Significance
There are hundreds to thousands of copies of mitochondrial DNA (mtDNA) in each human cell in contrast to only two copies of nuclear DNA. High-frequency pathogenic mtDNA mutations have been found in patients with classic mitochondrial diseases, premature aging, cancers, and neurodegenerative diseases. In this study we investigated the distribution of heteroplasmic mutations, their pathogenic potential, and their underlying evolutionary forces using genome sequence data from the 1000 Genomes Project. Our results demonstrated the prevalence of low-frequency high-pathogenic-potential mtDNA mutations in healthy human individuals. These deleterious mtDNA mutations, when reaching high frequency, could provide a likely source of mitochondrial dysfunction. Managing the expansion of deleterious mtDNA mutations could be a promising means of preventing disease progression.
Abstract
A majority of mitochondrial DNA (mtDNA) mutations reported to be implicated in diseases are heteroplasmic, a status with coexisting mtDNA variants in a single cell. Quantifying the prevalence of mitochondrial heteroplasmy and its pathogenic effect in healthy individuals could further our understanding of its possible roles in various diseases. A total of 1,085 human individuals from 14 global populations have been sequenced by the 1000 Genomes Project to a mean coverage of ∼2,000× on mtDNA. Using a combination of stringent thresholds and a maximum-likelihood method to define heteroplasmy, we demonstrated that ∼90% of the individuals carry at least one heteroplasmy. At least 20% of individuals harbor heteroplasmies reported to be implicated in disease. Mitochondrial heteroplasmy tend to show high pathogenicity, and is significantly overrepresented in disease-associated loci. Consistent with their deleterious effect, heteroplasmies with derived allele frequency larger than 60% within an individual show a significant reduction in pathogenicity, indicating the action of purifying selection. Purifying selection on heteroplasmies can also be inferred from nonsynonymous and synonymous heteroplasmy comparison and the unfolded site frequency spectra for different functional sites in mtDNA. Nevertheless, in comparison with population polymorphic mtDNA mutations, the purifying selection is much less efficient in removing heteroplasmic mutations. The prevalence of mitochondrial heteroplasmy with high pathogenic potential in healthy individuals, along with the possibility of these mutations drifting to high frequency inside a subpopulation of cells across lifespan, emphasizes the importance of managing mitochondrial heteroplasmy to prevent disease progression.
Hundreds to thousands of copies of mitochondrial DNA (mtDNA) are present in each single human cell, in contrast to only two copies of nuclear DNAs. These mtDNAs can differ from each other as a result of inherited or somatic mutations. The coexistence of multiple mtDNA variants in a single cell or among cells within an individual is called heteroplasmy (1). Mitochondrial heteroplasmy has been shown to be implicated in a large spectrum of human diseases. Besides classical mitochondrial diseases such as mitochondrial myopathy, myoclonic epilepsy with ragged red fibers, and mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes, mitochondrial heteroplasmy also plays roles in complex disorders, including type 2 diabetes mellitus, aging, cancer, and late-onset neurodegenerative diseases (1–7). Of the over 500 mtDNA point mutations reported so far that are implicated in disease, ∼55% were observed at known heteroplasmic sites (7). The coexistence of mutant and wild-type mtDNAs requires the pathogenic mutation to reach a frequency threshold before it could evince itself as clinical phenotypes (mitochondrial threshold effect) (4, 8).
Mitochondrial heteroplasmy is common in healthy human populations. Before the application of next-generation sequencing (NGS) technologies, most studies focused on the mtDNA control region and revealed that 6∼11.6% of the population carry heteroplasmy in this region (9–11). The advent of NGS technologies enables the inquiry of mitochondrial heteroplasmy at the genome-wide scale. Several studies using these approaches allowed detection of medium- and high-frequency heteroplasmy with minor allele frequency (MAF) higher than 9%, and it was found that 25∼65% of the general population have at least one heteroplasmy across the entire mitochondrial genome (12–14). However, deeper sequencing depth at the order of thousands is required for confident identification of low-frequency heteroplasmy (MAF in the range of 1%∼10%) (15, 16). Without considering these low-frequency heteroplasmy, the population prevalence of mitochondrial heteroplasmy is underestimated (12–14). Moreover, a preliminary study with ultra-deep sequencing (4,158∼20,803×) of two ∼300-bp mtDNA regions was able to find heteroplasmies with very low frequency (>0.2%) in all tested healthy samples (17). Further investigation with a large sample size and deep sequencing coverage across the whole mitochondrial genome is needed to reveal the universal prevalence of mtDNA heteroplasmy in healthy human populations.
Despite the widespread presence of heteroplasmy in the healthy population, its pathogenic potential has not been well characterized, and the population prevalence of pathogenic heteroplasmy might be underestimated. It has been recognized that mitochondrial heteroplasmy across the genome increases with age (9, 18–20) and acquires unique patterns in tumors (21, 22). A recent epidemiological study indicates that pathogenic mtDNA mutations might be more common in the general population than previously appreciated (23). This study investigated 10 common pathogenic mtDNA mutations in over 3,000 healthy individuals and revealed that at least 1 in 200 individuals harbors a mutation that could potentially cause disease (23); this is much higher than epidemiological estimates of the prevalence of mtDNA diseases, which is only ∼1 in 5,000 (24). This discrepancy is likely due to the mitochondrial threshold effect, because most of the pathogenic mutations exist as heteroplasmy and are compensated by the wild-type mtDNA (4, 23). The population prevalence of pathogenic mtDNA mutations should be much higher if more reported pathogenic mutations are examined in a large sample.
