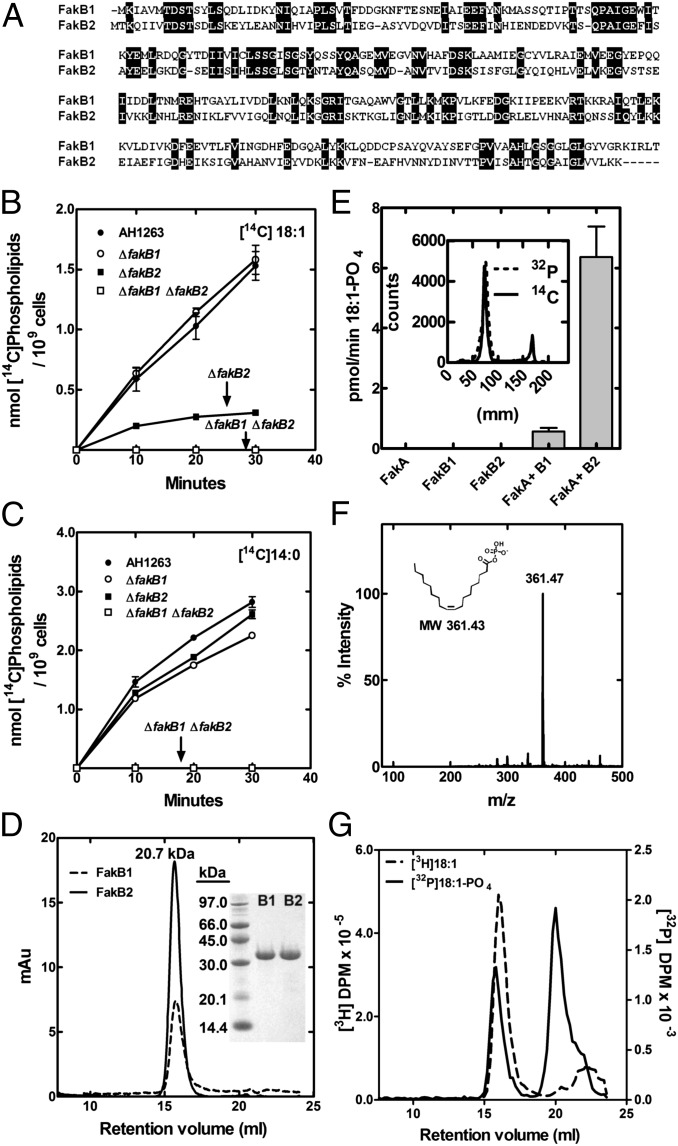
Fig. 2.
Sequence and function of FakB, the FA binding component of Fak. (A) Alignment of FakB1 (SAUSA300_0733) and FakB2 (SAUSA300_1318) proteins from S. aureus USA300. (B) Metabolic labeling experiment showing the rate of incorporation [1-14C]oleic acid into phospholipids in wild-type strain AH1263 and its ΔfakB1, ΔfakB2, and ΔfakB1 ΔfakB2 double-knockout derivatives. (C) Metabolic labeling experiment showing the rate of incorporation [14C]14:0 into phospholipids in wild-type strain AH1263, and its ΔfakB1, ΔfakB2, and ΔfakB1 ΔfakB2 double-knockouts. (D) Analytical size-exclusion chromatography of FakB1 and FakB2, showing a single species corresponding to 20.7 kDa. (Inset) Coomassie-stained electrophoretic gel analysis of purified FakB1 and FakB2. (E) Fak assay of different combinations of FakA and FakB1/FakB2 using [14C]18:1 and 1% Triton X-100. (Inset) Product of Fak reaction with FakB2 using either [14C]18:1 or [γ-32P]ATP with 18:1 delivered in 10 mg/mL BSA. Products were separated by TLC on Silica Gel G layers developed in in [chloroform/methanol/acetic acid (90/10/10) (vol/vol/vol)] and imaged using Bioscan detector. (F) Mass spectrum of the product of the Fak reaction using FakB2 and 18:1. (G) Binding of FakB2 to [3H]18:1 acid and 18:1-[32P]PO4. After incubation with the labeled ligand, FakB2 was separated on Superdex S200 10/300 size-exclusion column; 0.2 mL fractions were collected and analyzed by scintillation counting.
