Significance
Autophagy recycles cytoplasmic components to facilitate cellular homeostasis and adaptation in response to nutrient deprivation. Impaired autophagy regulation is linked to numerous metabolic and aging-related disorders. One important regulator of autophagy is the Target of Rapamycin (TOR) kinase, which assembles into two complexes: TOR Complex 1 (TORC1) and TOR Complex 2 (TORC2), where TORC1 is a well-established negative regulator of autophagy. Here we characterize a pathway wherein TORC2 and its downstream target kinase, Ypk1, act as positive regulators of autophagy by promoting the activation of the general amino acid control response during amino acid starvation. Thus TOR functions in two distinct complexes to provide a tunable autophagy response according to the cellular metabolic state.
Keywords: Atg8, Gcn4
Abstract
The highly conserved Target of Rapamycin (TOR) kinase is a central regulator of cell growth and metabolism in response to nutrient availability. TOR functions in two structurally and functionally distinct complexes, TOR Complex 1 (TORC1) and TOR Complex 2 (TORC2). Through TORC1, TOR negatively regulates autophagy, a conserved process that functions in quality control and cellular homeostasis and, in this capacity, is part of an adaptive nutrient deprivation response. Here we demonstrate that during amino acid starvation TOR also operates independently as a positive regulator of autophagy through the conserved TORC2 and its downstream target protein kinase, Ypk1. Under these conditions, TORC2-Ypk1 signaling negatively regulates the Ca2+/calmodulin-dependent phosphatase, calcineurin, to enable the activation of the amino acid-sensing eIF2α kinase, Gcn2, and to promote autophagy. Our work reveals that the TORC2 pathway regulates autophagy in an opposing manner to TORC1 to provide a tunable response to cellular metabolic status.
Autophagy is an evolutionarily conserved process that recycles cytoplasmic contents and organelles in eukaryotic cells (1–3). During normal cell growth, a basal rate of autophagy serves as a quality control mechanism to maintain cellular homeostasis and longevity (4, 5). However, in response to energetic stress, such as nutrient deprivation, autophagy flux markedly increases to generate biosynthetic precursors to facilitate cellular adaptation and survival (1–3, 6). Accordingly, impaired regulation of autophagy is linked to a number of metabolic and aging-related disorders including cancer, heart failure, and a number of neurodegenerative diseases (7, 8). Autophagy is catalyzed by the action of numerous highly regulated autophagy-related (Atg) proteins, many of which assemble at discrete peri-vacuolar structures, termed “phagophore assembly sites” (PAS) (9, 10). The PAS initiate the de novo assembly of double-membrane–bound vesicles, termed “autophagosomes,” which occurs at endoplasmic reticulum (ER) exit sites that facilitate membrane trafficking from the ER (11, 12). Growth of the autophagosome is linked to the sequestration of cargo and, upon completion of autophagosome formation, the outer membrane of the autophagosome fuses with the lytic compartment (vacuole or lysosome), exposing the inner membrane and cargo to the degradative action of resident hydrolases (1, 13).
Distinct signaling pathways regulate autophagy in response to nutrient availability, including the Protein Kinase A (PKA) and Target of Rapamycin Complex 1 (TORC1) pathways, which converge to independently control the activity of components that initiate autophagosome formation (14). The central component of TORC1 is the evolutionarily conserved TOR kinase, which assembles into two structurally and functionally distinct protein complexes, TORC1 and TORC2, where TORC1 is uniquely inhibited by the macrolide antibiotic rapamycin (15). The autophagy field primarily uses rapamycin treatment or nitrogen starvation, both of which inhibit TORC1 activity, to study autophagy induction (1–3, 16, 17). However, amino acid starvation, specifically that required auxotrophically, induces autophagy in a manner independent of TORC1 activity and instead requires the general amino acid control (GAAC) response regulated by the eIF2α kinase, Gcn2 (17, 18). During amino acid limitation, unconjugated tRNAs bind to and activate Gcn2, which acts as a sensor for intracellular amino acid levels (19). Activated Gcn2 indirectly promotes translation of the transcription factor Gcn4, which in turn induces the expression of genes important for both amino acid biosynthesis and autophagy (20–22).
In this study, we demonstrate that during amino acid starvation the rapamycin-insensitive TORC2 is a positive regulator of autophagy through its downstream target kinase, Ypk1, and is required for Gcn2 activation. We further demonstrate that TORC2/Ypk1 signals to Gcn2 via the Ca2+-dependent phosphatase calcineurin, which we establish is a negative regulator of the GAAC response and autophagy. Together, these findings identify a functional relationship between TORC2 and autophagy, whereby TORC2/Ypk1 signaling functions to promote autophagy upon amino acid limitation by regulating the activity of calcineurin and the GAAC response.
Results
TORC2/Ypk1 Signaling Is Required for Amino Acid Starvation-Induced Autophagy.
