Significance
Heretofore, ligand recognition at each subunit interface of the acetylcholine binding protein (AChBP) has been found to be independent of the other interfaces, representing a disconnection between the properties of the AChBP and the full receptor that it is intended to model. These results comprise the first examples of cooperative binding with the extracellular domain, providing insights into the structural basis for interactions between subunits. Within a single series of congeneric molecules, both positively and negatively cooperative behaviors toward AChBP are manifest. Hence, a distinct mode of binding to the agonist-competitive antagonist site is established in the AChBP protein.
Keywords: allostery, crystallography, nicotinic receptor
Abstract
The nicotinic acetylcholine receptor (nAChR) and the acetylcholine binding protein (AChBP) are pentameric oligomers in which binding sites for nicotinic agonists and competitive antagonists are found at selected subunit interfaces. The nAChR spontaneously exists in multiple conformations associated with its activation and desensitization steps, and conformations are selectively stabilized by binding of agonists and antagonists. In the nAChR, agonist binding and the associated conformational changes accompanying activation and desensitization are cooperative. AChBP, which lacks the transmembrane spanning and cytoplasmic domains, serves as a homology model of the extracellular domain of the nAChRs. We identified unique cooperative binding behavior of a number of 4,6-disubstituted 2-aminopyrimidines to Lymnaea AChBP, with different molecular variants exhibiting positive, nH > 1.0, and negative cooperativity, nH < 1.0. Therefore, for a distinctive set of ligands, the extracellular domain of a nAChR surrogate suffices to accommodate cooperative interactions. X-ray crystal structures of AChBP complexes with examples of each allowed the identification of structural features in the ligands that confer differences in cooperative behavior. Both sets of molecules bind at the agonist-antagonist site, as expected from their competition with epibatidine. An analysis of AChBP quaternary structure shows that cooperative ligand binding is associated with a blooming or flare conformation, a structural change not observed with the classical, noncooperative, nicotinic ligands. Positively and negatively cooperative ligands exhibited unique features in the detailed binding determinants and poses of the complexes.
Nicotinic acetylcholine receptors (nAChRs) function as allosteric pentamers of identical or homologous transmembrane spanning subunits. Ligand binding at two or more of the five intersubunit sites, located radially in the extracellular domain, drives a conformational change that results in the opening of a centrosymmetric transmembrane channel, internally constructed among the five subunits (SI Appendix, Fig. S2A) (1–4). Up to five potential agonist-competitive antagonist sites on the pentamer are found at the outer perimeter of the subunit interfaces. Amino acid side-chain determinants on both subunit interfaces dictate selectivity among the many subtypes of nAChRs. The interconversion between resting, active, and desensitized states occurs in the absence of ligands, and partial occupation of the binding sites suffices for agonist activation of the receptor and its antagonism (5–7). Cooperativity of agonist association and its coupling to channel gating likely play important roles in the dynamics of nicotinic responses and in sharpening the concentration and temporal windows for activation.
As revealed in functional studies, most nAChRs are hetero-oligomeric, where the sites of ligand occupation are not identical (1–4). This arrangement arises when a common α-subunit pairs with one or more nonidentical subunit partners, termed non–α-subunits (7, 8). Nonidentity of the subunit interface complementary to the α-subunit may also give rise to heterogeneity in binding constants typically seen for antagonists and mask partially the degree of agonist cooperativity. An exception to this is the α7-neuronal nAChR composed of five identical subunits and exhibiting a high degree of cooperativity for agonist activation (9). Recently, sequence alignments identified genes coding for pentameric ligand-gated ion channels in prokaryotes led to the resolution of the first structure by X-ray crystallography on 3D crystals of a pentameric receptor protein from Erminia chrysanthemi (ELIC) (10) and Gloeobacter violaceus (GLIC) (11, 12) and provided high-resolution structures of the two end point states of the cooperative gating mechanism in the same pentameric ligand-gated ion channel (GLIC) (13). Recently, the first structure of a eukaryotic member of the family, the anionic glutamate receptor from Caenorhabditis elegans (GluCl), was solved at atomic resolution (14), revealing remarkable identity of 3D structure with GLIC.
The acetylcholine binding protein (AChBP) was characterized from mollusks (15–17) and consists of only a homologous extracellular domain of the nAChR. Assembled as a homomeric pentamer, AChBP exhibits a similar profile of ligand selectivity toward the classical nicotinic agonists and antagonists of quaternary amine, tertiary and secondary amine (alkaloid), imine, and peptide origin that bind nicotinic receptors (18–25). If looked at solely on the basis of ligand-binding capacities, AChBP could be considered as a distinct subtype of nAChR. Although its homomeric composition and ligand selectivity best resemble the α7-subtype of nAChR, when the concentration dependence of ligand occupation has been examined, no evidence of cooperativity emerged (21). Accordingly the cooperative behavior for both activation and desensitization of receptors, seen for the classical nicotinic agonists with nAChRs, might arise from a cooperative torsional motion driven by the transmembrane spanning domain of the receptor (26).