Single fakA genes are found in bacteria in the Firmicute family (Fig. S3), but multiple fakB genes are common. The two FakB proteins of S. aureus were expressed and purified (Fig. 2D, Inset). These proteins were monomers by gel-filtration chromatography (Fig. 2D). This conclusion was confirmed by analytical ultracentrifugation (Table S1) and was consistent with the crystal structures of this protein family (7, 8). FakA plus a FakB reconstituted Fak activity in vitro (Fig. 2E). Neither FakA nor FakB alone resulted in product formation, but [14C]acyl-PO4 was generated when both proteins were present (Fig. 2E). FakB2 was more active in experiments using [14C]18:1 as a substrate than FaKB1. Acyl-PO4 was initially identified as the reaction product by its comigration in TLC with a [14C]acyl-PO4 standard generated using PlsX and [14C]acyl-ACP (10) (Fig. 2E, Inset). [14C]Acyl-PO4 was produced in assays containing [14C]18:1 and acyl[32P]PO4 was formed in Fak assays containing [γ-32P]ATP (Fig. 2E, Inset). The FakA-FakB2 assay was scaled up and the product positively identified as 18:1-PO4 by mass spectrometry (Fig. 2F). The incubation of FakB2 with [3H]18:1 showed that [3H]18:1 coeluted with FakB2 by gel-filtration chromatography (Fig. 2G), illustrating FakB2 bound free FA. Fractionation of the incubations containing FakB2 and acyl[32P]PO4 by gel-filtration chromatography showed that FakB2 also bound acyl[32P]PO4 (Fig. 2G). FakB2 did not possess acyl-ACP thioesterase activity nor did we detect an interaction between FakB2 and [14C]18:1-ACP by gel-filtration chromatography. Thus, Fak activity required FakA + FakB. FakA provided the ATP binding domain and FakB the FA binding component.
FA Selectivity of FakBs.
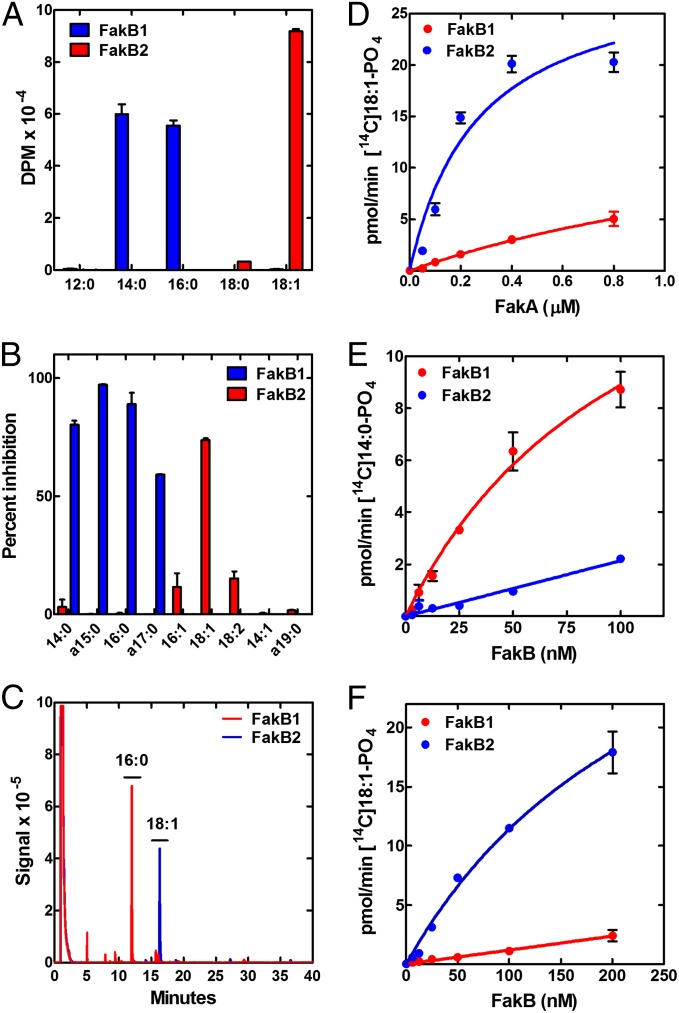
The two FakBs of S. aureus have distinct FA binding specificities. The first assay examined the binding of 5 [14C]FAs to the two FakBs (Fig. 3A). Each protein was incubated with the respective FA and the amount bound was determined by isolating the labeled FakB by gel-filtration chromatography (Fig. 3A). FakB1 bound 14:0 and 16:0 saturated FA, whereas FakB2 did not. In contrast, FakB2 bound 18:1, which was not bound to FakB1. A competition experiment was deployed to expand the analysis to nonradioactive acyl chains. FakB1 was incubated with [3H]14:0 along with