Characterizing the pathogenic potential of mitochondrial heteroplasmy in healthy individuals and its underlying evolutionary forces will further our understanding of the roles of mtDNA variations in aging, tumorigenic, and neurodegenerative processes. In this study, we addressed this issue by analyzing deep sequencing data of mtDNA for 1,085 healthy individuals sampled from 14 global populations in the 1000 Human Genomes Project (25). First, we quantified the prevalence of mitochondrial heteroplasmy, especially disease-associated heteroplasmy, in this healthy cohort. We further characterized the pathogenicity of mitochondrial heteroplasmy with computationally predicted and experimentally reported pathogenic effects. Moreover, we scrutinized the patterns of genomic distribution and site-frequency spectrum for mitochondrial heteroplasmy and elucidated the major evolutionary forces underlying these patterns. We demonstrated that pathogenic mitochondrial heteroplasmy is prevalent in healthy individuals, likely due to insufficient purifying selection in removing them. The implication of our results in health management was also discussed.
Results
Mitochondrial Heteroplasmy Is Prevalent in the Normal Human Population.
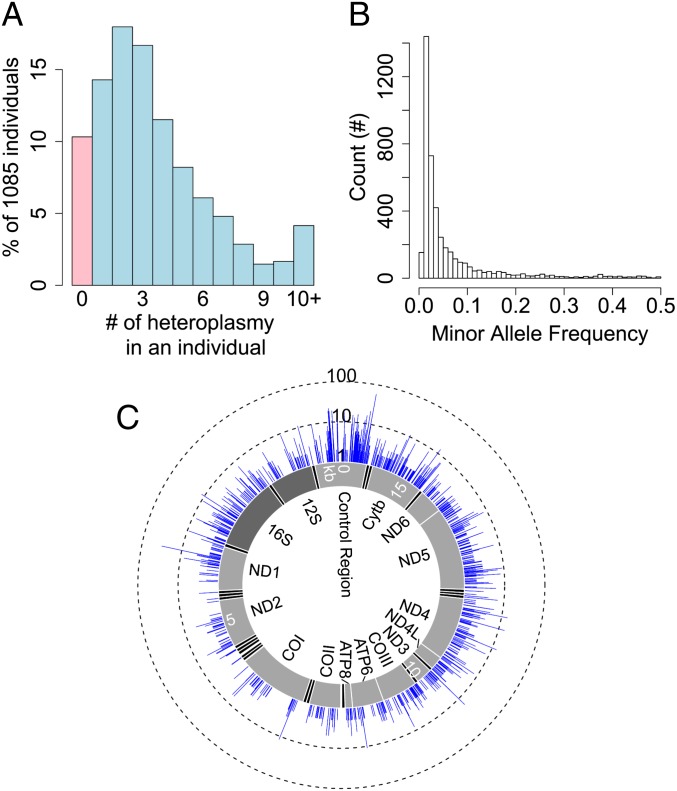
The average depth of coverage for 1,085 individuals sequenced by ILLUMINA or SOLID in the 1000 Genomes Project is 1,805× (SI Appendix, Fig. S1), allowing the identification of low-frequency heteroplasmies. We applied a combination of stringent thresholds to define heteroplasmy with high confidence and estimated the frequency of heteroplasmy with a maximum likelihood (ML) method. In total, we identified 4342 heteroplasmies. There were nine individuals included in our analysis that were additionally sequenced by LS454, providing an opportunity of verifying the accuracy of our computational pipeline. For the 22 heteroplasmies identified in these nine individuals with the ILLUMINA data, all of them were observed in the LS454 data with similar frequency (SI Appendix, Fig. S2), reassuring the reliability of our computational procedure. With the 4,342 heteroplasmies identified in 1,085 individuals, 973 individuals (89.68%) have at least one heteroplasmy. In an extreme case, an individual (HG00740) carried 71 heteroplasmies (Fig. 1A). The population prevalence of heteroplasmy depends on the criteria of defining heteroplasmy. The higher the cutoff for MAF, the lower the prevalence. However, even with MAF cutoffs of 5% and 10%, heteroplasmy is observed in 63.50% and 44.42% of the individuals, respectively (SI Appendix, Fig. S3).
Fig. 1.
Distribution of heteroplasmy in the sample. (A) The percentage of individuals carrying a specific number of heteroplasmy. The category of individuals who do not carry any heteroplasmy is highlighted in pink. (B) Histogram for minor allele frequency of heteroplasmy. (C) The genomic distribution of heteroplasmies and their incidences in the sample. The inner layer represents the mitochondrial genome with tRNA genes highlighted in black, rRNA genes in dark gray and protein-coding genes in light gray. The blue layer indicates the number of individuals (in a total of 1,085) carrying heteroplasmy at a specific site. The number of individuals is shown at a common logarithm scale.
The majority of heteroplasmies are present at low frequency (Fig. 1B). The median ML-estimated MAF is 2.71%. The skew to low frequency is similar to the site frequency spectrum of population polymorphism but less severe (SI Appendix, Fig. S4A). These heteroplasmies were observed at 2,531 mtDNA sites across different regions in mtDNA, and 1,757 (69.42%) of these sites are heteroplasmic in only one individual (Fig. 1C and SI Appendix, Fig. S4B). Among all heteroplasmic sites, 36.67% were also observed to be polymorphic in the population (permutation test, P < 1.00e-5; SI Appendix, Fig. S4 C and D). There is a positive correlation between the population incidence of heteroplasmy and the population MAF of polymorphic sites (linear regression R2 = 0.2358, P < 2.20e-16; SI Appendix, Fig. S5). In a previous study, a relative mutation rate for each site in the mitochondrial genome was defined as the absolute frequency of mutation occurrence in a phylogenetic tree constructed with global human samples (26). Using this dataset, we showed that heteroplasmic sites have significantly higher relative mutation rates than homoplasmic sites (Wilcoxon rank-sum test, P < 2.20e-16; SI Appendix, Fig. S6A), and that the incidence of heteroplasmy is positively correlated with relative mutation rate (linear regression R2 = 0.3702, P < 2.20e-16; SI Appendix, Fig. S6B). These observations further confirmed the reliability of our pipeline in identifying heteroplasmies and indicated that high mutation rate might be a major driving force for the population prevalence of heteroplasmy.