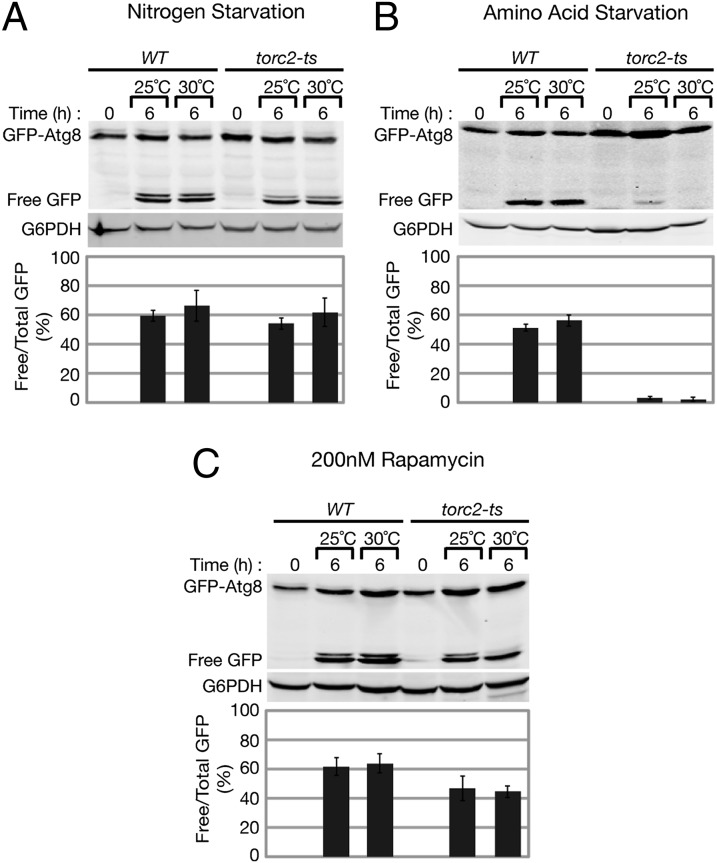
We asked whether TORC2 functions to regulate autophagy by using a temperature sensitive (ts) variant of the essential TORC2 component Avo3 (23), referred to here as torc2-ts. We first examined autophagy in torc2-ts cells under conventional nitrogen starvation conditions (low ammonium sulfate levels in combination with the absence of amino acids, as described in Materials and Methods). Cells were grown at 25 °C in rich media to mid log phase, and autophagy was induced by transferring cells to nitrogen starvation media for 6 h at both permissive (25 °C) and nonpermissive (30 °C) temperatures. We monitored autophagy using the established GFP-ATG8 reporter under the control of the endogenous ATG8 promoter (pRS416-prATG8-GFP-ATG8) (24). GFP-Atg8 associates with autophagosomes, and the Atg8 domain is degraded in the vacuole to create free GFP, allowing for visualization and quantification of autophagosomal degradation in the vacuole, or autophagosomal flux (24–26). We quantified autophagy flux by Western blot analysis by calculating the ratio of free GFP to total GFP signal (Materials and Methods). Autophagy flux was comparable in both wild-type and torc2-ts cells at permissive and nonpermissive temperatures (Fig. 1A), indicating that TORC2 is not required for the induction of autophagy following nitrogen starvation.
Fig. 1.
TORC2 is required for amino acid starvation induced autophagy. Wild-type (WT) and torc2-ts (PLY1141) cells expressing pRS416 prATG8-GFP-ATG8 were grown at 25 °C to log phase and then transferred to (A) nitrogen starvation media, (B) amino acid starvation media, (C) or treated with 200 nM rapamycin in SCD growth media for 6 h at both 25 °C (permissive) and 30 °C (nonpermissive). Cells were analyzed at indicated time points by whole-cell protein extraction and Western blot analysis. Membranes were probed with α-GFP and α-G6PDH (Zwf1) primary antibodies. Quantification of autophagy flux is shown as a ratio of free GFP to total GFP (GFP-Atg8 and free GFP) after 6 h of starvation (Materials and Methods).
We next examined the role of TORC2 in autophagy under amino acid starvation conditions (Materials and Methods). Autophagy flux in torc2-ts was significantly decreased compared with wild type at 6 h of starvation (Fig. 1B). This autophagy flux defect was observed at both permissive and nonpermissive temperatures, consistent with previous observations that torc2-ts cells are also impaired for TORC2 function at the permissive temperature (23). These data indicate a role for TORC2 as a positive regulator of autophagy flux during amino acid starvation. In addition to a defect in autophagy flux, torc2-ts cells exhibited an elevated basal level of GFP-Atg8 before starvation (Fig. 1 A–C), suggesting that TORC2 may also negatively regulate Atg8 expression and/or turnover. Treatment of torc2-ts cells under growing conditions with rapamycin increased autophagy flux to levels comparable to wild-type cells at permissive and nonpermissive temperatures (Fig. 1C), consistent with our observations that TORC2 function is not required for autophagy under nitrogen starvation conditions. These results confirm that the defect in flux in torc2-ts cells is not due to intrinsic defects in the autophagy machinery but rather in signaling used during amino acid starvation to induce autophagy. Our observations indicate that TORC1 and TORC2 act independently to regulate autophagy under different starvation conditions.
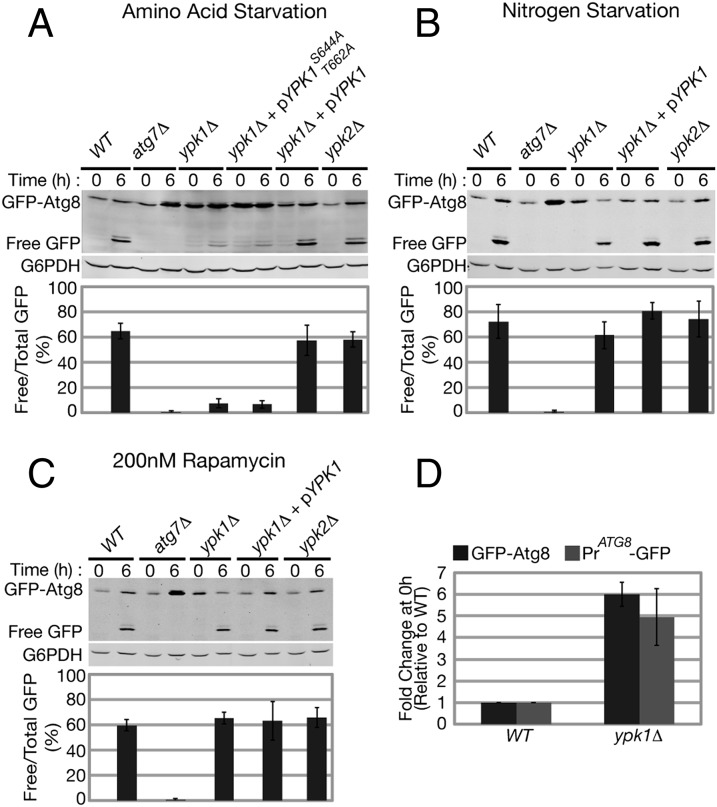
We asked whether the partially redundant AGC kinases Ypk1 and Ypk2, which function downstream of TORC2, are required for autophagy (27, 28). Similar to torc2-ts cells, autophagy flux in ypk1Δ cells was also severely compromised following amino acid starvation compared with wild-type cells (Fig. 2A). Like torc2-ts cells, ypk1Δ cells exhibited elevated basal levels of GFP-Atg8 under normal growing conditions before starvation (Fig. 2 A–D). Expression of a plasmid-born wild-type YPK1 gene under the control of its endogenous promoter fully restored autophagy flux and suppressed accumulation of GFP-Atg8 in ypk1Δ cells (ypk1Δ + pYPK1; Fig. 2A), indicating that the observed defects in autophagy are specific to loss of Ypk1. In contrast to Ypk1, loss of Ypk2 did not result in autophagy defects, as the levels of autophagy flux observed in ypk2Δ cells following amino acid starvation were similar to wild-type cells (Fig. 2A). However, the defect in autophagy flux in ypk1Δ cells is not as severe as that observed in torc2-ts cells (Figs. 1B and 2A) or in cells lacking the essential autophagy gene ATG7 (atg7Δ; Fig. 2A), both of which are completely defective in autophagy flux (17, 29). Thus, in ypk1Δ cells, Ypk2 may contribute to TORC2-dependent regulation of amino acid starvation-induced autophagy.
