Significance
Protein dynamics can conceptually be classified into three factors: folding, conformational transitions, and chemical events. Although couplings between every two of these factors have been well studied, explicit coupling among the three factors has not been examined due to its difficulty in direct experimental measurements. Toward this direction, calmodulin is a suitable model system. Here, constructing a computational model that integrates folding, ligand binding, and allosteric motions based on the energy landscape theory, we studied interplay among the three factors, in detail, for calmodulin domains. The analysis includes three conformational states and three binding states, resulting into nine states in total. We found multiple routes and their intriguing modulation by ligand concentration.
Keywords: metal, coarse grained, molecular dynamics, multiscale simulations, force
Abstract
Ligand binding modulates the energy landscape of proteins, thus altering their folding and allosteric conformational dynamics. To investigate such interplay, calmodulin has been a model protein. Despite much attention, fully resolved mechanisms of calmodulin folding/binding have not been elucidated. Here, by constructing a computational model that can integrate folding, binding, and allosteric motions, we studied in-depth folding of isolated calmodulin domains coupled with binding of two calcium ions and associated allosteric conformational changes. First, mechanically pulled simulations revealed coexistence of three distinct conformational states: the unfolded, the closed, and the open states, which is in accord with and augments structural understanding of recent single-molecule experiments. Second, near the denaturation temperature, we found the same three conformational states as well as three distinct binding states: zero, one, and two calcium ion bound states, leading to as many as nine states. Third, in terms of the nine-state representation, we found multiroute folding/binding pathways and shifts in their probabilities with the calcium concentration. At a lower calcium concentration, “combined spontaneous folding and induced fit” occurs, whereas at a higher concentration, “binding-induced folding” dominates. Even without calcium binding, we observed that the folding pathway of calmodulin domains can be modulated by the presence of metastable states. Finally, full-length calmodulin also exhibited an intriguing coupling between two domains when applying tension.
Protein folding and conformational dynamics have often been characterized by the energy landscape of proteins (1–5). The energy landscape is dependent on the molecular physiochemistry and thus is modulated by many factors, such as chemical modification and ligand binding. Ligand binding, in turn, is dependent on the conformation of proteins. Thus, folding, binding, and allosteric conformational dynamics are mutually correlated. Despite their obvious correlation in concept, it has been very challenging to characterize how they are indeed coupled for any single proteins. Here, we address, in depth, how these three types of dynamics, folding, binding, and allosteric conformational dynamics, are coupled from the energy landscape perspective for a specific protein, calmodulin (CaM).
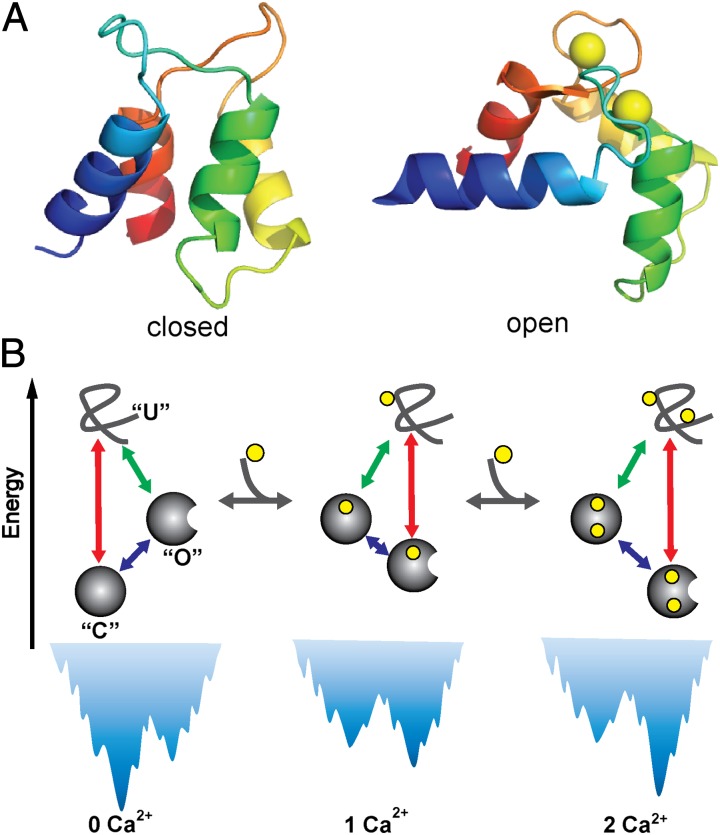
CaM is a ubiquitous calcium-binding messenger protein involved in signal transduction (6) and, more importantly here, has been a model protein to investigate folding, binding, and allostery. Full-length CaM has two nearly symmetric globular domains connected by a flexible central helix (7, 8). Each domain is composed of paired EF hands containing two Ca2+-binding sites (Fig. 1A). Upon binding to Ca2+, each CaM domain undergoes substantial conformational change from a closed state to an open state, exposing a hydrophobic patch that can bind with target proteins and regulate downstream processes (9). CaM has been frequently used as a model in studying the folding of multidomain proteins (10, 11), allosteric transitions (12–14), slow conformational dynamics around physiological temperatures (15–18), metal ion binding (19, 20), and correlation between inherent flexibility and protein functions (21, 22). For example, using structure-based coarse-grained (CG) simulations, Chen and Hummer elucidated the coexistence of an unfolded state, a closed state, and an open state around physiological temperatures for the C-terminal domain of CaM (CaM-C) without Ca2+ binding (15), which reconciles some seemingly contradictory experimental observations on the slow conformational dynamics of CaM.