Mitochondrial Heteroplasmy Is Overrepresented in Disease-Associated Sites.
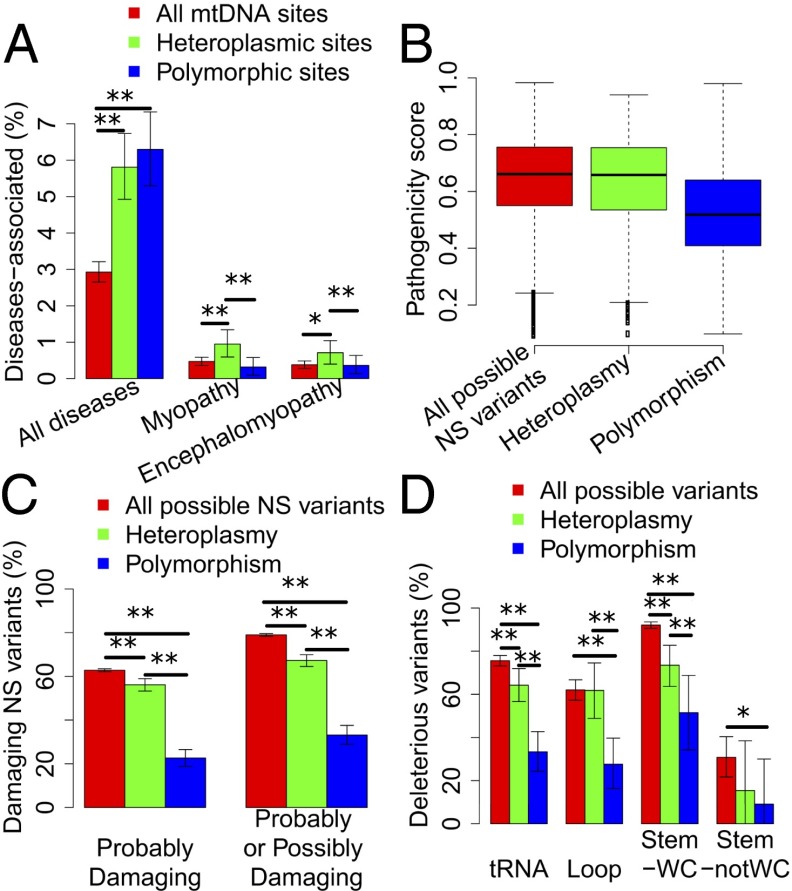
Of the 4,342 detected heteroplasmies, 301 (7.11%) are reported to be disease-associated (7) and 210 individuals (19.35% of 1,085) carried at least one disease-associated heteroplasmy. These observations prompted us to further investigate the disease implication for these heteroplasmies. Among the 13,639 mtDNA sites that satisfied quality control criteria and were examined in our study, 399 (2.93%) are disease associated (7). However, the corresponding number is 147 (5.81%) among the 2,531 heteroplasmic sites, which is significantly higher than expected by chance (χ2 test, P = 2.52e-12). The percentage of disease-associated sites among population polymorphic sites (6.30%) is also significantly more than random expectation (χ2 test, P = 1.44e-14) but is comparable to that of heteroplasmic sites (Fig. 2A). For the two disease categories that have the highest number of associated sites, mitochondrial myopathy and mitochondrial encephalomyopathy, heteroplasmy is overrepresented, even compared with polymorphism. Among all of the sites examined, 64 (0.47%) have been reported to be associated with mitochondrial myopathy, and 52 (0.38%) with mitochondrial encephalomyopathy. Only one site is shared between the two diseases. Heteroplasmic sites are 2.02× [95% confidence interval (CI): 1.40∼2.68] more likely to be associated with mitochondrial myopathy than given by random expectation, and 2.97× (95% CI: 1.60∼8.65) more likely than polymorphic sites. Similarly, for mitochondrial encephalomyopathy, heteroplasmic sites are 1.87× (95% CI: 1.19∼2.57) more likely to be disease associated than random expectation, and 1.97× (95% CI: 1.04∼4.97) more likely than polymorphic sites (Fig. 2A). Additionally, among the heteroplasmic sites we identified, 10 are associated with Leber hereditary optic neuropathy, four with deafness or sensorineural hearing loss, two with Leigh syndrome, three with cardiomyopathy, three with diabetes mellitus, and two with Alzheimer’s or Parkinson disease.
Fig. 2.
Mitochondrial heteroplasmy is highly pathogenic. (A) The percentage of loci associated with diseases in all, heteroplasmic, and polymorphic sites. “All diseases” represents all diseases included in MITOMAP. Myopathy and encephalomyopathy are the two disease categories that have the highest number of mitochondrial loci reported to be associated with. (B) The box plot of MutPred pathogenicity scores for all possible NS variants in the mitochondrial genome, NS heteroplasmies, and polymorphisms. Heteroplasmies occurring in multiple individuals were counted only once. (C) The percentage of PolyPhen-2 predicted damaging variants in all possible NS variants, NS heteroplasmies, and polymorphisms. (D) The percentage of predicted deleterious tRNA variants in all possible variants, heteroplasmies, and polymorphisms. tRNA represents all regions of tRNA genes, including loop and stem regions; Loop represents the loop region; Stem-WC refers to the Watson–Crick pairing positions in the stem region; Stem-notWC refers to those that are not Watson–Crick paired. The error bar represents 95% CI from 10,000 bootstraps. **P < 0.01; *P < 0.05.