Fig. 2.
Ypk1 is required for amino acid starvation induced autophagy. WT, atg7Δ , ypk1Δ, and ypk2Δ cells expressing pRS416 prATG8-GFP-ATG8 [and, when indicated, plasmids expressing wild-type Ypk1 (pPL250) or Ypk1S644A/T662A (pPL491)] were grown at 30 °C to log phase and then transferred to (A) amino acid starvation media, (B) nitrogen starvation media, (C) or treated with 200 nM rapamycin in SCD growth media for 6 h at 30 °C. Analysis of GFP-Atg8 and quantification of autophagy flux were performed as described in Fig. 1. (D) WT and ypk1Δ cells expressing either pRS416 prATG8-GFP-ATG8 or pRS426- prATG8-GFP were grown at 30 °C to log phase and harvested, and Western blot analysis was performed as described in Fig. 1. GFP or GFP-Atg8 protein bands were normalized to G6PDH (Zwf1), and fold increase was calculated relative to WT.
TORC2 phosphorylates Ypk1 at two residues, Ser-644 and Thr-662, within the so-called “turn motif” and “hydrophobic motif,” respectively (28). Significantly, both defects in autophagy flux upon amino acid starvation and an accumulation of GFP-Atg8 before starvation were observed in ypk1Δ cells expressing a TORC2 phospho-null version of Ypk1 (ypk1 S644A T662A) (Fig. 2A). This observation indicates that TORC2 directly signals to its target kinase, Ypk1, to promote autophagy during amino acid starvation. An autophagy response equivalent to that of wild-type cells was observed when ypk1Δ cells were either starved for nitrogen (Fig. 2B) or treated with rapamycin (Fig. 2C). Taken together, these results demonstrate that TORC2 functions through Ypk1 in a pathway parallel to TORC1 to positively regulate autophagy during amino acid starvation.
To address the basis for the observed elevated levels of GFP-Atg8 before starvation in ypk1Δ cells, we used a GFP-only reporter under control of the endogenous ATG8 promoter (pRS426-prATG8-GFP). Compared with wild-type cells, a similar fold-increase was observed in ypk1Δ cells with the GFP-only reporter as was observed using the GFP-Atg8 reporter (Fig. 2D). Based on this result we conclude that ATG8 transcription and/or translation is aberrantly induced in ypk1Δ cells.
Calcineurin Inhibits Autophagy Downstream of TORC2/Ypk1 Signaling.
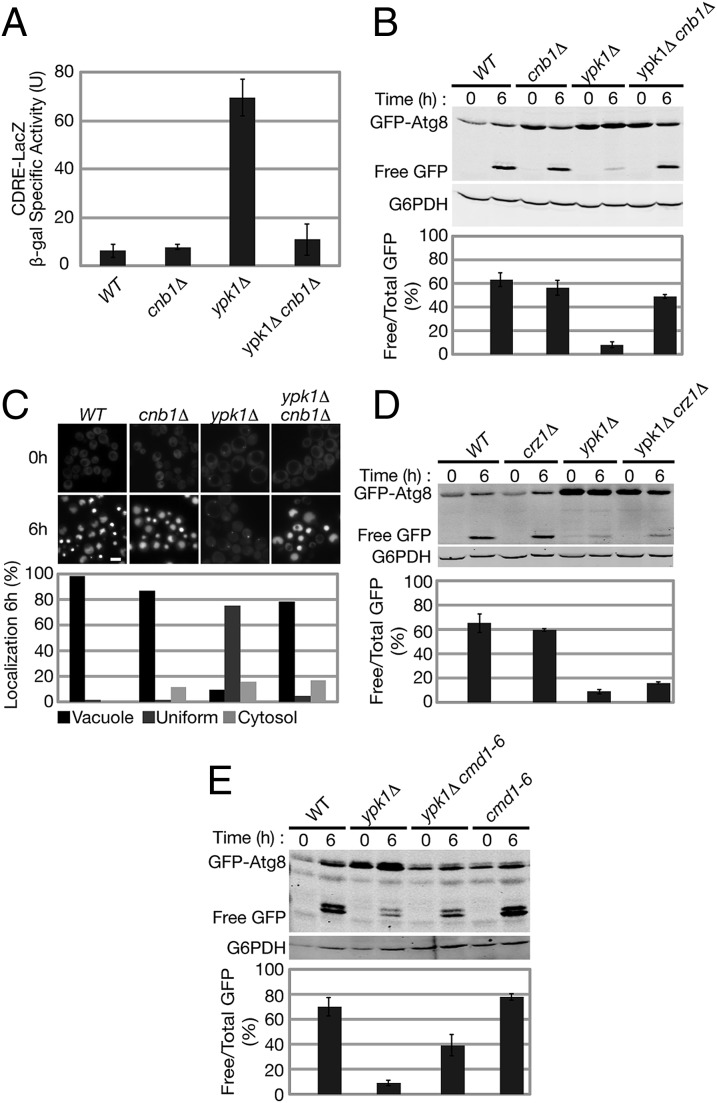
Previous studies have demonstrated that TORC2/Ypk1 signaling negatively regulates the activity of calcineurin, a highly conserved Ca2+/calmodulin-dependent Ser/Thr phosphatase, which consists of the catalytic subunit, calcineurin A, and an essential regulatory subunit, calcineurin B (28, 30, 31). Thus, we tested whether increased calcineurin activity in ypk1Δ cells contributed to autophagy inhibition by decreasing calcineurin activity through deletion of CNB1, which encodes the calcineurin B regulatory subunit (31). Using a calcineurin-dependent response element (CDRE) lacZ reporter (32), we observed that calcineurin activity increased greater than sevenfold in ypk1Δ cells, compared with wild-type cells, and this increase was dependent upon the presence of CNB1 (Fig. 3A), as previously reported (28). We examined autophagy flux in wild-type, ypk1Δ, and ypk1Δ cnb1Δ cells following amino acid starvation. Significantly, we observed that ypk1Δ cells deleted for CNB1 exhibit autophagy flux comparable to wild-type cells (Fig. 3B). These observations are consistent with a role for calcineurin as a negative regulator of amino acid starvation-induced autophagy downstream of TORC2/Ypk1 signaling.
Fig. 3.