We demonstrate here a set of ligands that bind to the AChBP in a cooperative fashion, whereby binding to a single subunit affects the binding energy at identical interfaces in the pentamer. Hence, interactions within the extracellular domain of this family of homologous pentameric proteins establish a circumferential linkage between subunit interfaces which results in cooperative behavior.
Results
Association of Substituted 2-Aminopyrimidines with AChBP Reveals Positive and Negative Cooperativity.
After preliminary assessment of other scaffolds, we focused our attention on 4,6-disubstituted 2-aminopyrimidines as being representative of drug-like molecules having a propensity to associate with neurological signaling receptors. These compounds showed selectivity for binding to Lymnaea stagnalis (Ls)-AChBP when characterized in a radioligand competition assay. The compounds could be divided in three groups of binding profiles based on Hill coefficients: ligands with nH < 1, nH ∼ 1, and nH >1 (Fig. 1). Nicotine competition with epibatidine served as our positive control and is an example of a noncooperative ligand for AChBP with nH = 1. If the binding of ligand at one site increases the affinity for ligand at a corresponding homologous sites on the pentamer, the ligand exhibits positive cooperativity (nH > 1) with AChBP. Conversely, if the binding of ligand at one site diminishes the affinity for ligand at another site, the protein exhibits negative cooperativity or site heterogeneity, nH < 1. Hence, partial occupation of sites in the pentamer diminishes allosterically with occupation of the remaining sites.
Fig. 1.
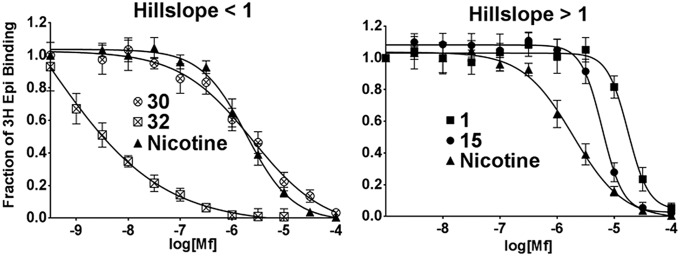
Representative titration profiles for 4,6-substituted 2-aminopyrimidine competition with 3H-epibatidine binding showing a range of dissociation constants (Kd) and Hill coefficients (nH) for ligand binding to Ls-AChBP. Measurements were carried out by a scintillation proximity assay and are reported as an average of at least three individual experiments (±SD).
Overall, a broad range of affinities are revealed in this series of congeneric compounds with apparent Kd values of 0.2 nM to >10 µM (Table 1 and SI Appendix, Table S4). Many compounds from the series showed Hill slope values close to one (Table 1; 28 and 37). However, compounds with large Hill coefficients were identified and were found to exhibit lower, but still respectable, affinities (Fig. 1; 1 and 15). In contrast, some ligands with shallow titration curves bound with exceptionally high affinity (Fig. 1; 30 and 32). There was significant tolerance to substitution at position 4 of the pyrimidine ring, accepting lipophilic alkyl (Table 1; 12–15) and aryl (Table 1; 28–30), as well as more polar groups (Table 1; 27 and 35). These substituents strongly influence the range of ligand Kd and Hill coefficients. A few heterocyclic aromatic rings substituted at 6-position were tested (Table 1; 9–10, SI Appendix, Table S4; 11), but no aryl group was found to give superior activity compared with the substituted phenyl ring.
Table 1.
Competition between substituted 2-aminopyrimidines against 3H-epibatidine binding to Ls-AChBP
 |
Dissociation constants (Kd) and Hill coefficients (nH) are reported as means (±SD).
Solved crystal structure.
Crystal Structures of Ls-AChBP with Substituted 2-Aminopyrimidines Shows Distinctive Tertiary Structural Changes at the Binding Interface.
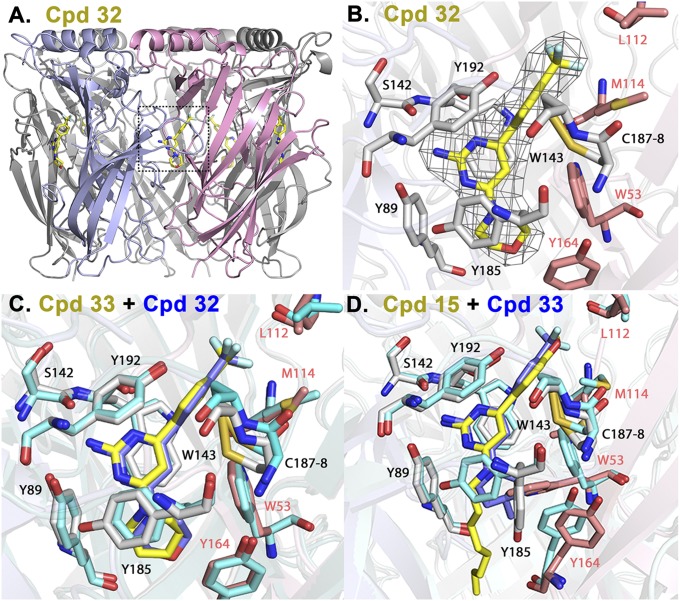
To gain structural insight, crystallization of several complexes was attempted. Crystal structures with Ls-AChBP in complex with ligands showing negative (32, Fig. 2 A and B; 33, Fig. 2C; chains A and B) and positive (15, Fig. 2D; chains D and E) cooperativity were refined to 3.0, 2.1, and 2.7 Å, respectively. For statistics on data collections, see SI Appendix. The Ls-AChBP complex with bound cooperative ligands reflected full occupation of the pentamer and a well-resolved electron density of all 10 subunits (Fig. 2 A and B). Ligand 15 has a clear electron density for the biaryl ring of the molecule, but a poor density of the alkyl chain possibly arising from multiple flexible conformations of the C loop. Several residues in F the loop (T155–E163) gave unresolved electron densities and were consequently excluded from the models. Additionally, in complex with ligand 15, residues 185–189 in the C loop encompassing the vicinal Cys–Cys bond were not seen in 9 of 10 subunits of the dimer of pentamers.