Nonsynonymous and tRNA Heteroplasmy Is Highly Pathogenic.
To explore the pathogenicity of mitochondrial heteroplasmy on a broader basis, we applied computational methods to predict the deleterious effect of nonsynonymous (NS) and tRNA mutations. For NS heteroplasmy, we first defined pathogenicity scores for all possible NS changes in the mtDNA with the MutPred algorithm (27, 28). The pathogenicity score ranges from 0 to 1 with a higher pathogenicity score indicating greater likelihood of being pathogenic. For all possible 24,206 NS changes in the mtDNA, the average pathogenicity score is 0.64 (SD = 0.15). For all 1,184 NS heteroplasmies in the dataset, the average score is 0.63 (SD = 0.16), similar to random NS mutations. In contrast, the average pathogenicity score for all 467 population polymorphisms is 0.52 (SD = 0.16), significantly lower than that of heteroplasmies and all possible variants (P = 4.24e-36 and 5.96e-55, respectively; Fig. 2B). To quantify the pathogenic potential of heteroplasmic variants in comparison with population polymorphic ones, we could choose a cutoff of pathogenicity score and define NS changes with scores higher than this cutoff as pathogenic. To avoid the arbitrary use of cutoffs, we applied a series of cutoffs from 0.6 to 0.8 and, in general, heteroplasmy is 1.87∼4.26× more likely to be pathogenic than polymorphism (SI Appendix, Fig. S7). As a verification, the pathogenicity of NS variants was also predicted using PolyPhen-2 (29, 30). PolyPhen-2 yielded comparable predictions with MutPred (SI Appendix, Fig. S8). The percentage of damaging heteroplasmic variants is significantly lower than random expectation, but significantly higher than that of polymorphic ones (Fig. 2C).
We further investigated the pathogenicity of heteroplasmy in tRNA genes. We used the pathogenic prediction for all possible variants in tRNA genes from a previous study which used evolutionary information in functional assessment (31). The percentage of pathogenic variants for heteroplasmy is 64.23%, significantly lower than that for all possible variants (75.58%, χ2 test, P = 0.0023) but significantly higher than that for polymorphism (33.33%, χ2 test, P = 2.63e-06). In other words, tRNA heteroplasmies are 1.93 (95% CI: 1.51∼2.63) times more likely to be pathogenic than polymorphisms. Similar trends were observed when we separated tRNAs into three regions: loop, Watson–Crick pairing positions in stem, and non-Watson–Crick pairing positions in stem (Fig. 2D).
Mitochondrial Heteroplasmy Is Subject to Purifying Selection.
The high pathogenicity of heteroplasmies and their strong association with diseases suggest that heteroplasmy might be under purifying selection. To test this hypothesis, we investigated the genome-wide distribution of heteroplasmies, their unfolded site frequency spectra and the relationship between pathogenicity and derived allele frequency (DAF).
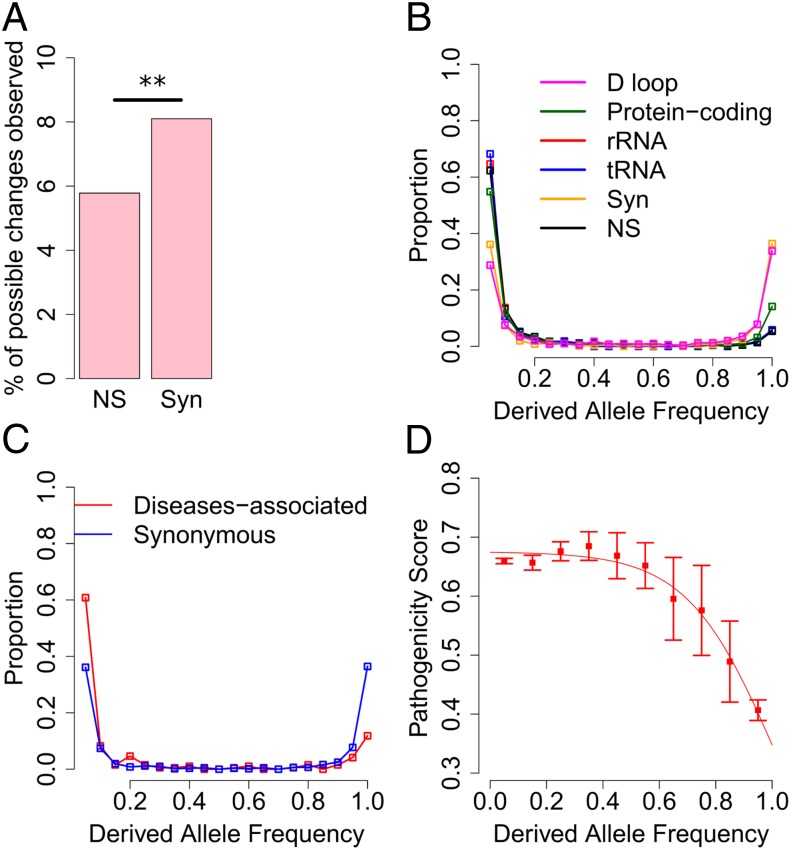
We first examined synonymous and NS variants in protein-coding genes: 5.78% of all possible NS changes in mtDNA were observed with heteroplasmy, which is significantly lower than that of synonymous changes (8.10%, χ2 test, P = 2.01e-10; Fig. 3A), indicating that NS heteroplasmies are subject to purifying selection. We further examined the site frequency spectrum of heteroplasmy. We found that the distribution of DAF for heteroplasmies in the control region is comparable to that for synonymous heteroplasmies. In comparison with these two types of sites, the distributions of DAF for NS, tRNA, and rRNA heteroplasmies are significantly shifted toward lower frequencies (Wilcoxon rank-sum test, P < 9.42e-16; Fig. 3B). Furthermore, heteroplasmies within the rRNA stem tend to have a lower DAF frequency than those in the loop and heteroplasmies in the tRNA stem and anticodon loop regions have significantly lower DAF than those in other tRNA regions (SI Appendix, Fig. S9). Intriguingly, heteroplasmy at disease-associated sites also exhibit significantly lower DAF than that of synonymous heteroplasmy (Wilcoxon rank-sum test, P = 2.07e-10; Fig. 3C). Taken together, these results suggest purifying selection is acting on functional heteroplasmies to keep them at low frequency.