Calcineurin inhibits autophagy downstream of TORC2/Ypk1. (A) WT, cnb1Δ, ypk1Δ, and ypk1Δ cnb1Δ cells containing plasmid pAMS363 expressing a 2xCDRE:lacZ fusion were grown at 30 °C to log phase and harvested. β-Galactosidase activity was measured (Materials and Methods) and is given in units of nanomoles of ONPG converted per minute per milligram of protein. (B) WT, cnb1Δ, ypk1Δ, and ypk1Δ cnb1Δ cells expressing pRS416 prATG8-GFP-ATG8 were grown at 30 °C to log phase and then transferred to amino acid starvation media for 6 h at 30 °C. Analysis of GFP-Atg8 and quantification of autophagy flux were preformed as described in Fig. 1. (C) WT, cnb1Δ, ypk1Δ, and ypk1cnb1Δ cells expressing endogenously tagged prATG8-2xyEGFP-ATG8 were grown as in B and transferred to amino acid starvation media supplemented with adenine. GFP-Atg8 localization was analyzed at 6 h of starvation using fluorescence microscopy (n > 200 cells for each strain). (Scale bar, 5 μm.) (D) WT, crz1Δ, ypk1Δ, and ypk1Δ crz1Δ and (E) WT, cmd1-6, ypk1Δ, and ypk1Δ cmd1-6 cells expressing pRS416 prATG8-GFP-ATG8 were grown and subjected to amino acid starvation, and autophagy flux was analyzed as in B.
To further test our conclusions, we examined the intracellular localization using fluorescence microscopy of GFP-Atg8 in wild-type, ypk1Δ, and ypk1Δ cnb1Δ cells, where ATG19 was also deleted from each strain to eliminate a selective form of autophagy termed “cytoplasm to vacuole targeting” (33). Following amino acid starvation, wild-type cells exhibited vacuolar localization of GFP-Atg8 as expected, whereas, in contrast, ypk1Δ cells displayed a more uniform cytosolic distribution of GFP-Atg8 (Fig. 3C), consistent with the observed defect in flux detected by Western analysis. Significantly, ypk1Δ cnb1Δ cells exhibited vacuolar localization of GFP-Atg8 similar to that of wild-type cells (Fig. 3C). Taken together, these data are in agreement with our biochemical analysis of autophagy flux. We also observed an accumulation of Atg8 punctate structures in ypk1Δ cells under rich nutrient conditions (Fig. 3C), suggesting the premature formation and/or accumulation of PAS sites within these cells. A lower number of Atg8 punctate structures, similar to wild-type, were observed in ypk1Δ cnb1Δ cells (Fig. 3C). Although the significance of altered numbers of Atg8 punctate structures is not clear, accumulation of PAS sites under growing conditions has been previously observed in a number of autophagy mutants (34), further linking Ypk1 function to the regulation of autophagy.
One function of activated calcineurin is to dephosphorylate and promote the nuclear localization of the zinc-finger transcription factor Crz1, which controls the transcription of numerous stress-responsive genes (35). To determine whether calcineurin functions through Crz1 to regulate autophagy, we examined autophagy flux in ypk1Δ crz1Δ cells following amino acid starvation. In contrast to wild-type levels of flux observed in ypk1Δ cnb1Δ cells, ypk1Δ crz1Δ cells were as defective as ypk1Δ cells for autophagy flux (Fig. 3D). This observation indicates that calcineurin suppresses autophagy by a mechanism that is independent of Crz1-dependent transcription.
Calcineurin is activated in a Ca2+-dependent manner by the conserved calcium sensing protein, Calmodulin (Cmd1) (36, 37). Thus, we tested whether increased calcineurin activity and impaired autophagy in ypk1Δ cells required the Ca2+-dependent activity of Cmd1. Because CMD1 is essential, we used a mutant allele, cmd1-6, which possesses reduced Ca2+ affinity (37), to selectively repress Ca2+-dependent signaling in ypk1Δ cells. Importantly, we observed that, in ypk1Δ cmd1-6 cells, autophagy flux was significantly (P < 0.01) greater than that observed in ypk1Δ cells following amino acid starvation (Fig. 3E). Thus, our data indicate that calmodulin functions as part of calcineurin signaling to negatively regulate autophagy during amino acid starvation.