Fig. 2.
X-ray crystal structures of ligands 32 and 33 (negative cooperativity, nH < 1) and 15 (positive cooperativity, nH > 1), in complexes with Ls-AChBP. (A) Radial view of Ls-AChBP pentameric structure in complex with 32. Full occupation of the 10 binding sites in the unit crystal of a dimer of pentamers was evident. A principal, C loop-containing, and complementary face are shown in gray and purple. (B) Expanded radial view of 32 in binding site, including ligand electron density. Side chain carbons of the principal and complementary subunits are shown in gray and purple, respectively. Ligand carbons are in yellow and fluorines in turquoise. (C) Overlay of 32 (blue) and 33 (yellow) crystal structures. Side-chain carbons for 32 are in turquoise. The overlay shows little or no variance in ligand pose or side-chain positions. (D) Superimposition of 15 (yellow) and 33 (blue) crystal structures. Side-chain carbons for 33 are in turquoise. The positively 15 and negatively 32/33 cooperative ligands show a similar positioning of the 4-substituted phenyl rings, but distinct poses or positions for the 2-aminopyrimidine ring and the substituents at position 4 of the pyrimidine ring. Marked changes in the orientation of the side chains of Y185, W53, and Y164 are evident for the positively and negatively cooperative ligands.
The structure of Ls-AChBP in complex with 33 superimposed on 32 with an RMS deviation of 0.38 Å for 1,212 Cα atoms (Fig. 2C) and on 15 with an RMS deviation of 0.34 Å for 1,132 Cα atoms (Fig. 2D). Binding orientations show a high degree of similarity at all five binding sites in each pentamer and a similar orientation of the ligands. All three compounds bind underneath a closed C loop at the interface between two subunits. Loop closure in the presence of bound ligands, as measured from the backbone carbonyl of W143 in the A loop to the γ-sulfur atom of the first vicinal Cys disulfide-linked residue in the C loop (C187 in Ls-AChBP), is 8.4 Å for 32 and 33 structures and 8.2 Å for 15. The ligands contact amino acids from both the principal face with residues from loops A (Y89), B (W143), and C (Y185 and Y192), and complementary face (loops D, W53; E, L112 and M114; and F, Y164). Parallel displaced π-stacking interactions with W143 are present in all structures. Importantly, the pyrimidine ring of 15, the ligand showing positive cooperativity, rotates by ∼26–36° compared with 32 and 33 showing negative cooperativity (SI Appendix, Fig. S5). The ring rotation results in its parallel alignment with Y192 side chain (2.9 Å and greater). The only potentially protonable nitrogen in the ligands at physiological pH is in the pyrimidine ring at the position N1 (pKa ∼ 6.7–6.8; computed by Marvin Sketch 5.12.3). It resides within hydrogen bonding distance of the carbonyl backbone oxygen of W143 (2.7–2.9 Å) and as close as 3.8–3.9 Å to the W143 side-chain aromatic ring. Other interactions in the complexes come from polar contacts formed by ligands N2 atoms with the hydroxyl group of the Y89 side chain from the B-loop (2.9 Å in 32 and 33; 2.7 Å in 15) and carbonyl oxygen of S142 (2.6–2.8) Å for all three ligands.
Morpholine (32) or 4-methyl-piperidine (33) substituents appear to associate with Y89 and Y185. Nitrogen atoms of these rings are positioned to stack with all seven atoms of the Y185 ring with distances ranging between 3.6 and 4.3 Å. Interactions of these substituents on the complementary subunit face are predominantly hydrophobic. The altered position of the indole of W53 in 32 and 33, compared with 15, is associated with a change in side-chain orientation of neighboring residues M114 and Q55. Phenyl rings substituted at the pyrimidine 6-position interact mainly with W143, Y192, and C188. Additionally, fluorines in trifluoromethyl substituent are in the vicinity of the T144 side chain and interact with multiple loop F residues, including L112 and M114, as well as neighboring water molecules, with interaction extending to M114 and R104 side chains.
