An article in PNAS by Hussain and Asgari (1) suggests that a viral small RNA (vsRNA) from the 3′ UTR of the Dengue virus (DENV) genome is important for the autoregulation of the viral replication in mosquito cells by acting as a microRNA (miRNA) targeting the NS1 region of the genome. The authors performed a sequence conservation analysis on the vsRNA-5 across the four serotypes. Unfortunately, the analysis was carried out using only one sequence from each serotype, which in the case of a genome segment from the variable region of the 3′ UTR may misrepresent the total virus diversity and, thus, its interpretation may be misleading.
To better examine the vsRNA-5 conservation, the sequences provided in the article by Hussain and Asgari (1) were first mapped to their corresponding complete DENV genome, and all of the publicly available complete DENV genome sequences (n = 3,372) were aligned and sequence logos were generated from the vsRNA-5 and the NS1 target region. The mapping revealed that the DENV-1 to -3–vsRNA sequences localized to the stem loop (SL) 2 of the 3′ UTR downstream of the stop codon. However, in contrast, the DENV-4–vsRNA-5 sequence mapped to a region 1,500 nt upstream of the stop codon, where no secondary structure has been described. Moreover, it has been shown that the homologous region is deleted in the DENV-4 3′ UTR (2). Therefore, this serotype should not have been used in the conservation analysis.
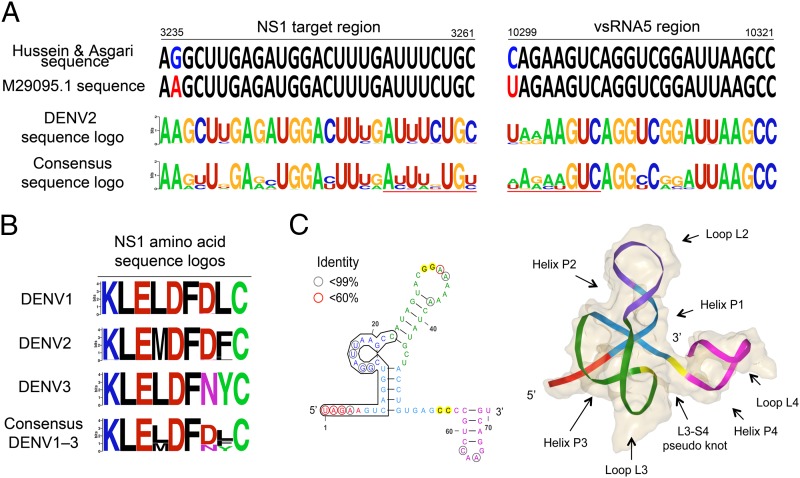
Interestingly, the mapping of the DENV-2–vsRNA-5 and NS1 sequences also revealed two substitutions in the strain used by Hussain and Asgari (1), 10299U > C and 3236A > G, respectively (Fig. 1A); the latter leads to an amino acid substitution K1047R in the NS1 protein. The DENV-2 NS1 and vsRNA-5 nucleotide sequence logos also revealed that these substitutions were found only in 2.4% and 9.3% of all published DENV-2 genomes, respectively.
Fig. 1.
Sequence conservation analysis on the regions involved in the suggested miRNA mechanism. (A) The substitutions 3236A > G and 10299U > C in the DENV-2 New Guinea C strain used by Hussain and Asgari (1) are highlighted in colors. Nucleotide sequence logos generated from the NS1 and vsRNA5 regions are also shown. The seed region involved in the miRNA binding is underscored in red. (B) Amino acid sequence logos for the NS1 target region show the conservation within and across serotypes. (C) Secondary and tertiary structure of the Xrn1-resistant structure in the SL2 and SL3 of DENV-2. Secondary structure suggested by Chapman et al. (3) based on chemical probing and conservation. The vsRNA5 sequence is highlighted in black. Positions with an identity lower than 99% across the 1,056 DENV-2 sequences are circled. The 3D RNA model was built based on the crystallography of the homologous xrRNA structure in the Murray Valley Encephalitis virus (Protein Data Bank ID: 4PQV) using ModeRNA, a software for comparative 3D RNA modeling (5).The RNA backbone is shown in different colors according to the secondary structure.
Furthermore, contrary to what we might have predicted if vsRNA-5 works as an miRNA, the NS1 and vsRNA-5 nucleotide sequence logos (Fig. 1A) show that the positions involved in the seed-region binding exhibited poor conservation across the three serotypes. The presence of highly conserved positions is more likely explained by the NS1 amino acid preservation within each serotype and the requirement of these nucleotides in the 3′ UTR for the formation of the recently described Xrn1-resistant RNA structure (3) (Fig. 1 B and C).
To further assess the conservation in the complementarity between the vsRNA-5 and its target sequence within each serotype, the minimal free energy for the binding in the 3,372 sequences was calculated using the ViennaRNA package (4). The average for the binding in DENV-2 (average ± SD: −20.64 ± 3.38 kcal/mol) was found to be significantly lower (P < 0.001) than the average minimal free energy in DENV-1 (−17.04 ± 1.43 kcal/mol) and DENV-3 (−12.01 ± 0.67 kcal/mol), suggesting a lack of conservation in the complementarity of the base pairing in these two serotypes, perhaps limiting the significance of this mechanism to DENV-2.
Taken collectively, these data—and the fact that the production of vsRNA has not been experimentally shown in other serotypes—suggest that the relevance of this mechanism might be limited to DENV-2.
Supplementary Material
Acknowledgments
I thank Mariano Garcia-Blanco and Eng Eong Ooi for critically reading this letter.
Footnotes
The author declares no conflict of interest.
References
- 1.Hussain M, Asgari S. MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc Natl Acad Sci USA. 2014;111(7):2746–2751. doi: 10.1073/pnas.1320123111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shurtleff AC, et al. Genetic variation in the 3′ non-coding region of Dengue viruses. Virology. 2001;281(1):75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife. 2014;3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: A tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39(10):4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]