Significance
CG cytosine methylation (mCG) is an important epigenetic marker present in most eukaryotic genomes that is maintained by an evolutionarily conserved DNA methyltransferase dubbed DNMT1 in mammals and MET1 in plants. Null mutation of DNMT1 or MET1 results in global loss of mCG and leads to embryonic death in mouse, inviability in human cancer cells, and wide-ranging developmental abnormality in Arabidopsis thaliana. This study characterizes global effects of null mutation of a MET1 gene in rice, a model plant for monocotyledons, through methylome, transcriptome, and small RNAome analyses. The findings of this study have implications for improving our understanding of the biological roles of cytosine methylation in monocots and, from an applied point of view, in epigenetic manipulation of cereal crops.
Keywords: Oryza sativa L., monocotyledons
Abstract
Cytosine methylation at CG sites (mCG) plays critical roles in development, epigenetic inheritance, and genome stability in mammals and plants. In the dicot model plant Arabidopsis thaliana, methyltransferase 1 (MET1), a principal CG methylase, functions to maintain mCG during DNA replication, with its null mutation resulting in global hypomethylation and pleiotropic developmental defects. Null mutation of a critical CG methylase has not been characterized at a whole-genome level in other higher eukaryotes, leaving the generality of the Arabidopsis findings largely speculative. Rice is a model plant of monocots, to which many of our important crops belong. Here we have characterized a null mutant of OsMet1-2, the major CG methylase in rice. We found that seeds homozygous for OsMet1-2 gene mutation (OsMET1-2−/−), which directly segregated from normal heterozygote plants (OsMET1-2+/−), were seriously maldeveloped, and all germinated seedlings underwent swift necrotic death. Compared with wild type, genome-wide loss of mCG occurred in the mutant methylome, which was accompanied by a plethora of quantitative molecular phenotypes including dysregulated expression of diverse protein-coding genes, activation and repression of transposable elements, and altered small RNA profiles. Our results have revealed conservation but also distinct functional differences in CG methylases between rice and Arabidopsis.
Cytosine methylation is an evolutionarily conserved epigenetic modification across biological kingdoms (1–3). However, fundamental differences exist for this epigenetic mark between animals and plants in many aspects (1–3). For example, in mammalian somatic cells, methylated cytosines (mCs) occur almost exclusively in a CG sequence context, and CG methylation (mCG) is maintained during DNA replication by a single CG methylase, DNA methyltransferase 1 (DNMT1). Cytosine methylation in plants is more complex, occurs in all sequence contexts (CG, CHG, and CHH; where H is A, T, or C), and is established and maintained by multiple enzymes that have distinct but also overlapping functions (2, 4–6). Nevertheless, mCG stands out as the most pervasive type (2). In the dicot model plant Arabidopsis thaliana, mCG is maintained by a major CG methylase, namely DNA methyltransferase 1 (MET1), the homolog of mammalian DNMT1 (2, 7–11). Null mutation of MET1 (the homozygous met1 mutant) virtually eliminates genome-wide mCG to 1.7% that of wild type (WT), that is, from 24.6% in WT to 0.42% in mutant (12, 13). Homozygous null met1 mutant can be obtained from selfed progenies of heterozygotes (MET1+/−) but was often at much lower frequencies than expected by Mendelian segregation (11). Homozygous Arabidopsis met1 plants show pleiotropic developmental abnormalities of variable penetrance, which aggravate in frequency and severity with inbreeding (11). Although conditional knockout DNMT1 mutants were established in human colorectal carcinoma cell lines (14) and loss-of-function DNA methyltransferase mutant (defective in DNA methylation 2) was generated in Neurospora (15), hitherto Arabidopsis met1 is the only null mutant of a major CG methylase that has been characterized at the genome-wide scale in higher eukaryotes.
Rice (Oryza sativa L.) is a staple grain for nearly one-half of the world’s population and a model plant for monocotyledons. Previous studies have identified two closely related putative MET1 genes in rice, OsMET1-1 and OsMET1-2 (16). Although the two genes are highly similar in sequence and structure and composed of all binding and catalytic domains required for a functional CG methylase, the transcripts of OsMET1-2 accumulated more abundantly than those of OsMET1-1 in all examined WT rice tissues, suggesting a major role of OsMET1-2 in maintaining mCG (17, 18). Moreover, a knockin mutant of OsMET1-1 failed to produce discernible developmental phenotypes (17), implicating a minimal and/or redundant function for it in maintaining mCG. In contrast, both knockin and retrotransposon (Tos17) insertion mutants of rice OsMET1-2 gene showed severe developmental defects in kernels, and exhibited no germination to only viviparous sprouting (18). In an independent effort, we were able to obtain and germinate seeds of the homozygous insertion mutant of OsMET1-2 into seedlings. Taking advantage of the situation, we studied the methylome, transcriptome, and small RNAome of the mutant and its WT. Major observations of this analysis include: (i) Homozygous mutant (OsMET1-2−/−) seeds could be produced by selfing heterozygotes (OsMET1-2+/−) at a frequency expected by Mendelian segregation; (ii) the homozygous mutant seedlings exhibited growth retardation that culminated in necrotic death; and (iii) genome-wide loss of mCG occurred in the mutant seedlings, which was accompanied by dysregulated expression of both protein-coding genes and transposable elements (TEs) as well as altered profiles of 21-nt and 24-nt small (sm)RNAs. Our results highlight the functional similarities and differences between the CG methylases from Arabidopsis and rice, which have implications for our further understanding of evolution and the function of cytosine DNA methylation (3) as well as manipulation of cereal epigenomes for crop improvement (19–21).