The flexible aliphatic chain of 15 at the 4 position in the pyrimidine does not yield discernable electron densities except for chain D. Lack of density likely reflects multiple conformations of the flexible C loop not constrained by the symmetry related molecule in the crystal structure. Rotation of the pyrimidine ring and presence of the alkyl chain in 15 brings the ligand in close contact with Y185 side chain (∼3.0 Å to N4 of the ligand) and causes the tyrosine ring to rotate toward the gorge interface presumably to avoid a clash with the alkyl chain of the ligand. Also, the indole of W53 on the complementary face rotates toward the subunit interface and is in contact with ligand N4 (3.5 Å). The phenyl ring in ligand 15 has a similar position as the aromatic ring in complexes 32 or 33, but its contacts are altered by methoxy- substituent interacting with T144 and with L102, L112, and M114 in the complementary face. Major differences for 15, compared with complexes of ligands showing negative cooperativity, are seen in Y185, Y164, M114, and W53 side-chain conformations.
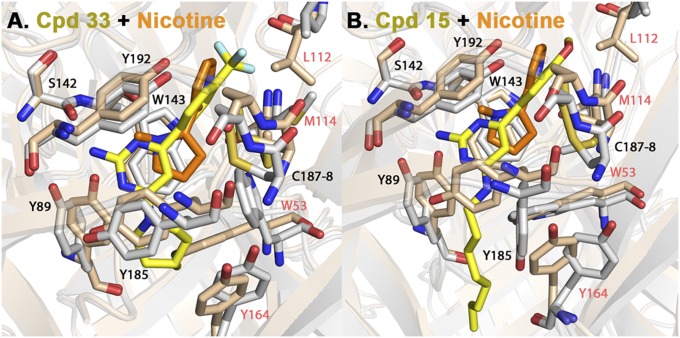
The interactions of 32/33 are compared with nicotine the in Ls-AChBP binding pocket [Protein Data Bank (PDB) ID code 1UW6] (18) in Fig. 3A. The pyrrolidine ring in nicotine only partially overlaps with pyrimidine ring of cooperative ligands, and nitrogens of these substituents are well aligned. In contrast to nicotine, the side chain of Y89 shifts to bring its hydroxyl group in hydrogen bonding distance of the pyrimidine nitrogens in the 32, 33, and 15 crystal structures. Striking differences in side-chain positions in nicotine complex compared with 32 and 33 are seen on the complementary face of the binding site that forms interactions through residues that are only partially conserved in AChBPs from different species. To accommodate the morpholine or methylpiperidine substituents in the 32/33 complex, the indole side chain in W53 changes its rotameric position. As a consequence, M114 is brought in contact with the 6-substituted trifluoromethyl phenyl group of the ligand. The position of the 32/33 phenyl ring is similar to the pyridine ring in nicotine. The trifluoromethyl group forces L112 to change its rotameric position. The presence of fluorines, however, affords additional hydrophobic stabilization with multiple residues on complementary subunit.
Fig. 3.
Superimposition of Ls-AChBP X-ray crystal structures in complex with 33 (A) and 15 (B) with nicotine (PDB code 1UW6) (in orange). 33 and 15 carbons are shown in yellow, and nicotine in orange. The protein side chains are shown in gray for 33 and 15 and pink for nicotine. Both ligands show distinctly different positions from the pyrrolidine and pyridine rings of nicotine, as well as the side-chain positions of residues in the C loop in the principal subunit (Y89 and Y185) and the complementary (W53 and Y164) subunit face.
Compared with the nicotine-AChBP complex, loop C in ligand 15 (chain D) shows significant variation, reflected in the Y185 side-chain conformation (Fig. 3B). Residues W53 and M114 in the 32/33 complexes, that represent most significant departures from nicotine, in the 15 X-ray structure have orientations similar to those in the nicotine complex. However, the indole ring of W53 side chain has more extensive contacts with the ligand 15 than with nicotine, through its substituent at the position 4 in the pyrimidine ring. To avoid steric occlusion with Y185 in the complex, the rotational state of Y164 on complementary face changes as well.
Quaternary Structural Changes of Ls-AChBP.