a cold competitor. The most effective competitors for [3H]14:0 binding were the saturated FA (Fig. 3B). The anteiso branched-chain FA produced by S. aureus displaced labeled [3H]14:0 from FakB1. In contrast, FakB2 binding to [3H]18:1 was effectively competed with 18:1, and to a lesser extent by 16:1 and 18:2, but not by the saturated FAs. The purified FakB proteins had a bound FA (Fig. 3C), and the compositions were determined by denaturing the proteins and analyzing the derivatized FA by gas chromatography. FakB1 was purified with 16:0 bound and FakB2 had bound 18:1. Taken together, these data showed that FakB1 saturated FA, whereas FakB2 prefers 18:1.
Fig. 3.
FA binding selectivity of FakB1 and FakB2. (A) Binding of [14C]FA to purified FakB1 and FakB2. Proteins incubated with FA, unbound FA removed using desalting column. (B) Competitive binding of different fatty acids to FakB1 and FakB2. Unlabeled FA incubated with protein at 10-μM concentration with 1 μM [3H]14:0 (FakB1) or 1 μM [3H]18:1 (FakB2). Increased inhibition of [3H]FA binding corresponds to increased affinity. (C) Overlay of representative gas-chromatograms of FA bound to FakB1 (blue) and FakB2 (red) purified from E. coli. FAs were identified by their comigration with standards. (D) Fak assay with constant concentrations of FakB1 (0.35 μM) or FakB2 (0.2 μM) in the presence of variable FakA. [14C]18:1 was delivered in 1% Triton X-100. (E) Fak assay using liposomes containing [14C]14:0 and constant FakA (0.2 μM). (F) Fak assay using liposomes containing [14C]oleic acid and constant FakA (0.2 μM).
The ability of the individual FakBs to support the Fak reaction was tested. First, the dependence of the reaction on FakA was determined for the two FakBs using [14C]18:1 as substrate (Fig. 3D). In this experiment, FakB2 was considerably more active than FakB1. To determine if this reflected the FA binding selectivity, each of the FakBs was evaluated in the presence of a fixed FakA concentration with both [14C]14:0 and [14C]18:1 as substrates. FakB1 was significantly more efficient at promoting the phosphorylation of [14C]14:0 than FakB2 (Fig. 3E). Conversely, FakB2 was considerably more active than FakB1 when [14C]18:1 was the substrate (Fig. 3F). Thus, the reactivity of the FakBs in the Fak assay reflected their FA binding specificities.