Fig. 3.
Purifying selection on mitochondrial heteroplasmy. (A) The prevalence of synonymous and NS heteroplasmies, which is defined as the percentage of all possible (synonymous or NS) changes that is observed to be heteroplasmic. **P = 2.01e-10 in χ2 test. (B) The distribution of DAF for heteroplasmies in different mtDNA genomic regions. (C) The distribution of DAF for disease-associated and synonymous heteroplasmies. (D) The average pathogenicity score in each bin of DAF. Error bar represents 1 SE. The red line represents model-fitting with a logistic function of . R2 = 0.9794, P < 9.76e-06.
The effect of purifying selection on removing deleterious heteroplasmy suggests a possible reverse correlation between the level of pathogenicity and the frequency of a heteroplasmy. Consistent with this expectation, as depicted in Fig. 3D, heteroplasmies with low derived frequency inside an individual tend to have high pathogenicity scores. This negative relationship can be modeled with a logistic function (R2 = 0.9794, P < 9.76e-06). From the regression, we inferred that the pathogenicity scores are comparable among heteroplasmies with DAF less than 60% and declines as DAF exceeds 60%. This pattern indicates that pathogenic heteroplasmies must reach high frequency before they are selected against, and 60% in general might be a good estimate of the threshold for pathogenic heteroplasmic mutations to express deleterious effect. Consistent with the impact of purifying selection on removing deleterious heteroplasmy, our results also show that though heteroplasmies observed in a few individuals (1∼4) have comparable pathogenicity scores (mean = 0.64, SD = 0.16), those observed in more than five individuals have significantly lower pathogenicity scores (mean = 0.43, SD = 0.25, Wilcoxon rank-sum test, P = 8.74e-04; SI Appendix, Fig. S10). When we examined the DAF of pathogenic tRNA heteroplasmies, we also found that 81.90% of heteroplasmies with DAF less than 5% are pathogenic, whereas only 30% of heteroplasmies with DAF larger than 95% are pathogenic (Fisher’s exact test, P = 0.0010).
Purifying Selection Is Less Efficient on Heteroplasmy Than on Polymorphism.
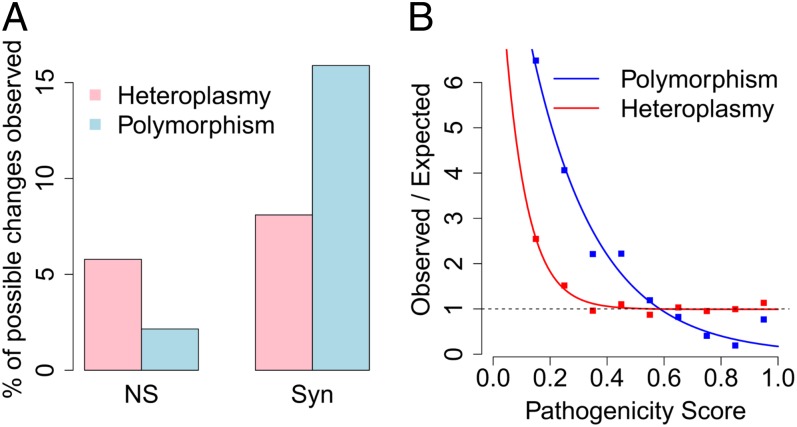
Although heteroplasmic sites show evidence of purifying selection, we hypothesized that purifying selection on heteroplasmy is much weaker than that on polymorphism due to the low frequencies of most heteroplasmies inside individual cells. Indeed, consistent with this hypothesis, the difference between the percentages of synonymous and NS variants is much bigger for polymorphisms than heteroplasmies (χ2 test, P < 2.2e-16; Fig. 4A). To further quantitatively compare the effect of natural selection on these two types of mtDNA variants, we defined a selection function by dividing the observed distribution of pathogenicity scores for all NS heteroplasmies (or polymorphisms) by the expected distribution of pathogenicity scores from all possible NS variants. This quantitative method has been previously applied to mitochondrial polymorphisms (28). In the absence of natural selection, mutations are similar to random draws from all possible changes in the genome, so the selection function is expected to be equal to a constant, 1. Consistent with previous study (28), the selection function of polymorphism can be modeled by a simple function of exponential decay (R2 = 0.9758, P = 4.71e-06; Fig. 4B). The parameterizations in our data are similar to the previous study (28). We also confirmed that the observed value for polymorphisms with very high pathogenicity scores (>0.9) deviates from the exponential fit, indicating that forces other than purifying selection might have acted on these variants (28).
Fig. 4.