Fig. 1.
(A) Three-dimensional structure of calmodulin domain at closed [Protein Data Bank (PDB) code: 1cfd] and open states (PDB code:1cll). Calcium ions are represented by yellow spheres. (B) Schematic of coupling among folding, calcium binding, and allosteric motions for the CaM domain. Due to the conformational transitions between open and closed states, in addition to the direct folding pathway (red solid arrow), folding to the most stable state may involve an alternative pathway via a metastable state (green arrow plus blue arrow). The calcium binding can modulate the relative stability of the conformational states and therefore the population of folding pathways. O, C, and U represent open, closed, and unfolded states, respectively.
More recently, Rief and coworkers studied the Ca2+-dependent folding of CaM based on a new generation technique of single-molecule force spectroscopy, which can probe the reversible folding/unfolding transitions with near equilibrium conditions (10, 23, 24). Their results revealed that at high Ca2+ concentrations, the folding pathway of the CaM domain proceeds via a transition state capable of binding Ca2+ ions, demonstrating the coupling between Ca2+ binding and CaM folding. All these computational and experimental works provided unprecedented understanding of many aspects of the folding and allosteric transitions of CaM. However, a full picture of the coupling among folding, Ca2+ binding, and allosteric motions, as schematically shown in Fig. 1B, is still lacking. Particularly, two fundamental issues arising from the allostery and Ca2+-binding characteristics of CaM remain elusive: (i) How does the allosteric feature of the energy landscape contribute to the folding complexity? And (ii) how can the folding mechanism of CaM be modulated by Ca2+ binding?
Motivated by previous computational and experimental studies (15, 23), in this work we investigated the folding coupled with Ca2+ binding and allosteric motions of the isolated CaM domains as well as the full-length CaM. To do so, we first integrated computational tools developed for folding, ligand binding, and allosteric motions together. The proposed CG protein model was used for the subsequent series of molecular dynamics (MD) simulations. First, corresponding to Rief’s experiments, we performed MD simulations of isolated CaM domains with pretensions, which gave consistent results with the experiments and, in addition, provided the direct structural assignment on the experimentally observed states. Second, at a higher temperature, without pretension we performed reversible folding/unfolding simulations for a wide range of Ca2+ concentrations. The conformational and ligand-binding energy landscape revealed as many as nine distinctive states. Then, we analyzed the binding-coupled folding reactions in terms of the nine states, finding multiple routes and their modulation by Ca2+ concentrations. Interestingly, as the Ca2+ concentration increases, the CaM domain folding mechanism switches from “combined spontaneous folding and induced fit” to “binding-induced folding,” which accords with the scenario deduced from single-molecule force spectroscopy experiments. Finally, the effects of tension on the conformational fluctuations of the full-length CaM are discussed.
Results and Discussion
Computational Modeling of Folding, Binding, and Allostery.
To study interplay among folding, binding, and allosteric motions of proteins in depth, we integrated the corresponding three computational models developed earlier. For folding, we used the atomic interaction-based coarse-grained (AICG) model (25, 26) that captures both sequence and native-structure information within a simple framework of perfect-funnel approximation (27). For allosteric conformational motions, we used a multibasin model that concisely realizes conformational transitions among multiple given structures (28).
For binding, we took an implicit ligand-binding model, in which we approximate the ligand-binding states as two states, unbound and bound, and impose Monte Carlo-based transitions between the two states (29). Specifically, calcium binding, in the current case, is assumed to be diffusion limited and occurs with the rate kon[Ca2+], where kon is the second-order rate constant and [Ca2+] is the calcium concentration. The calcium dissociation rate is given as , where is the intrinsic off rate, ΔVbind is the binding energy that depends on the conformations of the binding site, and kBT is the thermal energy (SI Text).