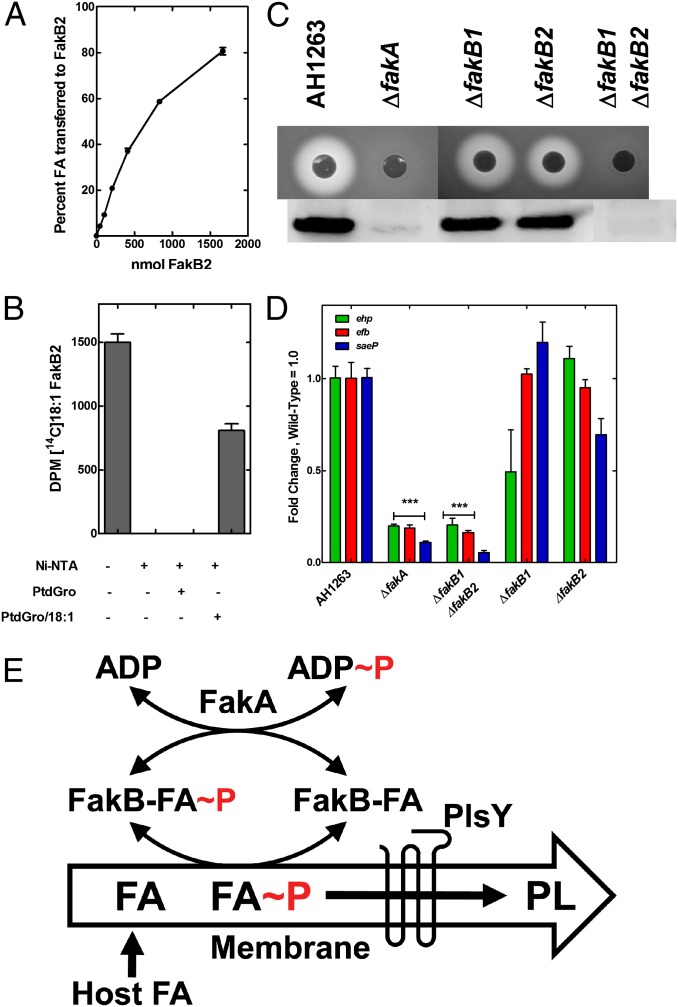
A key property of the FakB FA binding proteins was their ability to exchange bound FA for FA dissolved in PtdGro liposomes. There was a concentration-dependent increase in the amount of label associated with the protein in incubations of FakB2 with liposomes containing [14C]18:1 (Fig. 4A). In the converse experiment, FakB2 was loaded with [14C]18:1 and the labeled ([14C]18:1)FakB2 was isolated by gel-filtration chromatography. The labeled FA did not exchange off ([14C]18:1)FakB2 in the presence of PtdGro liposomes alone as indicated by the complete recovery of input radioactivity in the protein fraction (Fig. 4B). However, when the PtdGro liposomes contained unlabeled 18:1, [14C]18:1 was transferred from FakB2 to the liposome (Fig. 4B). These data indicated that FakB protein always had a bound FA, but was able to exchange with another FA or acyl-PO4 presented as free FA (Fig. 2G), or in either detergent micelles (Fig. 2E) or in liposomes (Fig. 4B).
Fig. 4.
Functions of Fak in FA uptake and virulence factor transcription. (A) Transfer of FA from liposome to FakB2. Liposomes (phosphatidylglycerol/[14C]18:1/biotin-phosphatidyl-ethanolamine 80/10/10) incubated with different amounts of FakB2. Liposomes removed by addition of streptavidin magnetic beads. The 100% value = 0.96 nmol 18:1. (B) Transfer of FA from FakB2 to liposomes. FakB2 with bound [14C]18:1 incubated with liposomes with and without FA. FakB2 was removed from solution by the addition of Ni-NTA agarose. (C) Lack of α-hemolysin production in Fak-negative strains. Blood agar plates inoculated with strain AH1263 (wild-type) and its knockout derivatives indicate the level of α-hemolysin secretion as rings of red cell lysis. Immunoblotting of the culture supernatants from the same series of strains with rabbit anti–α-hemolysin antisera shows that Fak activity is required for α-hemolsyin secretion. (D) Transcript abundance in triplicate samples measured by quantitative RT-PCR using gmk as calibrator in strain AH1263 and its knockout derivatives. Transcript levels were significantly reduced in ΔfakA and ΔfakB1 ΔfakB2 strains based on the Student t test (***P < 0.0001). (E) Pathway for exogenous FA incorporation in S. aureus. Host FA are flipped to the inner leaflet by the pH gradient where they can exchange with FakB(acyl-PO4). This deposits acyl-PO4 in the membrane where it is used by the PlsY acyltransferase to initiate phospholipid synthesis. FakB(FA) is phosphorylated by FakA to generate FakB(acyl-PO4) to initiate another round of exchange.