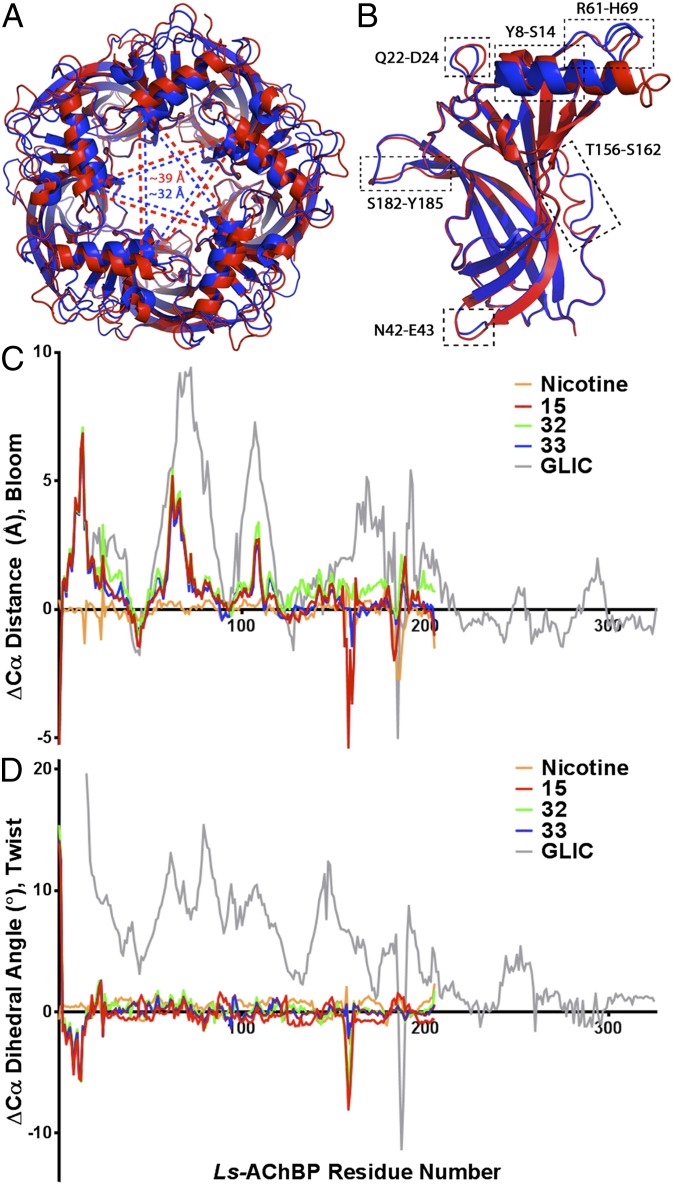
Superimposing Ls-AChBP in the Apo form with the complexes of substituted 2-aminopyrimidines showing cooperativity (ligands 15, 32, and 33) also revealed a major change in quaternary structure (Fig. 4A). Distances between Cα for T13 in the apical α-helices of diametrically opposed subunits showed 7 Å greater transsubunit spans than for the Apo reference structures or the nicotine complex. By superimposing a single chain in each pentamer (Fig. 4B), differences in tertiary structure could be observed for six regions (RMSD ∼ 5 or greater; Swiss-PdbViewer): residues 8–14 (N-terminal α-helix) and loops 22–24, 43–44 (loop connecting β1 and β2), 61–69 (η1 and its continuation), 156–162 (region in β8 N-terminal to loop F), and 182–185 (part of β9 and loop C). Based on a similar analysis for nicotine and carbamylcholine Ls-AChBP crystal structures (SI Appendix, Fig. S2), differences observed for four regions appeared to be unique for cooperative ligand complexes (regions 8–14, 22–24, 42–43, and 61–69). To quantify observed variations, two types of calculations were performed using 1UX2 as a reference structure (for detailed description of calculations, see SI Appendix).
Fig. 4.
Global differences in X-ray crystal structures of Ls-AChBP bound cooperative ligands in comparison with crystal structure of Ls-AChBP in its Apo form (PDB code 1UX2) and GLIC (PDB code 4NPP). (A) Top (apical) view on superimposed (UCSF chimera) Apo pentamer (in blue) and with bound 15 (in red). Dashed lines (blue and red, respectively) indicate the most significant differences in quaternary structures quantified by measuring distances between T13 backbone α carbon of distant subunits. (B) Superimposition (PyMOL) of Ls-AChBP Apo, chain D (in blue) and 15 complex, chain D (in red). Major differences in the quaternary structures of the AChBP are marked with dashed rectangles (RMS value of ∼0.5 or greater). (C) Chart of differences of Cα distances (n = 5) of diametrical subunits observed in cooperative ligands relative to Apo in comparison with GLIC (GLIC, closed form, is used as a reference). Nicotine used as a control. Observed differences reflect blooming profile of the protein complexes. (D) Plot of differences of Cα dihedral angles (n = 5) observed in cooperative ligands relative to Apo in comparison with GLIC (GLIC closed form used as a reference). Nicotine used as a control. Observed differences reflect twisting profile of the protein complex.
Differences in distances between every α-carbon (Cα) of diametrically opposed subunits were calculated relative to the reference structure (Fig. 4C). Additionally, differences in dihedral angles for every Cα residue were calculated (using three reference points) relative the reference structure (Fig. 4D). Values were compared with those obtained for the nicotine complex and the open and closed states of prokaryotic GLIC protein (13). For the α-carbon distances (Fig. 4C), significant differences compared with nicotine complex were observed for residues 1–120, with the largest differences emerging in the N-terminal helix. Interestingly, these differences showed similar patterns in linear sequence to those observed for GLIC (13) and are described as a blooming motion of the protein extracellular domain. Region 155–162 in complex 15 shows a significant distance reduction compared with nicotine. This region is a highly flexible segment of the protein, as is the case for complexes 32 and 33, where this region is omitted due to limiting densities in the crystal structure. Although short sequence regions in the crystal with positively cooperative ligand 15 diverge from crystals of 32/33 with negative cooperativity (e.g., residues 182–189), owing to differences between ligands 32 and 33 themselves, interpretation of the fine structural differences between 15 and 32 and 33 will require additional study. Based on the rearrangements observed at the interface of the orthosteric agonist/antagonist pocket (SI Appendix, Fig. S3) the greatest deviations from nicotine (up to ∼4 Å) are evident for Y185 and C187-8 in the crystal structure of the 32 and 33 complexes.