Less-efficient purifying selection on mitochondrial heteroplasmy than on polymorphism. (A) The prevalence of synonymous and NS heteroplasmies in comparison with that of synonymous and NS polymorphisms. (B) The selection function for heteroplasmy (or polymorphism) defined by dividing the observed distribution of pathogenicity scores for heteroplasmy by the expected distribution of pathogenicity scores from all possible NS variants. The dashed line represents the expected value, 1, for selection function under neutral evolution. The exponential fit for polymorphism is . R2 = 0.9758, P = 4.71e-06. The exponential function for heteroplasmy is . R2 = 0.9650, P = 4.65e-06.
The selection function for heteroplasmy also follows an exponential decay (R2 = 0.9650, P = 4.65e-06; Fig. 4B). Interestingly, in contrast to polymorphism, it has an additional constant very close to 1, indicating that purifying selection is too weak to effectively remove pathogenic heteroplasmies, likely due to their low frequency inside the cells. Using the selection functions for both polymorphisms and heteroplasmies, the relative effect of selection on two different amino acid variations could be assessed by a ratio of the exponential functions for the two pathogenicity scores (28). For example, a population polymorphic variant with a pathogenicity score of 0.8 is subject to ∼2× stronger purifying selection than a polymorphic variant with a score of 0.6. In comparison, the strength of purifying selection on two heteroplasmies with pathogenicity scores of 0.8 and 0.6 is almost the same. This quantitative comparison further confirms that purifying selection on heteroplasmy is much less efficient than that on population polymorphism in removing deleterious mutations.
Discussion
Next-generation sequencing technologies enable the detection of mitochondrial heteroplasmy at the genome-wide level with unprecedented resolution. However, specificity of detection and accuracy of quantification can only be achieved when sequencing errors and technical artifacts are carefully controlled for. A set of criteria for detecting heteroplasmy with modern sequencing technologies have been developed in a few pioneering studies (13, 16, 21, 32). Integrating criteria that have been proven to be effective (SI Appendix, Table S2), our computational pipeline filtered low-quality bases and unreliable mappings, especially minimizing the complications of nuclear mitochondrial sequences (NumtS) (33); it also used double-stranded validation, which required heteroplasmy to be detected in both strands with support from multiple reads. Furthermore, our computational pipeline estimated the frequency of heteroplasmy with a maximum likelihood method by taking into account sequencing error and yielded a log likelihood ratio (LLR) indicating the confidence of true positive heteroplasmy. The applications of these tested criteria ensure the correct detection and accurate quantification of heteroplasmy. The reliability of our computational pipeline was confirmed by examining nine individuals sequenced by both ILLUMINA and LS454 (SI Appendix, Fig. S2 and Dataset S1). Moreover, the biologically meaningful patterns of mitochondrial heteroplasmy observed in our study also augment the reliability of our computational pipeline. The complete list of heteroplasmies identified in our study can be found in Dataset S2. Additionally, we did not observe consistent and significant population or sex difference in heteroplasmy patterns (SI Appendix, Figs. S11 and S12).
The prevalence of mitochondrial heteroplasmy at genome-wide scale has been explored in a few studies with smaller sample size and shallower sequencing depth. From the 1000 Genomes Pilot Project, 163 individuals were sequenced to 37.7∼3,535× coverage and 45% were observed to possess heteroplasmic sites with MAF mostly larger than 10% (12). Another study sequenced 114 individuals with ILLUMINA to a mean coverage of 67× and 17 individuals to a mean coverage of 211×. Among these 131 individuals, 24.43% were detected to possess heteroplasmy with MAF larger than 10% (13). Moreover, a study used a 454 Genome Sequencer FLX system and sequenced 40 HapMap individuals to a mean coverage of 120×, and 65% individuals were found to have heteroplasmies with MAF higher than 9% (14). With a MAF cutoff of 10%, the prevalence of heteroplasmy is 44.42% in our dataset (SI Appendix, Fig. S3), which is within the range of previous estimates and very close to the estimate from the 1000 Genomes Pilot Project (12). Our study benefits from higher coverage and is able to detect heteroplasmy with MAF as low as 1%. With a much larger sample size, we estimate that the prevalence of heteroplasmy in the healthy population is at least 90%. Because the majority of heteroplasmy is present at very low frequency (Fig. 1B), it is very likely that heteroplasmy is universal to all healthy individuals. Results from a recent study conducted on a small sample support this idea (17).
The high pathogenic potential of mitochondrial heteroplasmy is consistently demonstrated with experimentally observed disease-associated mutations, computationally predicted functional effect, and the presence of weak negative selection. First, experimentally reported diseases-associated mtDNA mutations are overrepresented in both polymorphic and heteroplasmic sites (Fig. 2A). This pattern has been previously observed in a study with a much smaller sample size (13), and it suggests that heteroplasmic and polymorphic variants are either only mildly deleterious or not yet effectively removed by purifying selection. Because polymorphic variants have gone through generations of purifying selection, their overrepresentation in disease-associated sites is likely resulted from their mild deleterious effect. In contrast, because heteroplasmic variants have a much shorter time frame for natural selection, they are likely subject to weaker purifying selection and have higher pathogenic potential. However, the overrepresentation of polymorphism and heteroplasmy in disease-associated sites may also reflect the research bias toward using known polymorphic sites in disease studies. Second, we artificially created all possible variants in the mitochondrial genome and computationally predicted their pathogenic effects, which serve as a pathogenicity benchmark before being subject to purifying selection. In comparison with this theoretical expectation, heteroplasmy has slightly lower pathogenicity, whereas polymorphism has much lower effect (Fig. 2 B–D), which is consistent with the fact that polymorphism has been subject to generations of purifying selection and only variants with mild deleterious effect could survive; it also suggests that though purifying selection also acts on heteroplasmy, its strength may be weak and therefore the pathogenic effect of heteroplasmy is very close to the theoretical expectation without purifying selection. Last, we observed convincing signals of purifying selection on heteroplasmy and demonstrated its weaker strength than that on polymorphism, further supporting the high pathogenic potential of heteroplasmy.