Fak Regulation of Virulence Factor Transcription.
Transposon insertions in fakA were first isolated based on their increased resistance to an antimicrobial peptide (11). More recently, insertions in fakA (vfrB) were identified in a screen of a S. aureus knockout collection for the absence of a α-hemolysin production on blood agar plates (5). These data indicated FakA was important to support virulence factor production; therefore, we screened our mutant panel for α-hemolysin production to determine if FakA alone was responsible for the regulation of α-hemolysin production (Fig. 4C). The severe attenuation in α-hemolysin production was evidenced from indicator plates and Western blots in both ΔfakA and ΔfakB1 ΔfakB2 double-mutants. The individual ΔfakB1 and ΔfakB2 strains both produced α-hemolysin. These data meant that FakA and FakB function together as a Fak to control virulence factor production. Triplicate Affymetrix arrays of S. aureus strain AH1263 compared with its ΔfakA derivative showed 26 up-regulated and 19 down-regulated genes (>twofold; P < 0.05) (Table S2). Of note was that transcription of numerous virulence related genes (including α-hemoslysin, hla) were uniformly attenuated in the ΔfakA knockout (Table S2). Many of these virulence factors (saeP, ehp, efb, and so forth) are also known to be regulated by the SaeRS system (12, 13). The transcription of the early secreted antigenic target 6 kDa protein (ESAT-6) protein secretion system for the export of EsxA and EsxB was activated in the ΔfakA strain (Table S2). The ESAT-6 system is best known in Mycobacterium tuberculosis where the secreted components are involved in the escape from phagocytic compartment into the cytosol (14). EsxA and EsxB are also involved in S. aureus virulence, although the mechanism is not established (15). ESAT-6 operon transcription is inhibited by SaeRS signaling (16), and its up-regulation was consistent with a connection between the genes altered by SaeRS and FakA. Quantitative RT-PCR verified that these three SaeRS-regulated genes (12, 13) were expressed at significantly lower levels in ΔfakA and ΔfakB1 ΔfakB2 strains, and the single FakB deletions had intermediate phenotypes (Fig. 4D). These data confirmed the connection between the activity of Fak and the expression of virulence factors.
Discussion
Role of Fak in Host FA Incorporation.
This work identifies the FA kinase enzyme system in bacterial lipid metabolism that is responsible for activation of host FA for their incorporation into the phospholipids of Gram-positive pathogens (Fig. 4E). Extracellular FA first translocates to the inner aspect of the bilayer by the spontaneous flipping of the protonated FA (17, 18). There is no evidence for a protein-mediated FA transporter in bacteria, and FA flipping is driven by the membrane pH gradient (17, 18). The fundamental biochemical property of FakB that explains its role in exogenous FA incorporation is its ability to exchange a bound ligand (FA or acyl-PO4) for a ligand embedded in a phospholipid bilayer. This type of exchange reaction is catalyzed by eukaryotic FA binding proteins (19–21). FakA phosphorylates FakB(FA) and the resulting FakB(acyl-PO4) exchanges its bound acyl-PO4 for a membrane FA, and the cycle repeats (Fig. 4E). The utilization of acyl-PO4 by PlsY for phospholipid synthesis depletes the membrane of acyl-PO4 to promote the uptake and phosphorylation of exogenous FA. In the absence of extracellular FA, there are two sources for acyl-PO4 bound to FakB. FakB(FA) may be phosphorylated by FakA, or the FakB(FA) may exchange with membrane acyl-PO4 produced by PlsX. The biochemistry and mutant phenotypes show that FakA and FakB are both required for Fak, exogenous FA incorporation into phospholipid and for the regulation of virulence factor production.