TORC2/Ypk1 Signaling Is Required for the GAAC Response During Amino Acid Starvation.
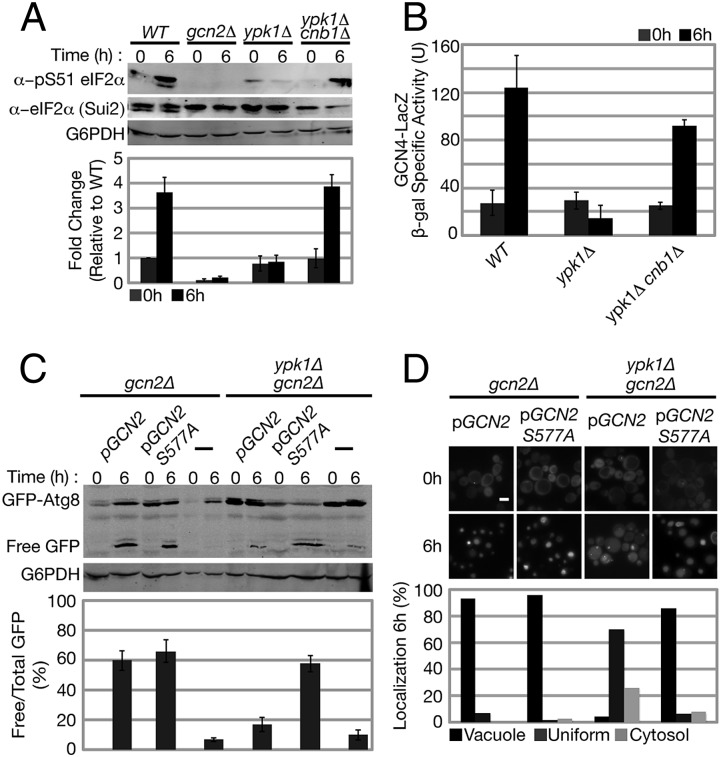
Previous studies have demonstrated that amino acid starvation-induced autophagy requires the GAAC response, as autophagy flux is impaired in gcn2Δ or gcn4Δ cells under amino acid but not nitrogen starvation conditions (18). Given this similar specificity of both TORC2 and Ypk1 mutants to amino acid starvation, we examined whether TORC2/Ypk1 signaling regulates the GAAC response and thus autophagy during amino acid starvation. Specifically, we examined the activity of Gcn2, the sole eIF2α kinase in yeast responsible for activation of the GAAC response (19). Using a phospho-specific antibody, we monitored phosphorylation of eIF2α on Ser-51, which becomes phosphorylated during amino acid starvation in a Gcn2-dependent manner (19, 22). As expected, we observed that wild-type cells exhibited a fourfold increase in eIF2α phosphorylation at Ser-51 during amino acid starvation (Fig. 4A). In contrast, this increase in phosphorylation at Ser-51 was completely blocked in ypk1Δ cells (Fig. 4A). Significantly, we observed that, in ypk1Δ cnb1Δ cells, eIF2α phosphorylation at Ser-51 was similar to wild-type levels during amino acid starvation (Fig. 4A). These results indicate that TORC2/Ypk1-mediated repression of calcineurin activity is essential for induction of Gcn2 activity and autophagy during amino acid starvation.
Fig. 4.
TORC2/Ypk1 are required for the GAAC response during amino acid starvation. (A) WT, gcn2Δ, ypk1Δ, and ypk1Δ cnb1Δ cells were grown at 30 °C to log phase and then transferred to amino acid starvation media for 6 h at 30 °C. Gcn2-dependent phosphorylation of eIF2α at Ser-51 was determined by Western blot using α-phospho eIF2α Ser-51, α-Sui2 (eIF2α), and α-G6PDH antibodies. Quantification represents the ratio of phospho eIF2α Ser-51 and total eIF2α (Sui2) signal with fold change relative to WT at growing conditions (t = 0 h). (B) WT, ypk1Δ, and ypk1Δ cnb1Δ cells expressing the GCN4 derepression lacZ reporter plasmid (p180) were grown and starved as described in A. β-Galactosidase activity was measured (Materials and Methods) and is given in units of nanomoles of ONPG converted per minute per milligram of protein. (C and D) gcn2Δ and ypk1Δ gcn2Δ cells harboring pRS415 prATG8-GFP-ATG8, and, when indicated, plasmids expressing either wild-type GCN2 or GCN2C S577A, were grown and starved as described in A. (C) Analysis of GFP-Atg8 and quantification of autophagy flux were preformed as described in Fig. 1. (D) GFP-Atg8 localization was analyzed using fluorescence microscopy as described in Fig. 3C (n > 200 cells for each strain). (Scale bar, 5 μm.)
Activated Gcn2 indirectly promotes the translation of the transcription factor Gcn4, which targets genes important for both amino acid biosynthesis and autophagy (20–22). To test whether this regulation is relevant for Ypk1-dependent autophagy regulation, we examined GCN4 translational derepression in wild-type, ypk1Δ, and ypk1Δ cnb1Δ cells following amino acid starvation, using a reporter composed of the four upstream regulatory ORFs of GCN4 fused to LacZ (38). We observed that ypk1Δ cells exhibit a severe decrease in LacZ expression relative to wild-type cells, indicating a defect in Gcn4 activation. In contrast, the level of LacZ expression in ypk1Δ cnb1Δ cells was similar to that observed in wild-type cells (Fig. 4B). Thus, taken together with our observations on Gcn2 activity, these results suggest that TORC2/Ypk1 signaling positively regulates the GAAC response via inhibition of calcineurin activity.