Results and Discussion
Null Function of MET1-2 Impairs Seed Development and Vegetative Growth and Causes Swift Seedling Lethality in Rice.
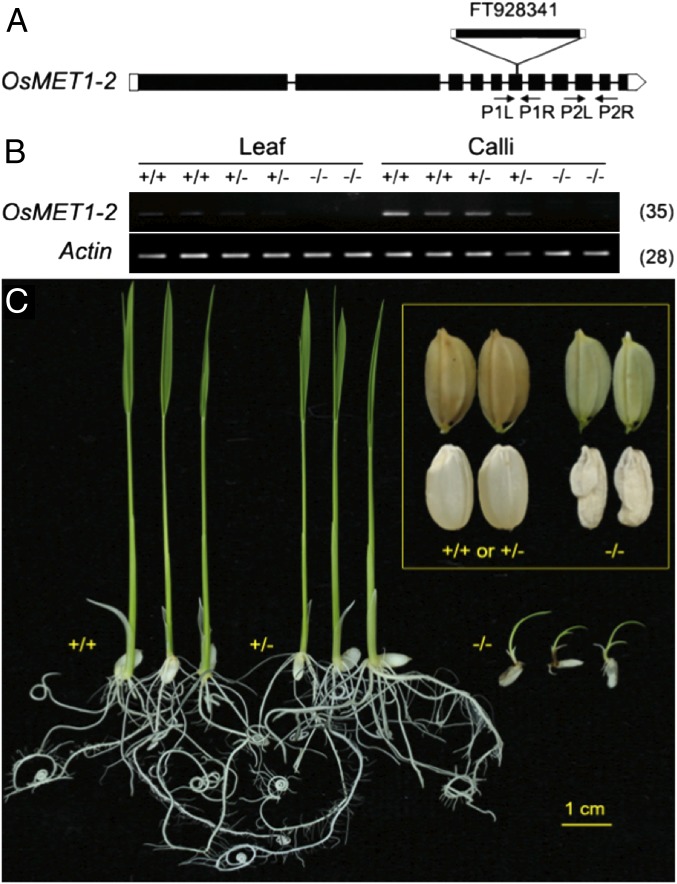
We isolated homozygous OsMET1-2−/−, a loss-of-function mutant of the standard laboratory cultivar Nipponbare of rice due to insertion of a copy of retrotransposon Tos17. We verified that Tos17 was inserted into the sixth exon (at position Chr07:4422236 nt) of OsMET1-2 (LOC_Os07g08500) (Fig. 1A) by both locus-specific PCR amplification, Sanger sequencing and Illumina whole-genome resequencing. Because the sixth exon of OsMET1-2 encodes motif IV, which contains the invariant prolylcysteinyl doublet that constitutes the functional active site of all known C5-MTases (16), this insertion of Tos17 (4.3 kb) was expected to fully knock out its function. Furthermore, the steady-state transcripts of OsMET1-2 were undetectable in both shoot and callus of the homozygous mutant (the only obtainable tissues) but were readily detected in the callus of its immediately segregated WT and heterozygote (OsMET1-2+/−) siblings (Fig. 1B). Together, these analyses confirmed the complete null nature of OsMET1-2 mutation (rather than hypomorphic) in OsMET1-2−/−. The heterozygote had no discernibly altered phenotypes relative to WT based on our examination of a large number of individual plants under both controlled conditions and field trials, suggesting gametophytes with an OsMET1-2−/− genotype had no strong phenotypic effect. This differed from findings in Arabidopsis, in which heterozygotes of MET1+/− resulted in immense epigenetic diversification of gametes, and produced phenotypic segregation among plants derived from the same heterozygotic plants (10). We suspect the lack of transgenerational phenotypic effects in the OsMET1-2+/− heterozygote plants is due to functional complementation by the other OsMET1 gene (i.e., OsMET1-1) during gametogenesis. The self-pollinated heterozygotes reproducibly produced abnormally developed seeds with severe shrinking in both embryo and endosperm (Fig. 1C), indicating the seed mutant genotype had immediate phenotypic effects. The abnormal seeds were produced from selfed heterozygotes at a frequency of ca. 22% based on 10,155 seeds of >300 panicles scored, suggesting a single loss-of-function mutation was responsible for this phenotype, which segregated close to Mendelian expectation. The OsMET1-2−/− seeds could not germinate under conventional conditions, but germinated well on half-strength, hormone-free Murashige & Skoog medium. Nevertheless, growth of the seedlings was significantly retarded, and they could only grow to ca. 3 cm in height with sporadic, deformed tiny roots, and then growth of the seedlings was totally halted and underwent necrotic death within 2 wk (Fig. 1C), consistent with their extremely abnormal microscopic structures (SI Appendix, Fig. S1). Interestingly, the callus could be induced from the abnormal seeds, suggesting mitosis in the mutant was not severely affected. This is different from observations in human colorectal cancer cells, in which complete inactivation of the CG methylase (DNMT1) led to mitotic catastrophe and growth arrest (14). However, we could not rule out the possibility that backup methylation was rapidly gained in the callus, for example via similar mechanisms as in Arabidopsis met1 (11), which warrants further investigation. Locus-specific pyrosequencing confirmed that all seedlings germinated from the abnormal seeds were homozygous for the Tos17 insertion at the OsMET1-2 gene, that is, of genotype OsMET1-2−/− (SI Appendix, Fig. S2). This seedling-terminal phenotype of a loss of function of OsMET1-2 contrasted starkly with Arabidopsis met1, which, although produced from segregated heterozygotes at significantly lower than expected frequencies and the surviving plants have severe pleiotropic developmental defects, is are capable of completing the whole cycle of ontogenesis including reproduction (9–11).