It seems reasonable to conclude that Fak is essential for phospholipid synthesis in organisms like Lactobacillus johnsonii and M. pneumoniae that lack FASII and depend completely on the host for FA. However, the absence of a growth phenotype or lipidomics abnormalities in either the ΔfakA or ΔfakB1 ΔfakB2 strains suggest that Fak does not contribute to the rate of S. aureus phospholipid synthesis when exogenous FA are absent. S. pneumoniae uses the same enzymes to initiate phospholipid synthesis as S. aureus (Fig. 1A); however, the fakA gene in this organism (SP0443) is reported to be essential (22–24). The essential function in S. pneumoniae is unknown but is likely to be because of the regulatory properties of Fak rather than its role in phospholipid synthesis. Finally, M. tuberculosis also has a FakA (Rv2974c) (Fig. S1) and a FakB (Rv2417c). M. tuberculosis fakA expression is up-regulated during infection (25) and fakA knockouts are attenuated in macrophage survival (26). The role for Fak in this organism appear unrelated to phospholipid synthesis because M. tuberculosis does not have a PlsX or PlsY.
Fak and Dha Kinases.
The functions of Fak are reminiscent of the analogous Dha kinase components that function in Dha phosphorylation and the control of gene expression (27). E. coli DhaA kinase is composed of three proteins: a phosphotransferase component (DhaM), a nucleotide-binding protein (DhaL), and a Dha-binding protein (DhaK) (28–31). In C. freundii the DhaL and DhaK are fused to form a functional kinase that uses ATP rather than the phosphotransferase system (6). All of these proteins have a related fold, called the EDD domain (32). Proteins in this group include the EIIA component of the mannose transporter, Dha kinase components, and the DegV protein family. This versatile fold has been repurposed for the multiple functions in the overall Dha reaction, and has been repurposed in Firmicutes to carry out the Fak reaction. The FakA amino-terminal domain is clearly similar to the ATP binding domain (Dak2) of C. freundii Dha kinase (27, 33). The FakA carboxyl-terminal domain is also predicted to adopt an EDD-like fold; however, we did not detect FA binding to FakA. The DegV family (32) EDD fold is related to the DhaL Dha-binding protein/domain of Dha kinases (33) and all members of this family characterized to date are FA binding proteins (7, 8). Interestingly, the DhaL and DhaK proteins also regulate transcription by binding to DhaR, an AAA+ type enhancer (34). Binding to DhaR is regulated by the attached ligand (ATP/ADP for DhaL and Dha/DhaP for DhaK). Thus, the EDD fold has been adopted by a number of proteins to phosphorylate a variety of substrates and control gene expression in different settings.
Fak Regulation of Virulence Factor Transcription.
An important finding was the connection between Fak and the control of virulence factor gene expression. The initial observation that ΔfakA mutants were defective in α-hemolysin production (5) is not because of FakA alone, but rather Fak activity is the responsible entity. Our analysis indicates that multiple virulence factors whose transcription is regulated by Fak activity are related to the genes regulated by the SaeRS two-component system (12, 13, 16), suggesting that Fak may function in the SaeRS biochemistry. It is tempting to speculate that FakB(acyl-PO4) acts as a phosphorelay component much like acetyl-phosphate in Gram-negative bacteria, where its role in the phosphorylation of response regulator receiver domains is established (35). Additional experiments will be needed to determine if the Fak effect on the SaeRS regulon is direct or indirect, but it is clear that Fak is a regulatory factor that is critical to supporting the transcription of these important virulence factors. There are two sources for FakB(acyl-PO4). First, it can arise from FakA phosphorylation of FakB(FA), or second FakB(FA) can exchange with membrane-associated acyl-PO4 (Fig. 4E). Our previous work shows that the elimination of acyl-PO4 production by FASII using AFN-1252 to block FabI, there is a strong down-regulation of these same SaeRS-controlled virulence genes in S. aureus (13). This result suggests that FASII, in addition to FakA, provides acyl-PO4 to support virulence factor transcription. A curious finding was the high degree of specificity exhibited by FakB2 for 18:1, a very abundant mammalian FA. However, S. aureus does not produce unsaturated FA, suggesting the possibility that FakB2 functions as a sensor for the presence of host 18:1.