Relative differences of dihedral angles show small torsional or twist motions seen for residues 1–23 with as much as a 5° shift for R11. Also, residues 157–160 exhibit a similar tendency with changes up to 8°. Data for 32 and 33 overlay very well and do not differ much from 15 across the entire sequence, especially for the abovementioned regions with greatest torsional motions that are not observed for nicotine. However, the patterns of dihedral angle changes are far smaller than seen in the GLIC structure and AChR models (13, 26, 27).
An alternative presentation of the data is shown in SI Appendix, Fig. S2, and all values were projected onto a centrosymmetric axis in the pentamer, and they reflect relative distances from a reference point at the transmembrane region interface. The blooming amplitude develops from cytoplasmic toward the extracellular, apical region, resulting in increased intersubunit distances between diametrically opposed, nonadjacent subunits. Accordingly, the blooming quaternary structure is evident for all three members of the 2-aminopyrimidine series and emphasizes the importance of the conformational differences in quaternary structure associated with the state changes. These sequence patterns of quaternary rearrangements reflected in distances between diametric subunits are in close correspondence in residue positions with those found in the open and closed channel forms of GLIC (13).
Discussion
Our studies establish a previously unknown level of conformational communication between AChBP subunits accompanying ligand binding. We report a series of selective AChBP ligands exhibiting negative, as well as positive cooperativity, a type of allosteric behavior in which binding interactions in an oligomeric protein take place between distant subunit interfaces. Selected ligands showing marked differences in cooperativity were used to obtain three atomic resolution, X-ray crystal structures of ligands with different Hill slope values in their complexes with Ls-AChBP. The lower affinity ligands showing positive cooperativity, along with the high affinity of the negatively cooperative ligands, serve to achieve full occupation of ligand in the crystal structures. These cooperative interactions occur in the absence of the transmembrane, channel gating domain required of the nAChR to elicit subunit interactions and cooperativity. Rather cooperativity in AChBP is confined to the extracellular domain of the nAChR reference structure and is mediated circumferentially around the cylindrical pentamer.
The differences in side-chain positions at the subunit interface (W53, Y164, and Y185) that distinguish between positively and negatively cooperative ligands also diverge from the nicotine side-chain positions. Nicotine also adds Y89 to the perturbed side chains. Changes in quaternary structure noted in reference to the gating transition of prokaryotic GLIC, as blooming (13, 27), are not seen for nicotine and carbamylcholine, where the Apo form of the receptor is used as a reference state. However, it should be recognized that the apo-receptor contains Hepes buffer occupying the reference site (16). Hence, at this stage, an nAChBP unoccupied by a ligand is not available in a crystallized form.
Nevertheless, we should ask what distinguishes the substituted 2-aminopyrimidines from the conventional ligands hitherto fore characterized. They may fit into three categories: (i) quaternary amines stabilized by a cation-π interaction between the cation and a nest of surrounding aromatic side chains. Such interactions are evident in the crystal structures of acetylcholine, carbamylcholine, and other quaternary complexes (16, 18), and the energetics have been established through mutagenesis studies modifying the polarizability, and electronegativity of the aromatic side chains in the binding pocket (28, 29); (ii) secondary and tertiary amines (19–21) and imines (21), which, when in a protonated state, hydrogen bond to the backbone carbonyl of W143. The protonated dihydropyridine-bound state for the benzylidene anabaseines has been demonstrated by difference spectroscopy (30); and (iii) peptides, such as the α-conotoxins and α-neurotoxins, stabilized by multiple interactions on the C loop and interfacial residues on both the principal and complementary faces (31–34). The binding interactions of the 2-aminopyrimidines do not appear to follow the above patterns and therefore these structures constitute a distinct fourth group of ligands. The nitrogens in the 2-aminopyrimidine ring are not very basic, suggesting a far higher energy requirement for the ligand to bind in a protonated state than that found for secondary and tertiary amines and the imines. Hence, the 2-aminopyrimidines may be considered as electron-rich ring systems capable of ring stacking with the side chains of Y192 and W143. The pyrimidine ring nitrogens are not as exposed as the bicyclic ring in epibatidine and the pyrrolidine nitrogen in nicotine affording a proper directionality for hydrogen bonding. Thus, the global conformational changes primarily manifested by blooming appear characteristic of the substituted 2-aminopyrimidines and are not seen with carbamylcholine and nicotine (Fig. 4 and SI Appendix, Fig. S2). To date, a clear state change has only been documented for the substituted 2-aminopyrimidines.