The prevalence of pathogenic heteroplasmic mtDNA mutations in the general population due to inefficient purifying selection has important clinical implication. Although only ∼1 in 5,000 people suffers from mitochondrial diseases (24), the incidence of pathogenic mtDNA mutations could be much higher because of the mitochondrial threshold effect that masks the deleterious effect of low-frequency pathogenic mutations. A study of 10 common pathogenic mtDNA mutations revealed an incidence of at least 1 in 200 subjects (23). For these 10 mutations, the prevalence of heteroplasmy in our samples is 1 in 155 (95% CI: 83–556). When we included all identified disease-associated mtDNA mutations (7), the incidence of pathogenic heteroplasmies is 19.35%, or 1 in 5 individuals (95% CI: 4.62–5.87). Given the likely underestimation of disease–mtDNA mutation association and the observed prevalence of heteroplasmic mtDNA mutations with high predicted pathogenic scores in this study, the real frequency of pathogenic mitochondrial heteroplasmy could be much higher than this estimation.
Multiple underlying mechanisms have been proposed to modulate the expansion of deleterious mtDNA mutations at the cellular level. According to computational modeling of the relaxed replication of mtDNA in both dividing and nondividing cells, even with random genetic drift alone, the typical lifespan of an individual is more than enough for low-frequency heteroplasmy to reach high frequency or even homoplasmy in a small population of cells (34–36). On average it only takes ∼70 generations of cell divisions to reach homoplasmy from a new mutation; that is only ∼25 y for epithelial cells, which experience three cell turnovers per year (34). In postmitotic tissues, such as skeletal muscle and neurons, the mean time to homoplasmy is ∼40 y (35, 36). Besides random genetic drift during intracellular mitochondrial turnover and cell divisions (34, 35, 37), natural selection with replicative or survival advantage has also been proposed to either accelerate or decelerate the spread of pathogenic mutations (38–40). Extensive experimental observations have recorded abundant clonally expanded mtDNA mutations in human tissues, especially in aged individuals (40, 41). More importantly, both computational modeling and experimental evidence support that mutation accumulated with age results mostly from the clonal expansion of mutations that existed early in life, rather than de novo mutations later in life (35, 37, 41, 42). All individuals included in the 1000 Genomes Project were healthy at the time of sample collection (25). The prevalence of pathogenic mitochondrial heteroplasmy in healthy individuals observed in this study raises the concern that they could expand to high frequency in a fraction of cells later in life, exceed the critical phenotypic threshold, and lead to age-related diseases. Future studies are needed to unravel the mechanisms of clonal expansion of pathogenic heteroplasmy, to elucidate the roles of mitochondrial heteroplasmy in complex disorders, and to develop effective strategies in managing these mutations to prevent the progression into disease.
Materials and Methods
Sequencing Data.
Sequencing reads mapped to the mitochondrial genome in the 1000 Genomes Project phase 1 data were downloaded from the 1000 Genomes Project data server. Our analysis focused on 1,085 unrelated individuals from 14 populations, which were sequenced using either ILLUMINA or SOLID platforms. There were nine individuals sequenced by two methods (ILLUMINA and LS454). These individuals were used to confirm the reliability of our computational pipeline with ILLUMINA data. See SI Appendix, Table S1 for more detailed information.
Computational Pipeline for Calling Heteroplasmy and Polymorphism.
An expanded version of the methods is in SI Appendix. Briefly, sequencing reads retrieved from the 1000 Genomes Project data sever were remapped to the human genome, both nuclear and mitochondrial genomes, using GSNAP (43). Only reads uniquely mapped to the mitochondrial genome were recorded to minimize the complications of NumtS (33). We further filtered the data and defined “usable sites” based on the following three quality control criteria: (i) Phred quality score ≥20 for used bases; (ii) 10× coverage of qualified bases on both positive and negative strands; (iii) 95% individuals satisfy criteria i and ii. Together, 13,639 mtDNA sites satisfied these quality control criteria and were examined in our study. A candidate heteroplasmic site was defined with the following two criteria: (i) the raw frequency for the minor allele is no less than 1% on both strands; and (ii) all alleles have support from at least two reads on each strand.
For each candidate heteroplasmic site, we further applied a ML method to accurately estimate the frequency of the major allele while taking into account sequencing error (32, 44). For example, for all bases mapped to the positive strand of a site, bases are the major alleles and bases are the minor alleles. Each base has respective sequencing quality, corresponding to the probability of sequencing error . The underlying parameter of interest is the frequency of the major allele . The likelihood function could be written as follows:
We estimated under two models: heteroplasmy (m1) and homoplasmy (m0), and a LLR was calculated as . A high-confidence heteroplasmy was defined as candidate heteroplasmy with LLR no less than 5 (32). With all these criteria (see SI Appendix, Table S2 for a brief list), a total of 4,342 heteroplasmies were defined; among them, 153 have a minor allele frequency estimated by the ML method to be smaller than 1%, even though we required that the raw frequency for the minor allele is no less than 1% on both strands. After detecting heteroplasmy, consensus sequences were assembled for each individual and compared among all individuals to identify polymorphic sites. A consensus sequence for each individual was assembled using the alleles present at homoplasmic sites, and the major alleles at heteroplasmic sites. Sites were classified as polymorphic if there was more than one allele present in the population.