Materials and Methods
AH1263 ΔfakA was described previously, and a complete strain list is provided in Table S3. FakA and FakB proteins were purified using amino-terminal His-tags from E. coli BL21(DE3). FA uptake experiments were performed using 1-14C-labeled 14:0 and 18:1 FAs to determine incorporation into phospholipids. FA kinase assay contained 0.1 M Tris⋅HCl (pH 7.5), 20 mM MgCl2, 10 mM ATP, 1% Triton X-100, 20 μM [1-14C]oleic acid or myristic acid, 0.2 μM FakA, and 0.2 μM FakB1 or 0.2 μM FakB2 in a 60-μL final volume. The reaction was initiated with the addition of the Fak enzymes and incubated at 37 °C for 20 min. Next, 40 μL of the reaction was spotted on DE81 disks and washed three times with ethanol containing 1% acetic acid to remove the unreacted FA. Quantitative real-time PCR was performed as previously described (13). Full details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Amanda Nourse of the Molecular Interaction Analysis Facility for the ultracentrifugation analysis, the Protein Production Facility for purified proteins, and Matthew Frank for liposome preparation. This work was supported by National Institutes of Health Grant GM034496 (to C.O.R.), Cancer Center Support Grant CA21765, and the American Lebanese Syrian Associated Charities.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408797111/-/DCSupplemental.
References
- 1.Brinster S, et al. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458(7234):83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 2.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci USA. 2011;108(37):15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balemans W, et al. Essentiality of FASII pathway for Staphylococcus aureus. Nature. 2010;463(7279) doi: 10.1038/nature08667. E3, discussion E4. [DOI] [PubMed] [Google Scholar]
- 4.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol Microbiol. 2014;92(2):234–245. doi: 10.1111/mmi.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose JL, Daly SM, Hall RR, Bayles KW. Indentification of the vfrAB operon in Staphylococcus aureus: A novel virulence factor regulatory locus. Infect Immun. 2014;82(5):1813–1822. doi: 10.1128/IAI.01655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siebold C, Arnold I, Garcia-Alles LF, Baumann U, Erni B. Crystal structure of the Citrobacter freundii dihydroxyacetone kinase reveals an eight-stranded α-helical barrel ATP-binding domain. J Biol Chem. 2003;278(48):48236–48244. doi: 10.1074/jbc.M305942200. [DOI] [PubMed] [Google Scholar]
- 7.Schulze-Gahmen U, Pelaschier J, Yokota H, Kim R, Kim SH. Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima, reveals its function as a fatty acid-binding protein. Proteins. 2003;50(4):526–530. doi: 10.1002/prot.10305. [DOI] [PubMed] [Google Scholar]
- 8.Nan J, et al. Structure of a fatty-acid-binding protein from Bacillus subtilis determined by sulfur-SAD phasing using in-house chromium radiation. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 5):440–448. doi: 10.1107/S0907444909007756. [DOI] [PubMed] [Google Scholar]
- 9.Meier M, Sit RV, Quake SR. Proteome-wide protein interaction measurements of bacterial proteins of unknown function. Proc Natl Acad Sci USA. 2013;110(2):477–482. doi: 10.1073/pnas.1210634110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y-J, et al. Acyl-phosphates initiate membrane phospholipid synthesis in Gram-positive pathogens. Mol Cell. 2006;23(5):765–772. doi: 10.1016/j.molcel.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Li M, et al. Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob Agents Chemother. 2009;53(10):4200–4210. doi: 10.1128/AAC.00428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol. 2012;194(19):5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons JB, et al. Perturbation of Staphylococcus aureus gene expression by the enoyl-acyl carrier protein reductase inhibitor AFN-1252. Antimicrob Agents Chemother. 2013;57(5):2182–2190. doi: 10.1128/AAC.02307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Leon J, et al. Mycobacterium tuberculosis ESAT-6 exhibits a unique membrane-interacting activity that is not found in its ortholog from non-pathogenic Mycobacterium smegmatis. J Biol Chem. 2012;287(53):44184–44191. doi: 10.1074/jbc.M112.420869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci USA. 2005;102(4):1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson M, Aly KA, Chen YH, Missiakas D. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol. 2013;90(4):734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garlid KD, Orosz DE, Modrianský M, Vassanelli S, Jezek P. On the mechanism of fatty acid-induced proton transport by mitochondrial uncoupling protein. J Biol Chem. 1996;271(5):2615–2620. doi: 10.1074/jbc.271.5.2615. [DOI] [PubMed] [Google Scholar]