The changes in AChBP quaternary structure add another dimension to considering subunit interactions. The blooming and torsional conformational changes noted with AChBP show analogies with those observed in GLIC (13, 27) in relation with correspondences of protein sequence (Fig. 4 A–C and SI Appendix, Fig. S2). The amino terminal helix, regions between residues 58 and 70 and between 106 and 110, all show increased distances between diametrically opposed subunits. The sharper negative peaks around residues 156 and 185 likely involve local perturbations of the C and F loops proximal to the ligand binding site resulting in local compaction of the structure, measured in Cα distances. The comparison of the dihedral bond angles should reflect a torsional or twist motion as seen between the two states of GLIC (13, 27). Changes in dihedral angle positions are small (SI Appendix, Fig. S2C), but involve the same set of residues. Hence, when the binding of the 2-aminopyrimidines are compared with the pH-dependent conformations of GLIC, the dominant common change is found with the Cα distances between diametric subunits (blooming) (SI Appendix, Fig. S2B) rather than the dihedral angles for torsional movement (twist) (SI Appendix, Fig. S2C).
The interfacial binding sites residing under the C loop of AChBP and the nAChR appear surprisingly accommodating for the binding of ligands of a different structure. For example, Stornaiaolo and colleagues reported on large planar, aromatic molecules binding under the C loop in a stacked sandwich fashion and extending the C loop (35).
The cooperative ligands binding to AChBP add a new dimension to ligands interaction with the extracellular domain of the pentameric ligand-gated ion channels. We are currently examining the structural determinants of selectivity of this ligand family with other AChBP’s and the homomeric α7 nAChR. With homomeric nAChRs, selectivity for the primary agonist could be altered through partial site occupation by the 2-aminopyrimidines showing negative cooperativity. In the case of the predominant heteromeric receptors where the binding interfaces will differ, such ligands may possibly serve as positive or negative allosteric modulators at sites distinct from those occupied by agonist and competitive antagonist (36). Such appears to be the case for the benzodiazepines (36, 37) and other sedative agents (38) that act in this manner with the GABA receptor (39, 40). Accordingly, new dimensions for achieving pharmacologic selectivity for particular nAChR subtypes may result with the cooperative nAChR ligands possessing electron-rich substituted 2-aminopyrimidines.
Materials and Methods
Synthetic schemes and procedures are described in SI Appendix. Ls-AChBP was expressed and purified as previously described (30). A full description of protocols, screening methods (17) and crystallization details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Kwok-Yiu Ho and Wenru Yu for tissue culture and radioligand screening expertise, and members of the beamline staff and The Advanced Light Source, Lawrence Berkeley National Laboratory. Drs. Manish B. Shah, Lanfeng Wang, John G. Yamauchi, Stewart J. Edelstein, Marco Ceccjhini, and Jaroslaw Maderski provided engaging discussions and valued suggestions. The work was supported by National Institutes of Health Grant GM 18360-40 (to P.T.), the Tobacco-related Disease Research Program 21FT_0024 (to K.K.), and the Kavli Institute for Brain and Mind to (J.-P.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4QAA, 4QAB, and 4QAC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410992111/-/DCSupplemental.
References
- 1.Changeux JP. 50 years of allosteric interactions: The twists and turns of the models. Nat Rev Mol Cell Biol. 2013;14(12):819–829. doi: 10.1038/nrm3695. [DOI] [PubMed] [Google Scholar]
- 2.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3(2):102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 3.Thompson AJ, Lester HA, Lummis SC. The structural basis of function in Cys-loop receptors. Q Rev Biophys. 2010;43(4):449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 4.Corringer PJ, et al. Structure and pharmacology of pentameric receptor channels: From bacteria to brain. Structure. 2012;20(6):941–956. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Heidmann T, Changeux JP. Fast kinetic studies on the interaction of a fluorescent agonist with the membrane-bound acetylcholine receptor from Torpedo marmorata. Eur J Biochem. 1979;94(1):255–279. doi: 10.1111/j.1432-1033.1979.tb12893.x. [DOI] [PubMed] [Google Scholar]
- 6.Neubig RR, Cohen JB. Equilibrium binding of [3H]tubocurarine and [3H]acetylcholine by Torpedo postsynaptic membranes: Stoichiometry and ligand interactions. Biochemistry. 1979;18(24):5464–5475. doi: 10.1021/bi00591a032. [DOI] [PubMed] [Google Scholar]
- 7.Sine SM, Taylor P. The relationship between agonist occupation and the permeability response of the cholinergic receptor revealed by bound cobra alpha-toxin. J Biol Chem. 1980;255(21):10144–10156. [PubMed] [Google Scholar]
- 8.Sine SM, Taylor P. Relationship between reversible antagonist occupancy and the functional capacity of the acetylcholine receptor. J Biol Chem. 1981;256(13):6692–6699. [PubMed] [Google Scholar]
- 9.Bertrand D, et al. Positive allosteric modulation of the alpha7 nicotinic acetylcholine receptor: Ligand interactions with distinct binding sites and evidence for a prominent role of the M2-M3 segment. Mol Pharmacol. 2008;74(5):1407–1416. doi: 10.1124/mol.107.042820. [DOI] [PubMed] [Google Scholar]