These three computational models can be naturally combined into one model although, to our knowledge, this has not been realized before. Unless otherwise mentioned, we used this integrated CG model throughout this work.
Calmodulin Domain Under Constant Mechanical Extension.
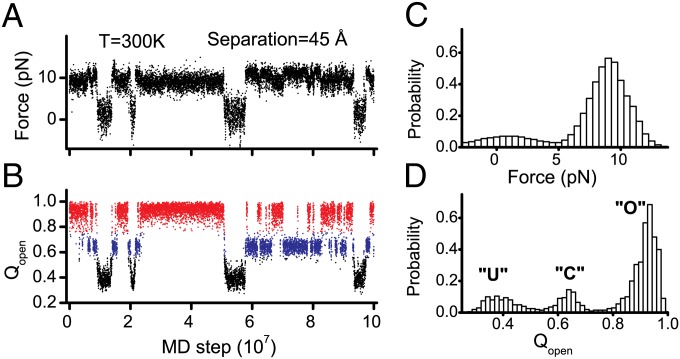
Force is a powerful single-molecule probe to explore the conformational fluctuations of proteins (30, 31). Following recent single-molecule force spectroscopy experiments (23), we conducted MD simulations of the isolated CaM domains under constant mechanical extensions at [Ca2+] = 1.7 μM. The two terminal residues were connected to two dummy beads with constant separations (SI Text). By choosing appropriate separations (45 Å), we observed reversible folding/unfolding transitions at 300 K. The obtained force trajectories and corresponding distributions of force demonstrate that the conformational ensemble of the CaM domains apparently involves two states (Fig. 2 A and C and Fig. S1), i.e., a folded state and an unfolded state, which highly resemble the force trajectories from single-molecule force spectroscopy experiments (23). However, when we plot the time series of a structural reaction coordinate, Qopen or Qclosed (definitions in Materials and Methods), we clearly see three major states (Fig. 2 B and D and Fig. S1): the unfolded state (black in Fig. 2B), the closed state (blue), and the open state (red). These results suggest that there are hidden transitions in force trajectories that cannot be directly probed by force, because the force difference between the open and closed states is largely buried by intrinsic fluctuations of force values (Fig. 2C). Previous computational work also showed that the three conformational states of CaM-C can coexist around physiological temperatures (15). The unambiguous identification of the conformational transitions among the three states by MD simulations makes it possible to investigate the interplay among folding, binding, and allosteric motions.
Fig. 2.
(A and B) Representative trajectories for the isolated CaM-C monitored by force (A) and Qopen (B) for the pretension simulations with Ca2+ concentrations of 1.7 μM. Color code: red, open state; blue, closed state; black, unfolded sate. (C and D) Distributions of force value (C) and Qopen (D).
Calmodulin Domain Near Denaturation Temperature.
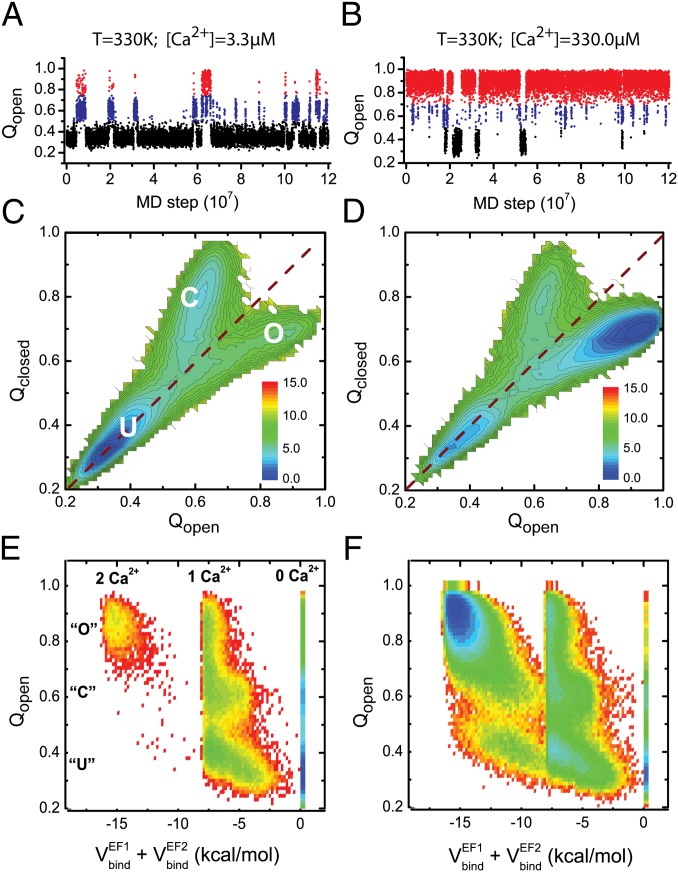
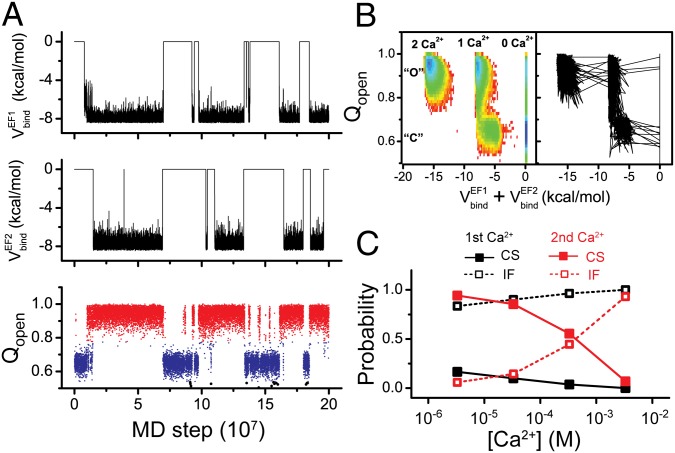