To test our conclusion, we asked whether artificial activation of the GAAC response in ypk1Δ cells is sufficient to restore amino acid starvation-induced autophagy. To accomplish this, we used the Gcn2 S577A mutation, which causes an increase in tRNA binding and a partial constitutive activation of Gcn2 kinase activity under growing conditions (39). Accordingly, we introduced plasmids that expressed either wild-type GCN2 (pGCN2) or GCN2C-S577A (pGCN2C-S577A) into gcn2Δ and ypk1Δ gcn2Δ cells and analyzed autophagy flux following amino acid starvation. We observed that flux is comparable between gcn2Δ cells expressing either pGCN2 or pGCN2C-S577A. Importantly, unlike ypk1Δ gcn2Δ cells expressing pGCN2, autophagy flux upon expression of pGCN2C-S577A in ypk1Δ gcn2Δ cells was uniquely comparable to the gcn2Δ strain expressing either wild-type or constitutively active Gcn2 (Fig. 4C). We also observed that the total level of GFP-Atg8 protein is reduced in ypk1Δ gcn2Δ cells expressing pGCN2C-S577A (Fig. 4C). The basis of this latter observation is not known; however, this prompted us to independently assess autophagy in these strains using fluorescence microscopy to examine the localization of GFP-Atg8. Consistently, we observed that GFP-Atg8 possesses a uniform distribution in pGCN2 ypk1Δ gcn2Δ cells, whereas GFP-Atg8 was localized to the vacuole in pGCN2C-S577A ypk1Δ gcn2Δ cells following amino acid starvation (Fig. 4D). We conclude from these data that constitutive activation of the GAAC response is sufficient to restore autophagy flux in ypk1Δ cells during amino acid starvation.
Discussion
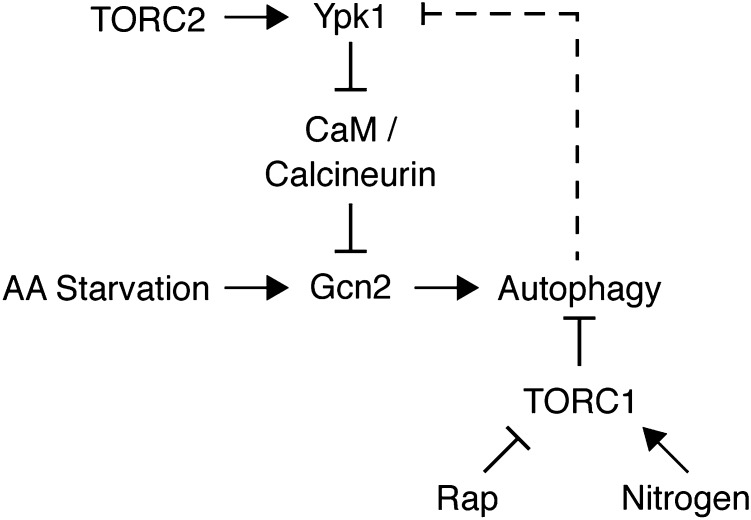
Our study establishes TORC2/Ypk1 signaling as an important positive regulator of amino acid starvation-induced autophagy. In total, our findings support a model wherein TORC2 and Ypk1 function to repress the activity of calcineurin, which we demonstrate is a negative regulator of Gcn2, and promote both the GAAC response and autophagy following amino acid starvation (Fig. 5). As Gcn2 is a well-established sensor of intracellular amino acid levels due to its ability to bind directly and be activated by uncharged tRNA, one possibility is that TORC2 and Ypk1 facilitate activation of Gcn2 under amino acid-limiting conditions. Alternatively, TORC2 and/or Ypk1 may also sense amino acid levels upstream of Gcn2, as has been proposed for mTORC2 in mammalian cells (40).
Fig. 5.
A model for TORC2 regulation of autophagy and the GAAC response. TORC2 and Ypk1 promote autophagy upon amino acid starvation in a pathway distinct from TORC1. Specifically, TORC2 and Ypk1 promote amino acid starvation-induced autophagy by negatively regulating Calmodulin (CaM)/Calcineurin, whose activity inhibits the GAAC response (Gcn2-Gcn4). Dashed line depicts potential autophagy-mediated degradation of Ypk1 protein upon nutrient limitation (see Discussion for details).
Although previous studies have emphasized the importance of rapamycin-sensitive TORC1 as a regulator of autophagy under nitrogen starvation conditions, our findings show crucial mechanistic differences and identify additional components that regulate autophagy in response to amino acid availability. In addition, our discovery that calcineurin acts as a negative regulator of the GAAC response expands the scope of cellular processes regulated by this phosphatase. In this context, a previous global affinity-capture study detected a physical interaction between Gcn2 and Cmp2, one of two isoforms encoding the catalytic subunit calcineurin A, suggesting that calcineurin may directly regulate Gcn2 (41). Interestingly, calcineurin has been shown recently to suppress oxidative stress-induced autophagy in cardiomyocytes (42). Thus, a similar role for calcineurin in negatively regulating autophagy may be evolutionarily conserved.
Induction of autophagy proteins and autophagy flux are independently regulated events, which are important for regulation of the autophagy response (17). Differential regulation of induction vs. flux may facilitate a tunable response for the regulation of both the size and the number of autophagosomes formed (18, 26). Interestingly, TORC2/Ypk1 mutants exhibit opposing defects in autophagy flux and transcriptional regulation of Atg8. Specifically, despite a defect in flux, inhibition of TORC2/Ypk1 signaling leads to increased expression of Atg8. Our findings suggest that TORC2/Ypk1 signaling regulates autophagy flux downstream of Atg8 recruitment to the PAS, as Atg8 puncta accumulate before and persist throughout amino acid starvation. Interestingly, a similar increase in steady-state Atg8 puncta has been observed upon deletion of a subset of ATG genes (Atg1, 2, 13, 17, and 18), further linking Ypk1 as a regulator of autophagy (10, 34).