Fig. 1.
Characterization of a rice MET1-2 mutant (OsMET1-2−/−). (A) Position of retrotransposon Tos17 insertion into the sixth exon of MET1-2 (not drawn to scale). P1L and P1R are primers used for identification of the three MET1-2 genotypes, WT (+/+), heterozygote (+/−), and homozygote (−/−), by pyrosequencing (SI Appendix, Fig. S2), and P2L and P2R are primers used for qRT-PCR. (B) qRT-PCR analysis of the expression of OsMET1-2 (OsMET1-2 is known as unexpressed) (19, 20) in the 11-d-old shoot and calli tissue of the three genotypes of OsMET1-2: WT, heterozygote, and homozygote. (C) Seedling (11-d-old) and kernel/seed phenotypes of the three MET1-2 genotypes immediately segregated from the same heterozygote plants.
Global Cytosine Hypomethylation in OsMET1-2−/−.
To test whether cytosine methylation was impacted in OsMET1-2−/−, we compared sensitivity of its genomic DNA vs. that of WT/heterozygote to methylation-sensitive endonuclease digestions, followed by locus-specific assay of methylation status by bisulfite sequencing. We found that compared with WT and heterozygote (between which no difference was seen), OsMET1-2−/− was hypersensitive to digestion by HapII (SI Appendix, Fig. S3A), implicating global loss of CG methylation at the 5′ CCGG sites in the mutant. This was confirmed by bisulfite sequencing of a body region of Tos17 (SI Appendix, Fig. S3B), an established locus in the rice genome for assaying cytosine methylation liability (22). These results prompted us to conduct a genome-wide investigation of cytosine methylation of the mutant. We generated single-nucleotide–resolution maps of cytosine methylation (methylome) for the shoot tissue of 11-d-old seedlings for the null mutant of MET1-2 (OsMET1-2−/−), its immediately segregated sibling heterozygote (OsMET1-2+/−), and WT (OsMET1-2+/+) by whole-genome bisulfite sequencing (SI Appendix, Materials and Methods).
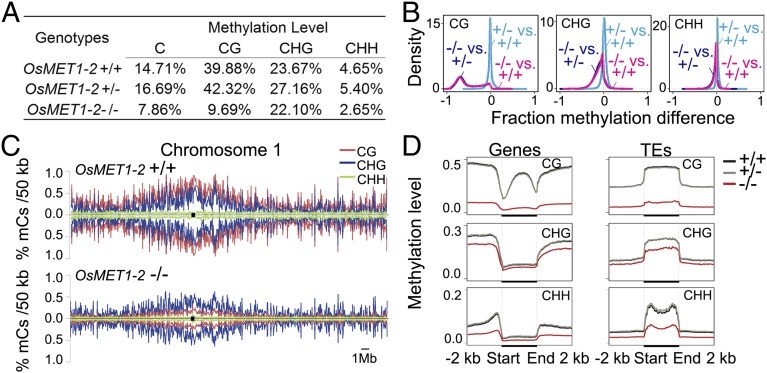
We found that the global mCG level in WT and heterozygote was similar (Fig. 2A), and both are in good agreement with previously published rice methylome data in the same genotype (cv. Nipponbare) of similar tissues (1, 19). In contrast, the genome-wide mCG level in OsMET1-2−/− was reduced by 76% compared with WT (Fig. 2A). We noted, however, that this reduction of mCG in the rice mutant was of a lesser magnitude than that of Arabidopsis met1 vs. its WT, in which mCG was reduced by 98.3% (SI Appendix, Table S1) (5, 12, 13). An intuitive reason for this difference between OsMET1-2−/− and Arabidopsis met1 and their respective WTs might be that, as mentioned above, Arabidopsis expresses mainly a single CG methylase (MET1) whereas rice expresses two, OsMET1-1 and OsMET1-2 (19). Although OsMET1-2 has been suggested to be the major CG methylase in rice (17, 18), it is unclear whether and to what extent OsMET1-1 is functionally redundant with OsMET1-2, especially when the latter is null-mutated. To test this possibility, we quantified expression of OsMET1-1 in the seedling shoot tissue in OsMET1-2−/− and WT by quantitative (q)RT-PCR. Indeed, we found a moderate but significant up-regulation of OsMET1-1 by ca. 2.5-fold in the mutant relative to WT; in addition, significant up-regulation (by ca. 4.5-fold) of one of the variation in DNA methylation (VIM) genes, LOC_Os05g01230, was also observed in the mutant (SI Appendix, Fig. S4). VIMs are known to function cooperatively with CG methylase to maintain CG methylation in mammals and likely have a conserved function in plants (2). Notably, the concomitant up-regulation of both OsMET1-1 and the VIM genes was also found in our RNA sequencing (RNA-seq) profiling data (described in subsequent sections). Thus, one explanation for the loss of mCG to a markedly less magnitude in OsMET1-2−/− than in Arabidopsis met1 relative to their respective WTs was likely the enhanced expression of the alternative CG methylase together with the VIM gene(s) in the rice mutant, which maintained the rest of the mCGs.