Next, to address thermally activated reversible folding/unfolding dynamics without mechanical force, we conducted MD simulations at 330 K for the isolated CaM domains with wide range of [Ca2+]. Representative trajectories of the structural similarity to the open state, Qopen, clearly show three major states (Fig. 3 A and B), in a similar way to the mechanically forced simulations. At a lower concentration, [Ca2+] = 3.3 μM, CaM-C is predominantly denatured (black), with a small fraction of time reaching the closed (blue) and open (red) states. Contrarily, CaM-C takes the open state with the highest probability at a higher concentration, [Ca2+] = 330.0 μM. Thus, calcium concentration modulates stability of CaM-C. We also plot free energy surfaces of CaM-C at the corresponding [Ca2+] (Fig. 3 C and D), which quantitatively show a shift in probabilities for the three states. Moreover, for the CaM-C, increasing Ca2+ concentrations shifts the location of the transition state, implying that the Ca2+ binding can be coupled with the folding processes. A more detailed coupling mechanism is discussed in subsequent sections.
Fig. 3.
(A and B) Representative trajectories for the isolated CaM-C monitored by Qopen for the simulations at 330 K with Ca2+ concentrations of 3.3 μM (A) and 330.0 μM (B). (C and D) Free energy profiles of the isolated CaM-C on the reaction coordinates Qopen and Qclosed at 330 K with Ca2+ concentrations of 3.3 μM (C) and 330.0 μM (D). The dashed lines correspond to the diagonal line of the 2D free energy surfaces. (E and F) Free energy profiles projected on the reaction coordinates Qopen and total binding energy of the two binding sites with Ca2+ concentration of 3.3 μM (E) and 330.0 μM (F) at 330 K for the CaM-C. The unit of the free energy is kBT.
In Fig. 3 A–D, there is no clear population of a partially folded state in which only one of the two EF hands is folded, suggesting high folding cooperativity between the two EF hands, a feature for most small proteins (32). The folding cooperativity can also be observed from the free energy landscapes projected on the reaction coordinates monitoring the folding extent of the component EF hands (Fig. S2), which shows that stability of one EF hand strongly depends on the folding of the other EF hand. Such results accord with previous experimental observations that EF-hand motifs tend to appear in pairs (19).
Interplay Among Folding, Binding, and Allostery.
Next, we try to characterize the detailed coupling mechanisms among folding, Ca2+-binding, and allosteric motions. Fig. 3 E and F shows the free energy profiles projected on Qopen that can monitor folding and allosteric motions and the binding energy that monitors Ca2+ binding, with [Ca2+] = 3.3 μM and [Ca2+] = 330.0 μM for the isolated CaM-C. Representative trajectories projected on the same reaction coordinates are shown in Fig. S3A. As in the previous plots, Qopen takes three peaks representing the unfolded, the closed, and the open conformations. The binding energy also shows three modes representing zero, one, and two calcium-bound states. Together, we observe maximally nine substates that distinguish the Ca2+-binding states and conformational states.