In addition to the Gcn2-Gcn4 pathway, both PKA and mitochondria have been linked to the regulation of amino acid starvation-induced autophagy (17). Specifically, perturbation of mitochondrial respiration was recently found to increase PKA activity and subsequently suppress autophagy following amino acid but not nitrogen starvation conditions (17). Similar to our observations with TORC2/Ypk1, mitochondria and PKA also regulate autophagy independently of TORC1 (17). Interestingly, impaired TORC2/Ypk1 signaling also results in mitochondrial dysfunction as a consequence of an aberrant increase in PKA activity (43). Thus, the mechanism of regulation between PKA and mitochondria appears to be bidirectional (17, 43, 44). Whether PKA and/or mitochondria are involved in TORC2/Ypk1-mediated regulation of the GAAC response and autophagy is presently unknown and represents another area for future investigation.
Although autophagy is important for maintaining cellular viability during nutrient stress, excessive autophagy can be deleterious to cells and lead to apoptosis or necrosis (45). As such, autophagy paradoxically serves as a mechanism for both the suppression as well as the proliferation and survival of tumor cells (46). Thus, mechanisms are likely to exist to downregulate autophagy following a prolonged period of induction. In this context, a recent report has demonstrated that Ypk1 becomes degraded by an autophagy-dependent process following nutrient limitation (47). Thus, one possibility is that, in the context of a wild-type cell, Ypk1 degradation following amino acid starvation may lead to increased calcineurin activity and the subsequent inhibition of both the GAAC response and autophagy (Fig. 5).
In general, a more complete understanding of the regulation of autophagy in response to various forms of cellular stress will be indispensable for the development of novel and effective cancer therapeutics. In this regard, the Gcn2-Atf4 (Gcn4) pathway is required for the survival and proliferation of tumor cells upon nutrient deprivation (48). Thus, in the context of our findings, an interesting possibility is that mTORC2, which is often upregulated in many cancers (49), may facilitate the activation of Gcn2 in cancer cells, thus triggering prosurvival pathways such as autophagy to allow cancer cells to thrive under low-nutrient conditions. As mTORC2 inhibitors continue to be developed for cancer treatment, specifically targeting the Gcn2 branch of TORC2 regulation could represent a novel avenue for the development of more effective therapies.
Materials and Methods
Yeast Strains, Media, and Plasmids.
All yeast strains used in this study are derivatives of W303α (leu2-3,112; ura3-1; his3-11,15;trp1-1; ade2-1; can1-100) and are listed in Table S1. Gene deletions were generated by PCR-based targeted homologous recombination replacing complete ORFs with kanMX6, HIS3MX6, or TRP1 cassettes as indicated (50). The previously described torc2-ts strain, LHY291 avo3-30 (23), was backcrossed 13 times to W303α to generate PLY1141. All yeast transformations were conducted using lithium acetate (51). All strains were grown to log phase (∼ OD600 = 1) in synthetic complete dextrose (SCD) media [0.8% yeast nitrogen base without amino acids, 2% (wt/vol) dextrose, pH 5.5] supplemented with amino acids as described previously (52). Strains were then exposed to nitrogen starvation media [0.17% yeast nitrogen base without (NH4)2SO4 and amino acids, 2% (wt/vol) dextrose], SCD media containing rapamycin (200 nM), or amino acid starvation media [0.05% yeast extract, 2% (wt/vol) dextrose] previously shown to be specifically limiting in auxotrophic amino acids (17, 18). Genomically modified prATG8-2xyEGFP-ATG8 strains were constructed using pRS306-2xyEGFP-ATG8 integrated into the URA3 locus as previously described (12). All plasmids used in this study are listed in Table S2. pRS415 prATG8-GFP-ATG8 (pPL590) was generated by digesting pRS416 prATG8-GFP-ATG8 (24) with XhoI and XbaI and inserting the prATG8-GFP-ATG8–containing cassette into pRS415 at the XhoI and XbaI sites.
Whole-Cell Extraction, Antibodies, and Western Blot Analysis.
Protein extracts were prepared using the NaOH cell lysis method (53), loaded onto SDS/PAGE gels and transferred to nitrocellulose membrane. Membranes were probed with α-GFP (1 μg/mL; Antibodies Inc.), α-G6PDH (Zwf1) (1:100,000; Sigma), α-phospho eIF2α S51 (1:1,000; Cell Signaling), and α-Sui2 (eIF2α) [1:1,000; a generous gift from Thomas E. Dever (National Institutes of Health, Bethesda)] primary antibodies and visualized using the appropriate secondary antibodies conjugated to IR Dye (1:5,000; LI-COR Biosciences). All Western blot images were quantified using Image Quant software (GE Healthcare). Autophagy flux was calculated as the ratio of free GFP to total GFP signal and is shown as a percentage. In cases where incomplete degradation of the free GFP band was observed, both bands of the subsequent doublet were counted as free GFP and included in the quantification of autophagy flux. Induction of Atg8 protein under growing conditions was determined using both pRS426-prATG8-GFP and pRS416-prATG8-GFP-ATG8 reporter constructs, and GFP or GFP-Atg8 protein bands were normalized to G6PDH (Zwf1). A fold increase in eIF2α S51 phosphorylation was determined by calculating the ratio of phospho eIF2α Ser-51 and total eIF2α (Sui2) signal with fold change relative to WT under growing conditions (t = 0 h). Averages are presented with means ± SD of at least three independent experiments. When indicated, significance was determined using a two-tailed unpaired Student t test to determine P value.
β-Galactosidase Activity Assay.