Fig. 2.
Genome-wide hypomethylation in OsMET1-2−/−. (A) Percentages (levels) of mCs of total Cs, CGs, CHGs, and CHHs in the three OsMET1-2 genotypes (+/+, +/−, and −/−) immediately segregated from the same heterozygote (+/−) plants. (B) Pairwise comparisons among the three OsMET1-2 genotypes, depicted as kernel density plots with trace colors deep pink (−/− vs. +/+), dark blue (−/− vs. +/−), and light blue (+/− vs. +/+). Only mC differences for 1-kb windows containing at least five informative sequenced cytosines were considered. (C) Distribution of mCs in WT and OsMET1-2−/− in chromosome 1 (for the rest of the chromosomes, refer to SI Appendix, Fig. S5). (D) Genome-wide average levels of mC in each sequence context (CG, CHG, and CHH) of genes and TEs, each with their contiguous upstream and downstream 2-kb flanks in the three MET1-2 genotypes.
Given that mCs of both CHG and CHH were also affected in Arabidopsis met1 (1, 13, 23), we compared mCs of these two sequence contexts in OsMET1-2−/− and WT. We found that, relative to WT, mCs of CHH were reduced by 43% whereas those of CHG were reduced by only 6.6% in OsMET1-2−/− (Fig. 2A). Although the extent of loss of mCHH was broadly similar to Arabidopsis met1, in which mCHH was decreased by 34.7%, mCHG was reduced to a much lesser extent in OsMET1-2−/− than in Arabidopsis met1, in which 37.97% was lost (13).
The genome- and chromosome-scale distributions of mCs are depicted by kernel density plots showing genome-wide pairwise comparison of mCs of all three sequence contexts (CG, CHG, and CHH) of the three genotypes (WT, heterozygote, and mutant) and by the chromosomal distribution of mCs of all three contexts in the mutant and WT, respectively. Compared with WT, conspicuously reduced mCG level was evident both at the whole-genome scale and across the entire length of each of the 12 rice chromosomes, whereas the heterozygote and WT were similar at both scales of comparison (Fig. 2 B and C and SI Appendix, Fig. S5). The general distribution trend of mCs in mutant vs. WT at both genome and chromosomal scales was in broad agreement with those between met1 and WT in Arabidopsis (1, 13, 23), although with less magnitude of mC loss in the rice mutant vs. WT.
Next, we separately examined mC levels of protein-coding genes and TEs in the three genotypes. We found that mCG of gene bodies was most severely affected and reduced by 86% in OsMET1-2−/− (from 27.35% in WT to 3.95% in OsMET1-2−/−), whereas mCG of TEs was reduced by 77% in OsMET1-2−/− (Fig. 2D and SI Appendix, Fig. S6 and Table S3). This is different from the findings in Arabidopsis, in which mCG of both genes and TEs is equally eliminated (5). One explanation for this phenomenon could be functional redundancy between the two OsMET1 genes in rice, with OsMET1-1 being capable of maintaining some “regained” mCG (see below) by other pathways (e.g., RNA-directed DNA methylation; RdDM) in OsMET1-2−/−, which preferentially targeted to heterochromatic regions mainly composed of TEs and other repeats (24).