- 18.Abumrad NA, Davidson NO. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92(3):1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falomir-Lockhart LJ, Franchini GR, Guerbi MX, Storch J, Córsico B. Interaction of enterocyte FABPs with phospholipid membranes: Clues for specific physiological roles. Biochim Biophys Acta. 2011;1811(7-8):452–459. doi: 10.1016/j.bbalip.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smathers RL, Petersen DR. The human fatty acid-binding protein family: Evolutionary divergences and functions. Hum Genomics. 2011;5(3):170–191. doi: 10.1186/1479-7364-5-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storch J, Thumser AE. Tissue-specific functions in the fatty acid-binding protein family. J Biol Chem. 2010;285(43):32679–32683. doi: 10.1074/jbc.R110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zalacain M, et al. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J Mol Microbiol Biotechnol. 2003;6(2):109–126. doi: 10.1159/000076741. [DOI] [PubMed] [Google Scholar]
- 23.van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012;22(12):2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skvortsov TA, Ignatov DV, Majorov KB, Apt AS, Azhikina TL. Mycobacterium tuberculosis transcriptome profiling in mice with genetically different susceptibility to tuberculosis. Acta Naturae. 2013;5(2):62–69. [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart GR, Patel J, Robertson BD, Rae A, Young DB. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1(3):269–278. doi: 10.1371/journal.ppat.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erni B, et al. Small substrate, big surprise: Fold, function and phylogeny of dihydroxyacetone kinases. Cell Mol Life Sci. 2006;63(7-8):890–900. doi: 10.1007/s00018-005-5518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi R, et al. Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2011;108(4):1302–1307. doi: 10.1073/pnas.1012596108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutknecht R, Beutler R, Garcia-Alles LF, Baumann U, Erni B. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 2001;20(10):2480–2486. doi: 10.1093/emboj/20.10.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siebold C, García-Alles LF, Erni B, Baumann U. A mechanism of covalent substrate binding in the X-ray structure of subunit K of the Escherichia coli dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2003;100(14):8188–8192. doi: 10.1073/pnas.0932787100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zurbriggen A, et al. X-ray structures of the three Lactococcus lactis dihydroxyacetone kinase subunits and of a transient intersubunit complex. J Biol Chem. 2008;283(51):35789–35796. doi: 10.1074/jbc.M804893200. [DOI] [PubMed] [Google Scholar]
- 32.Kinch LN, Cheek S, Grishin NV. EDD, a novel phosphotransferase domain common to mannose transporter EIIA, dihydroxyacetone kinase, and DegV. Protein Sci. 2005;14(2):360–367. doi: 10.1110/ps.041114805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberholzer AE, Schneider P, Baumann U, Erni B. Crystal structure of the nucleotide-binding subunit DhaL of the Escherichia coli dihydroxyacetone kinase. J Mol Biol. 2006;359(3):539–545. doi: 10.1016/j.jmb.2006.03.057. [DOI] [PubMed] [Google Scholar]
- 34.Bächler C, Schneider P, Bähler P, Lustig A, Erni B. Escherichia coli dihydroxyacetone kinase controls gene expression by binding to transcription factor DhaR. EMBO J. 2005;24(2):283–293. doi: 10.1038/sj.emboj.7600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol. 2010;13(2):204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.