Exponentially growing cells (∼OD600 = 1) were incubated at 30 °C in SCD media supplemented with amino acids, and, when indicated, transferred to amino acid starvation media for 6 h. β-Galactosidase activity was measured at 30 °C using the substrate O-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma) as described previously (54). β-Galactosidase activity is given in units of nanomoles of ONPG converted per minute per milligram of protein. Averages are presented with means ± SD of three independent experiments.
Fluorescence Microscopy.
Fluorescence microscopy was performed using a Nikon E600 fluorescent microscope and an Orca ER charge-coupled device camera (Hamamatsu) controlled by Micro Manager 1.2 Image J software. Strains were grown to log phase (∼OD600 = 1) in SCD media supplemented with amino acids. Strains were then subjected to amino acid starvation media supplemented with adenine to avoid starvation-induced vacuolar accumulation of red purine precursors characteristic of ade2 mutants, such as W303. Image capture and processing were done using ImageJ and Photoshop (Adobe).
Supplementary Material
Acknowledgments
We thank Dr. Thomas E. Dever, Dr. Martha Cyert, Dr. Alan Hinnebusch, Dr. Ronald Wek, and Dr. Trisha Davis for providing constructs, antibodies, and yeast strains and members of the T.P. and J.N. laboratory for critical discussion and comments. This work was supported by National Institutes of Health (NIH) Grant GM086387 (to T.P.), Grants R01GM062942 and R01GM097432 (to J.N.), and T-32 Training Grant in Molecular and Cellular Biology (to A.V.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406305111/-/DCSupplemental.
References
- 1.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1-2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 3.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280(36):31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 4.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12(9):842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 5.Ryter SW, Cloonan SM, Choi AM. Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013;36(1):7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS ONE. 2011;6(2):e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(19):1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in aging, disease and death: The true identity of a cell death impostor. Cell Death Differ. 2009;16(1):1–2. doi: 10.1038/cdd.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 11.Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: Detection of autophagosomes and their characterization. J Cell Biol. 1994;124(6):903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24(18):2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9(3-4):65–76. [PMC free article] [PubMed] [Google Scholar]
- 14.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106(40):17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powers T, Dilova I, Chen CY, Wedaman K. Yeast TOR signaling: A mechanism for metabolic regulation. Curr Top Microbiol Immunol. 2004;279:39–51. doi: 10.1007/978-3-642-18930-2_3. [DOI] [PubMed] [Google Scholar]
- 16.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 17.Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 2011;30(11):2101–2114. doi: 10.1038/emboj.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ecker N, Mor A, Journo D, Abeliovich H. Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy. 2010;6(7):879–890. doi: 10.4161/auto.6.7.12753. [DOI] [PubMed] [Google Scholar]
- 19.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15(8):4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia MH, et al. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics. 2000;3(2):83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- 21.Natarajan K, et al. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21(13):4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dever TE, et al. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 23.Aronova S, et al. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008;7(2):148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14(2):477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirisako T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147(2):435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151(5):1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamada Y, et al. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25(16):7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci USA. 2012;109(5):1536–1541. doi: 10.1073/pnas.1117563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanida I, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10(5):1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cyert MS, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci USA. 1991;88(16):7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuno T, et al. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991;180(2):1159–1163. doi: 10.1016/s0006-291x(05)81188-x. [DOI] [PubMed] [Google Scholar]
- 32.Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11(24):3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7(6):1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki K, Noda T, Ohsumi Y. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast. 2004;21(12):1057–1065. doi: 10.1002/yea.1152. [DOI] [PubMed] [Google Scholar]
- 35.Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 1999;13(7):798–803. doi: 10.1101/gad.13.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyert MS. Genetic analysis of calmodulin and its targets in Saccharomyces cerevisiae. Annu Rev Genet. 2001;35:647–672. doi: 10.1146/annurev.genet.35.102401.091302. [DOI] [PubMed] [Google Scholar]
- 37.Geiser JR, van Tuinen D, Brockerhoff SE, Neff MM, Davis TN. Can calmodulin function without binding calcium? Cell. 1991;65(6):949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- 38.Hinnebusch AG. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Barrio M, et al. Serine 577 is phosphorylated and negatively affects the tRNA binding and eIF2alpha kinase activities of GCN2. J Biol Chem. 2002;277(34):30675–30683. doi: 10.1074/jbc.M203187200. [DOI] [PubMed] [Google Scholar]
- 40.Tato I, Bartrons R, Ventura F, Rosa JL. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J Biol Chem. 2011;286(8):6128–6142. doi: 10.1074/jbc.M110.166991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 42.He H, et al. Calcineurin suppresses AMPK-dependent cytoprotective autophagy in cardiomyocytes under oxidative stress. Cell Death Dis. 2014;5:e997. doi: 10.1038/cddis.2013.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niles BJ, Joslin AC, Fresques T, Powers T. TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Reports. 2014;6(3):541–552. doi: 10.1016/j.celrep.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aun A, Tamm T, Sedman J. Dysfunctional mitochondria modulate cAMP-PKA signaling and filamentous and invasive growth of Saccharomyces cerevisiae. Genetics. 2013;193(2):467–481. doi: 10.1534/genetics.112.147389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 46.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: Therapeutic implications. Mol Cancer Ther. 2011;10(9):1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimobayashi M, Takematsu H, Eiho K, Yamane Y, Kozutsumi Y. Identification of Ypk1 as a novel selective substrate for nitrogen starvation-triggered proteolysis requiring autophagy system and endosomal sorting complex required for transport (ESCRT) machinery components. J Biol Chem. 2010;285(47):36984–36994. doi: 10.1074/jbc.M110.119180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye J, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29(12):2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masri J, et al. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res. 2007;67(24):11712–11720. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- 50.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 51.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 52.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 53.Dilova I, Aronova S, Chen JC, Powers T. Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1.Rtg3p-dependent target genes. J Biol Chem. 2004;279(45):46527–46535. doi: 10.1074/jbc.M409012200. [DOI] [PubMed] [Google Scholar]
- 54.Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.