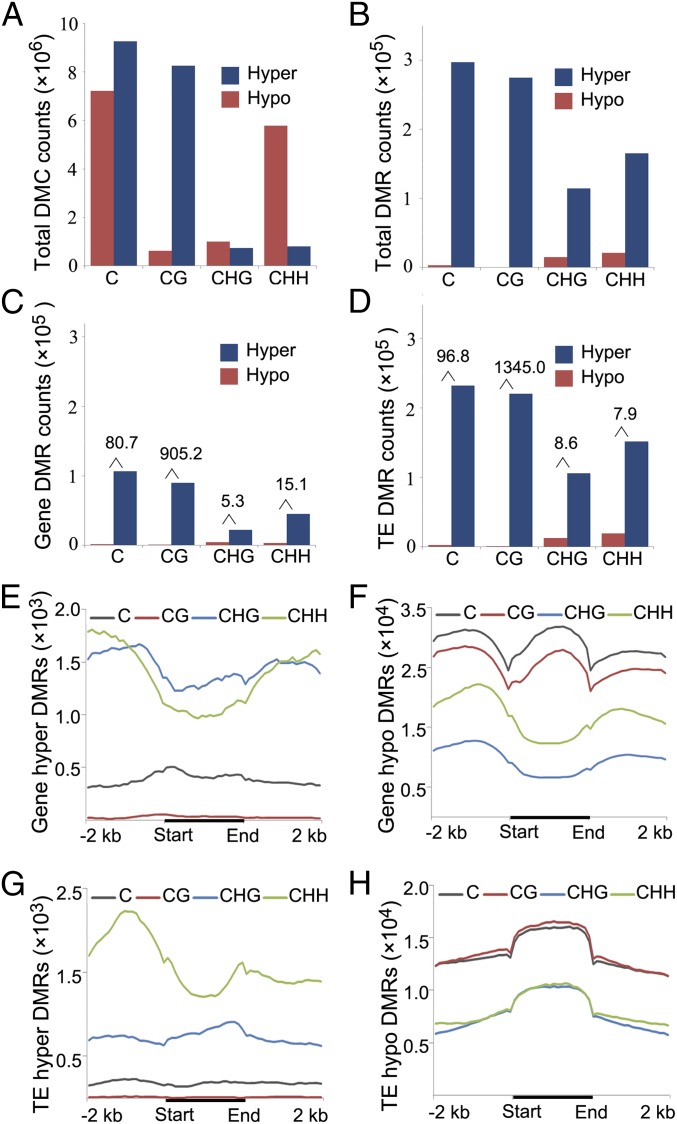
It was found in Arabidopsis met1 that massive loss of mCG was accompanied by ectopic gain of mCHG in gene bodies (13), and one mechanism leading to this outcome is due to transcriptional repression of the H3K9 demethylase IBM1 (Increase in BONSAI Methylation 1), which resulted in accumulation of H3K9me2 and hence recruitment of mCHG (25). To investigate whether regain of mC also occurred in OsMET1-2−/−, we compared genome-wide differentially methylated cytosines (DMCs) and differentially methylated regions (DMRs) between OsMET1-2−/− and WT. We found that compared with WT, a larger number of hypo-DMCs could be identified, 89% of which were of the CG context, as expected (Fig. 3A and SI Appendix, Table S4). Notably, hyper-DMCs were also found in each sequence context (but predominantly CHH) in the mutant (Fig. 3A and SI Appendix, Table S4). Nevertheless, because hyper-DMCs occurred sporadically and mainly landed in intrinsically mC-poor genomic regions, they did not alter the overall lower mC level of most regions in the mutant relative to WT. Observationally, this explains why the vast majority of DMRs are hypo-DMRs, with hyper-DMRs being rare in all sequence contexts in both genes and TEs along their entire length (Fig. 3 B–H and SI Appendix, Table S4). An interesting observation is that although discernibly more CHG hyper-DMRs mapped to genes and their flanks than to TEs, no enrichment to gene bodies was found (Figs. 2D and 3 E and G and SI Appendix, Fig. S7). In light of this, we analyzed the methylation status and expression of five JmjC domain-containing genes in OsMET1-2−/− and WT, which bear high homology to Arabidopsis IBM1 (26). We found that despite heavy loss of mCG along the entire length of these genes, their expression was not repressed like IBM1 in Arabidopsis met1, consistent with lack of aggregated CHG hyper-DMRs to gene bodies in the rice mutant (SI Appendix, Fig. S8) (25). Alternatively, the still-remaining mCs in OsMET1-2−/− (in contrast to Arabidopsis met1) may have been sufficient to tether heterochromatic binding proteins such that little mC regain is entailed.
Fig. 3.
Statistics of differentially methylated cytosines and differentially methylated regions between OsMET1-2−/− and WT (+/+). (A and B) Hyper- or hypomethylation DMCs and DMRs that occurred in all three cytosine contexts (for DMRs, a window size of 1 kb was adopted). (C and D) Enrichment of hyper- or hypomethylation DMRs in genes and TEs, respectively. The numbers above the columns represent the value of hypo-DMRs vs. hyper-DMRs. (E–H) Distribution of hyper- or hypomethylation DMRs along the body region, and their up- or downstream 2-kb flanks in protein-coding genes and TEs.
Transcriptional Dysregulation of Diverse Protein-Coding Genes in OsMET1-2−/−.
To investigate impacts of genome-wide loss of mCs on global expression of protein-coding genes (henceforth referring to genes), we conducted deep RNA-seq analysis on OsMET1-2−/− relative to its sibling WT and heterozygote (OsMET1-2+/−), each with two biological replicates, in the same tissue as used for the methylome. There were 1.98% (553 out of 27,993) of genes (based on rice gene annotation MSU7.0) that showed differential expression by Cuffdiff version 2.0.2 (27) between the heterozygote and WT (SI Appendix, Table S5). In contrast, expression of 13.37% (3,744 of 27,993) of genes was altered in OsMET1-2−/− compared with WT (SI Appendix, Table S5 and Datasets S1 and S2). This proportion (13.37%) of expression-altered genes in OsMET1-2−/− vs. WT far exceeded that in Arabidopsis met1 vs. WT, which was 2.05% (13). Together with the smaller reduction of methylation in OsMET1-2−/− vs. WT compared with Arabidopsis met1 vs. WT, it suggested that a higher proportion of genes is directly or indirectly associated with cytosine methylation for regulation in rice than in Arabidopsis, consistent with the more severe phenotypic effect in OsMET1-2−/− (Fig. 1C) than in Arabidopsis met1 (7, 9, 11).