- 10.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452(7185):375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 11.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457(7225):111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 12.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457(7225):115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 13.Sauguet L, et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci USA. 2014;111(3):966–971. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474(7349):54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smit AB, et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411(6835):261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 16.Brejc K, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411(6835):269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 17.Hansen SB, et al. Tryptophan fluorescence reveals conformational changes in the acetylcholine binding protein. J Biol Chem. 2002;277(44):41299–41302. doi: 10.1074/jbc.C200462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41(6):907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 19.Hansen SB, et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24(20):3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibbs RE, et al. Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor. EMBO J. 2009;28(19):3040–3051. doi: 10.1038/emboj.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rucktooa P, et al. Structural characterization of binding mode of smoking cessation drugs to nicotinic acetylcholine receptors through study of ligand complexes with acetylcholine-binding protein. J Biol Chem. 2012;287(28):23283–23293. doi: 10.1074/jbc.M112.360347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen SB, Talley TT, Radic Z, Taylor P. Structural and ligand recognition characteristics of an acetylcholine-binding protein from Aplysia californica. J Biol Chem. 2004;279(23):24197–24202. doi: 10.1074/jbc.M402452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brams M, et al. Crystal structures of a cysteine-modified mutant in loop D of acetylcholine-binding protein. J Biol Chem. 2011;286(6):4420–4428. doi: 10.1074/jbc.M110.188730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahsavar A, et al. Crystal structure of Lymnaea stagnalis AChBP complexed with the potent nAChR antagonist DHβE suggests a unique mode of antagonism. PLoS ONE. 2012;7(8):e40757. doi: 10.1371/journal.pone.0040757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulens C, et al. Use of acetylcholine binding protein in the search for novel alpha7 nicotinic receptor ligands. In silico docking, pharmacological screening, and X-ray analysis. J Med Chem. 2009;52(8):2372–2383. doi: 10.1021/jm801400g. [DOI] [PubMed] [Google Scholar]
- 26.Taly A, et al. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys J. 2005;88(6):3954–3965. doi: 10.1529/biophysj.104.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calimet N, et al. A gating mechanism of pentameric ligand-gated ion channels. Proc Natl Acad Sci USA. 2013;110(42):E3987–E3996. doi: 10.1073/pnas.1313785110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dougherty DA. Cys-loop neuroreceptors: Structure to the rescue? Chem Rev. 2008;108(5):1642–1653. doi: 10.1021/cr078207z. [DOI] [PubMed] [Google Scholar]
- 29.Xiu X, Puskar NL, Shanata JA, Lester HA, Dougherty DA. Nicotine binding to brain receptors requires a strong cation-pi interaction. Nature. 2009;458(7237):534–537. doi: 10.1038/nature07768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talley TT, et al. Spectroscopic analysis of benzylidene anabaseine complexes with acetylcholine binding proteins as models for ligand-nicotinic receptor interactions. Biochemistry. 2006;45(29):8894–8902. doi: 10.1021/bi060534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 2005;24(8):1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sine SM, Huang S, Li SX, daCosta CJ, Chen L. Inter-residue coupling contributes to high-affinity subtype-selective binding of α-bungarotoxin to nicotinic receptors. Biochem J. 2013;454(2):311–321. doi: 10.1042/BJ20130638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talley TT, et al. Alpha-conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as alpha3beta2 and alpha7 nicotinic acetylcholine receptors. J Biol Chem. 2006;281(34):24678–24686. doi: 10.1074/jbc.M602969200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulens C, et al. Structural determinants of selective alpha-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc Natl Acad Sci USA. 2006;103(10):3615–3620. doi: 10.1073/pnas.0507889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stornaiuolo M, et al. Assembly of a π-π stack of ligands in the binding site of an acetylcholine-binding protein. Nat Commun. 2013;4:1875. doi: 10.1038/ncomms2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter L, et al. Diazepam-bound GABAA receptor models identify new benzodiazepine binding-site ligands. Nat Chem Biol. 2012;8(5):455–464. doi: 10.1038/nchembio.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morlock EV, Czajkowski C. Different residues in the GABAA receptor benzodiazepine binding pocket mediate benzodiazepine efficacy and binding. Mol Pharmacol. 2011;80(1):14–22. doi: 10.1124/mol.110.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanson SM, Morlock EV, Satyshur KA, Czajkowski C. Structural requirements for eszopiclone and zolpidem binding to the gamma-aminobutyric acid type-A (GABAA) receptor are different. J Med Chem. 2008;51(22):7243–7252. doi: 10.1021/jm800889m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Changeux JP. The concept of allosteric modulation: An overview. Drug Discov Today Technol. 2013;10(2):e223–e228. doi: 10.1016/j.ddtec.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Smith GB, Olsen RW. Deduction of amino acid residues in the GABA(A) receptor alpha subunits photoaffinity labeled with the benzodiazepine flunitrazepam. Neuropharmacology. 2000;39(1):55–64. doi: 10.1016/s0028-3908(99)00104-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.