The Measure of Pathogenicity.
The pathogenicity scores for all possible NS changes, inferred based on the revised Cambridge Reference Sequence , were predicted with the MutPred algorithm (27); as a verification, their pathogenic effects were further predicted with PolyPhen-2 (29). Both methods yielded comparable results. The MutPred pathogenicity scores were retrieved from a previous study (28). A higher pathogenicity score indicates a higher likelihood that the NS change is pathogenic (27, 28). The pathogenic effect of tRNA mutations were obtained from a previous publication (31). Disease association information was obtained from MITOMAP (7).
Supplementary Material
Acknowledgments
We thank Mr. Paul Billing-Ross and Drs. Andrew Clark, Jason Locasale, Patrick Stover, and Lin Xu for their discussions and comments on the manuscript. This work was supported by various funds from Cornell University, an International Life Sciences Institute future leader award, National Science Foundation Grant MCB-1243588, and National Institutes of Health Grant 1R01AI085286 (to Z.G.). K.Y. is a Center for Vertebrate Genomics Scholar at Cornell University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403521111/-/DCSupplemental.
References
- 1.Chinnery PF, Hudson G. Mitochondrial genetics. Br Med Bull. 2013;106:135–159. doi: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51(5):440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 4.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat Rev Genet. 2012;13(12):878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpley MS, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151(2):333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keogh M, Chinnery PF. Hereditary mtDNA heteroplasmy: A baseline for aging? Cell Metab. 2013;18(4):463–464. doi: 10.1016/j.cmet.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Pesini E, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35(Database issue):D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossignol R, et al. Mitochondrial threshold effects. Biochem J. 2003;370(Pt 3):751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calloway CD, Reynolds RL, Herrin GL, Jr, Anderson WW. The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. Am J Hum Genet. 2000;66(4):1384–1397. doi: 10.1086/302844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Camargo MA, et al. No relationship found between point heteroplasmy in mitochondrial DNA control region and age range, sex and haplogroup in human hairs. Mol Biol Rep. 2011;38(2):1219–1223. doi: 10.1007/s11033-010-0220-1. [DOI] [PubMed] [Google Scholar]
- 11.Irwin JA, et al. Investigation of heteroplasmy in the human mitochondrial DNA control region: A synthesis of observations from more than 5000 global population samples. J Mol Evol. 2009;68(5):516–527. doi: 10.1007/s00239-009-9227-4. [DOI] [PubMed] [Google Scholar]
- 12.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87(2):237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sosa MX, et al. Next-generation sequencing of human mitochondrial reference genomes uncovers high heteroplasmy frequency. PLOS Comput Biol. 2012;8(10):e1002737. doi: 10.1371/journal.pcbi.1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13(5):R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto H, et al. Dynamics of mitochondrial heteroplasmy in three families investigated via a repeatable re-sequencing study. Genome Biol. 2011;12(6):R59. doi: 10.1186/gb-2011-12-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne BA, et al. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22(2):384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sondheimer N, et al. Neutral mitochondrial heteroplasmy and the influence of aging. Hum Mol Genet. 2011;20(8):1653–1659. doi: 10.1093/hmg/ddr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross JM, et al. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501(7467):412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9(9):e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larman TC, et al. Cancer Genome Atlas Research Network Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci USA. 2012;109(35):14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83(2):254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer AM, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63(1):35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 25.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soares P, et al. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84(6):740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25(21):2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira L, Soares P, Radivojac P, Li B, Samuels DC. Comparing phylogeny and the predicted pathogenicity of protein variations reveals equal purifying selection across the global human mtDNA diversity. Am J Hum Genet. 2011;88(4):433–439. doi: 10.1016/j.ajhg.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei I, Jordan DM, Sunyaev SR. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 76:7.20.1–7.20.41. [DOI] [PMC free article] [PubMed]
- 31.Kondrashov FA. Prediction of pathogenic mutations in mitochondrially encoded human tRNAs. Hum Mol Genet. 2005;14(16):2415–2419. doi: 10.1093/hmg/ddi243. [DOI] [PubMed] [Google Scholar]
- 32.Picardi E, Pesole G. Mitochondrial genomes gleaned from human whole-exome sequencing. Nat Methods. 2012;9(6):523–524. doi: 10.1038/nmeth.2029. [DOI] [PubMed] [Google Scholar]
- 33.Simone D, Calabrese FM, Lang M, Gasparre G, Attimonelli M. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics. 2011;12:517. doi: 10.1186/1471-2164-12-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coller HA, et al. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet. 2001;28(2):147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 35.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68(3):802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chinnery PF, Samuels DC. Relaxed replication of mtDNA: A model with implications for the expression of disease. Am J Hum Genet. 1999;64(4):1158–1165. doi: 10.1086/302311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne BA, et al. Mitochondrial aging is accelerated by anti-retroviral therapy through the clonal expansion of mtDNA mutations. Nat Genet. 2011;43(8):806–810. doi: 10.1038/ng.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz F, et al. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 2002;30(21):4626–4633. doi: 10.1093/nar/gkf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18(6):1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nekhaeva E, et al. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci USA. 2002;99(8):5521–5526. doi: 10.1073/pnas.072670199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 42.Khrapko K. The timing of mitochondrial DNA mutations in aging. Nat Genet. 2011;43(8):726–727. doi: 10.1038/ng.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chepelev I. Detection of RNA editing events in human cells using high-throughput sequencing. Methods Mol Biol. 2012;815:91–102. doi: 10.1007/978-1-61779-424-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.