Of the 3,744 affected genes, 47.5% (1,780) and 52.5% (1,964) showed significant up- and down-regulation in OsMET1-2−/− vs. WT, respectively (SI Appendix, Table S5 and Datasets S1 and S2). Gene ontology (GO) analysis showed that 1,247 up-regulated genes and 1,625 down-regulated genes could be assigned to GO categories. We found that the up-regulated genes were enriched for diverse GO terms in biological process (SI Appendix, Fig. S9). In contrast, the down-regulated genes showed fewer enriched GO terms (SI Appendix, Fig. S9). Notably, significant up-regulation of several DNA methylation/chromatin-related genes including MET1-1, VIM, CMT3, two DDM1 genes (OsDDM1a and OsDDM1b), and four DNA glycosylase genes was detected, which accorded well with the qRT-PCR analysis of these genes (SI Appendix, Fig. S4). These observations, together with the findings in Arabidopsis (11), suggested that rewired expression of multiple genes directly or indirectly related to cytosine methylation maintenance might reflect a conserved adaptive cellular response to massive loss of mCGs; expectedly, given sufficient time, this may result in tinkering of impaired intrinsic chromatin states, although the process may be stochastic and hence imprecise, as documented in Arabidopsis (11).
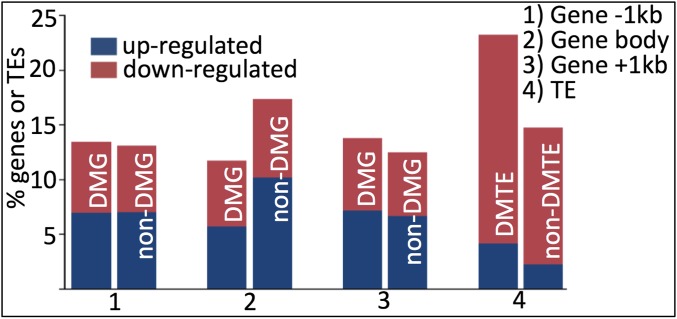
We next tested possible relationships between distribution of DMRs and expression alteration of the methylation-affected genes in OsMET1-2−/−. We categorized the affected genes into two groups, namely differentially methylated genes (DMGs) and non-DMGs (SI Appendix). We found that (i) with respect to 5′ or 3′ flanks, DMGs and non-DMGs were more or less equally represented in the expression-altered genes and (ii) with respect to body regions, non-DMGs were overrepresented in the expression-altered genes (Fig. 4). Together, these results suggest that the role of cytosine methylation in gene expression was primarily indirect in rice, conceivably through orchestrating activity of trans-acting transcription factors and chromatin modifiers whereby expression of their downstream genes can be altered irrespective of the methylation states of the target genes per se.
Fig. 4.
Proportion of differentially expressed protein-coding genes and transcribed TEs between WT (+/+) and OsMET1-2 (−/−) mutant that were DMGs or non-DMGs. For protein-coding genes, the body regions and their contiguous upstream and downstream 1-kb flanks were analyzed.
Both Activation and Repression of TEs in OsMET1-2−/−.
In total, 594 of the 2,716 (21.9%) transcribed TEs (henceforth called TEs) showed statistically significant differential expression between OsMET1-2−/− and WT based on the same RNA-seq data and statistical cutoff criterion as for protein-coding genes. Of these expression-altered TEs, 489 (82.3%) showed up-regulation and the remaining 105 (17.7%) were down-regulated (SI Appendix, Table S5). It should be noted that we may have detected many more TEs that showed expression changes in OsMET1-2−/− relative to WT based on our RNA-seq data; however, the changes were of low magnitude and did not reach the defined statistical threshold (SI Appendix, Fig. S10), and therefore were not taken into account further. In general, the collective expression level of all annotated TEs together is higher in the mutant than in the WT (SI Appendix, Fig. S10), clearly indicating a pivotal role of OsMET1-2 in repressing genome-wide transcriptional activity of TEs in WT rice. Next, we analyzed the correlation between TE expression and the methylation states for those TEs that showed significant up- or down-regulation in the mutant (SI Appendix, Fig. S10 and Table S5). We found that the mutant vs. WT differentially methylated TEs (DMTEs) were significantly greater in number than the non-DMTEs (Fig. 4). We should caution, however, that more complex relationships between mC and TE expression activity may exist in rice than in Arabidopsis due to differences in proportion and/or structural complexity of TEs in their respective genomes (28, 29).
Altered Small RNA Profiles in OsMET1-2+/− and Their Correlation with DMRs.
Given the intrinsic relationships between mC and smRNAs via the plant-specific RdDM pathway (30–33), we conducted small RNA-seq (smRNAome) of OsMET1-2−/− and its sibling WT/heterozygote using the same tissue as for the methylome and RNA-seq. We observed highly similar smRNA profiles of all size groups between heterozygote and WT. In contrast, the steady-state abundance of several smRNA size groups (e.g., 21-, 22-, 24-, and 25-nt) showed marked differences between OsMET1-2−/− and WT (SI Appendix, Fig. S11A). In particular, the 21- and 24-nt size groups showed overall over- and underrepresentation in the mutant vs. WT, respectively, which mirrored Arabidopsis met1 (13). We further compared abundance of the 21- and 24-nt smRNA groups in each of the 12 chromosomes of OsMET1-2−/− and WT. We found that abundance for both size groups of smRNAs was significantly altered in OsMET1-2−/− vs. WT for each of the 12 chromosomes (proportion test, P < 0.05), albeit to highly variable extents (SI Appendix, Fig. S11B). In particular, 21-nt smRNAs were significantly underrepresented in chromosome 4 but overrepresented in chromosome 9 in the mutant vs. WT, respectively (SI Appendix, Fig. S11B). In addition, although the 24-nt smRNAs showed overall down-regulation in the mutant (SI Appendix, Fig. S11A), they showed significant up-regulation in four chromosomes (8, 9, 11, and 12) (SI Appendix, Fig. S11B).
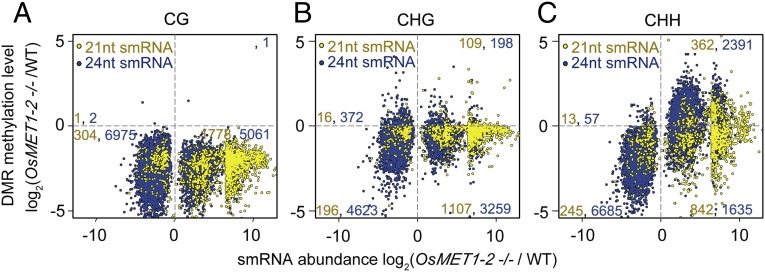
To explore possible relationships between regional abundance of 21- or 24-nt smRNAs and DMRs in each sequence context (CG, CHG, or CHH), we binned the abundance of 21- and 24-nt smRNAs into 1-kb sliding windows across the genome (as for DMRs) and compared for differences between OsMET1-2−/− and WT, which we defined as differential smRNA regions (DSRs). In total, 2,385 21-nt DSRs and 12,589 24-nt DSRs were identified between OsMET1-2−/− and WT, respectively. Next, we plotted both 21- and 24-nt DSRs, which at the same time were also CG DMRs in terms of relative smRNA abundance (x axis) and DNA methylation level (y axis), between mutant and WT (Fig. 5A). We found that virtually all DSRs (both 21- and 24-nt) were CG hypo-DMRs (Fig. 5A), consistent with genome-wide loss of mC in the mutant (Fig. 3B). Then, we plotted these DSRs that at the same time were either CHG DMRs or CHH DMRs in terms of relative smRNA abundance (x axis) and DNA methylation level (y axis) between mutant and WT (Fig. 5 B and C). We found that in either the CHG or CHH context the 21- and 24-nt DSRs contained both hypo- and hyper-DMRs but more hypo-DMRs (Fig. 5 B and C). We noted, however, that in CHH contexts, a significant shift from hypo- to hyper-DMRs occurred for both 21- and 24-nt DSRs but especially for 24-nt DSRs (Fig. 5C). In Fig. 5, those 21- and 24-nt DSRs and DMRs mapped to the I and III quadrants representing concomitant increase or decrease of smRNA abundance and changes in mC levels, respectively, which were termed concordant changing regions (CCRs); accordingly, those mapped to the II and IV quadrants were termed opposite changing regions (OCRs). We examined the relative proportions of CCRs vs. OCRs for 21- and 24-nt smRNAs in each context, CG, CHG, or CHH. For 21-nt smRNAs, we found that the ratios of CCRs vs. OCRs were <1 in all three sequence contexts (Fig. 5), suggesting a negative correlation between abundance of 21-nt smRNAs and mC level of all three contexts. For 24-nt smRNAs, we found that in the CG and CHG contexts, the ratios of 24-nt CCR vs. 24-nt OCR numbers were close to 1:1 (CG: 6,976 vs. 5,063; CHG: 4,821 vs. 3,631), whereas those in the CHH contexts were >5:1 (9,076:1,962) (Fig. 5 A and B). This suggests that the dynamics of 24-nt smRNA abundance and changes in CHH methylation were positively correlated. When the CCRs (CHH and smRNA) and OCRs (CHH and smRNA) were mapped back to the genome, they were found to reside in repeat-related regions (2,029; 80.74%) (SI Appendix, Fig. S12). Moreover, the level of mCG was higher in CCRs than in OCRs (SI Appendix, Fig. S13). This, together with the observation that the remaining mCG was more in TEs than in genes (Fig. 2D), suggests that the relatively higher mCG level in TEs than in genes in OsMET1-2−/− was maintained by both MET1-1 and the RdDM pathway. This is consistent with findings in Arabidopsis that both CHH methylation and 24-nt smRNAs are integral components of the RdDM pathway (11, 30, 34, 35). Given the interlaced nature of DNA methylation, histone modification, and chromatin remodeling (5, 36, 37), it will be of apparent interest to investigate these properties in the rice OsMET1-2 mutant.
Fig. 5.
Regional colocalization of differentially expressed 21- and 24-nt smRNAs with DMRs of each of the three cytosine sequence contexts, CG, CHG, and CHH. (A) Colocalization of differential smRNA regions that at the same time were also CG DMRs in terms of relative smRNA abundance (x axis; presented as log2FC of DSRs) and DNA methylation level (y axis; presented as log2FC of CG DMRs) between mutant and WT. (B and C) Colocalization of DSRs that at the same time were either CHG or CHH DMRs in terms of relative smRNA abundance (x axis; presented as log2FC of DSRs) and DNA methylation level (y axis; log2FC of CG DMRs) between mutant and WT. Yellow and blue dots denote 21- and 24-nt DSRs, respectively.
Materials and Methods
Plant Materials.
Heterozygous seeds (FT928341) of the Tos17 insertion mutant for the rice MET1-2 gene (OsMET1-2+/−) were obtained from the National Institute of Agrobiological Sciences (Tsukuba, Japan) and then selfed for five additional generations. The WT (OsMET1-2+/+), heterozygote (+/−), and homozygous mutant (−/−) plants were produced by immediate segregation of the OsMET1-2+/− plants. Shoots of 11-d-old seedlings were accumulated from all three genotypes for DNA and RNA isolation (SI Appendix, Materials and Methods).
Identification of Heterozygote and Homozygote MET1-2 Mutants.
Heterozygote (OsMET1-2+/−) and homozygote (−/−) plants were distinguished by locus-specific pyrosequencing (SI Appendix, Materials and Methods).
Whole-Genome Bisulfite Sequencing and Analysis.
Genomic DNA extracted from the 11-d-old shoots of the three OsMET1-2 genotypes (+/+, +/−, and −/−) was used for whole-genome bisulfite sequencing by the HiSeq 2000 (Illumina). For each genotype, >10 Gb of raw data were generated (SI Appendix, Materials and Methods).
RNA Sequencing and Analysis.
Total RNA from the 11-d-old shoots of the three OsMET1-2 genotypes was isolated for RNA-seq. For each genotype, two independent libraries were constructed and sequenced following the standard Illumina sequencing procedure on the HiSeq 2000. For each library more than 4 Gb of clean data (quality score 20 >90%) were produced and used for further expression analysis (SI Appendix, Materials and Methods).
Designation of DMCs, DMRs, and DMGs.
The differentially methylated cytosines, regions, and genes between any two genotypes were defined (SI Appendix, Materials and Methods).
Small RNA Sequencing, Analysis, and Designation of DSRs.
smRNA libraries for the 11-d-old shoots of the three OsMET1-2 genotypes were constructed. At least 15 million clean reads were generated for each genotype (SI Appendix, Materials and Methods).
Supplementary Material
Acknowledgments
We thank Drs. Hongzhi Kong and Jinxing Lin for help with anatomical studies and analysis. This work was supported by grants from the National Natural Science Foundation of China (30990243), the State Key Basic Research and Development Plan of China (2013CBA01404), and the Program for Introducing Talents to Universities (B07017).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the NCBI Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession nos. SRP043447–SRP043449).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410761111/-/DCSupplemental.
References
- 1.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 4.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 8.Genger RK, Kovac KA, Dennis ES, Peacock WJ, Finnegan EJ. Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol Biol. 1999;41(2):269–278. doi: 10.1023/a:1006347010369. [DOI] [PubMed] [Google Scholar]
- 9.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163(3):1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34(1):65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 11.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130(5):851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39(3):391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 15.Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20(15):4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlopoulou A, Kossida S. Plant cytosine-5 DNA methyltransferases: Structure, function, and molecular evolution. Genomics. 2007;90(4):530–541. doi: 10.1016/j.ygeno.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Yamauchi T, et al. Homologous recombination-mediated knock-in targeting of the MET1a gene for a maintenance DNA methyltransferase reproducibly reveals dosage-dependent spatiotemporal gene expression in rice. Plant J. 2009;60(2):386–396. doi: 10.1111/j.1365-313X.2009.03947.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamauchi T, Johzuka-Hisatomi Y, Terada R, Nakamura I, Iida S. The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol Biol. 2014;85(3):219–232. doi: 10.1007/s11103-014-0178-9. [DOI] [PubMed] [Google Scholar]
- 19.Zemach A, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107(43):18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen S, et al. Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proc Natl Acad Sci USA. 2012;109(50):20543–20548. doi: 10.1073/pnas.1217927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Springer NM. Epigenetics and crop improvement. Trends Genet. 2013;29(4):241–247. doi: 10.1016/j.tig.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 22.La H, et al. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc Natl Acad Sci USA. 2011;108(37):15498–15503. doi: 10.1073/pnas.1112704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics. 2012;13:300. doi: 10.1186/1471-2164-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigal M, Kevei Z, Pélissier T, Mathieu O. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 2012;31(13):2981–2993. doi: 10.1038/emboj.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu F, et al. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol. 2008;50(7):886–896. doi: 10.1111/j.1744-7909.2008.00692.x. [DOI] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: Where genetics meets genomics. Nat Rev Genet. 2002;3(5):329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Zhou DX. Rice epigenomics and epigenetics: Challenges and opportunities. Curr Opin Plant Biol. 2013;16(2):164–169. doi: 10.1016/j.pbi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bond DM, Baulcombe DC. Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 2014;24(2):100–107. doi: 10.1016/j.tcb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Melnyk CW, Molnar A, Bassett A, Baulcombe DC. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol. 2011;21(19):1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- 33.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151(1):194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13(24):2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 35.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz RJ, Zhang X. High-throughput approaches for plant epigenomic studies. Curr Opin Plant Biol. 2011;14(2):130–136. doi: 10.1016/j.pbi.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X. The epigenetic landscape of plants. Science. 2008;320(5875):489–492. doi: 10.1126/science.1153996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.