One can see that at a wide range of Ca2+ concentrations, one Ca2+ ion can bind to the CaM domain at the unfolded state with a high probability, suggesting that Ca2+ ions can bind to the CaM domains at the early stage of folding and therefore modulate the folding thermodynamics and pathways. Although maximally nine substates are allowed as discussed above, the events that two Ca2+ ions bind to the unfolded state are very rare with [Ca2+] = 3.3μM (Fig. 3E). As the Ca2+ concentration increases, the probability of Ca2+ binding to the unfolded state increases. Particularly, the state in which two Ca2+ ions bind to the CaM domain at the unfolded state also becomes probable. Similar results can be observed for the N-terminal domain of CaM (CaM-N) (Fig. S4), except that the Ca2+ ions have lower probability to bind to the unfolded state with the same concentration due to the lower Ca2+-binding affinity.
Folding Pathway Coupled with Binding and Allostery.
In terms of the nine states identified in the previous subsection, we next describe the folding pathway network that involves folding, Ca2+ binding, and allosteric motions. For this purpose, starting from random unfolded conformations with no bound Ca2+, we performed 200 independent folding/binding simulations of the CaM domains at 300 K for a range of Ca2+ concentrations, [Ca2+] = 3.3 μM ∼ 3.3 mM. In any of these conditions, the 2 Ca2+-bound open state is the most stable state for the CaM-C, which is set as the final state of the pathway analysis. We counted frequencies of folding/binding pathways from the initial to the final states.
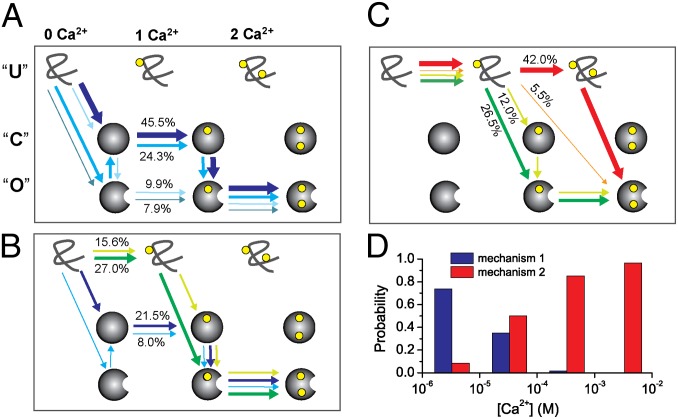
Fig. 4 A–C shows the folding pathways thus obtained for the CaM-C at 300 K with Ca2+ concentration of [Ca2+] = 3.3 μM (Fig. 4A), [Ca2+] = 33.0 μM (Fig. 4B), and [Ca2+] = 330.0 μM (Fig. 4C). The top left state corresponds to the Ca2+-unbound denatured state, set as the initial state, whereas the bottom right state is the 2 Ca2+-bound open state. A sequence of one-colored arrows represents one pathway. First, we see multiple folding/binding pathways coexisting for CaM-C for wide range of Ca2+ concentrations. The multiroute feature of the CaM domains is consistent with the kinetic partitioning picture (33), which was shown to be common for the structurally symmetric proteins and multidomain proteins (25, 33–35).
Fig. 4.
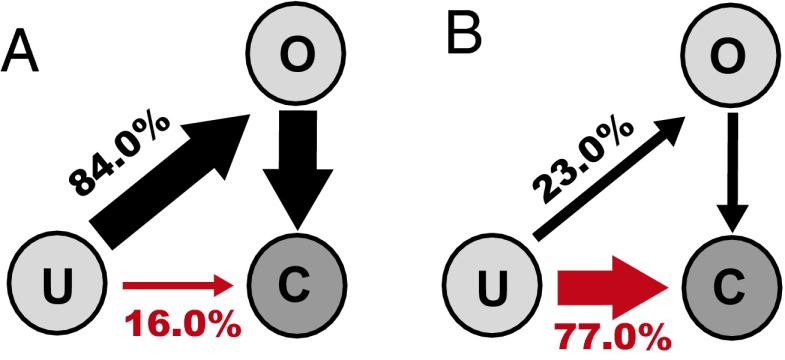
Folding pathways of the CaM-C with the Ca2+ concentration of 3.3 μM (A), 33.0 μM (B), and 330.0 μM (C) at 300 K without pretension. The closed state is represented by a solid sphere, and the open state by a sphere with a notch. The yellow sphere represents Ca2+ ions. The end point of the folding pathway was set as an open state with two Ca2+ ions bound. The probabilities of the pathways are represented by line breadth and percentage numbers, and different pathways are represented by arrow lines with different colors. Only the four most populated pathways are shown. For clarity, the positions of the U state were slightly left shifted. (D) Probability of the folding mechanisms, combined spontaneous folding and induced fit (mechanism 1, blue) and binding-induced folding (mechanism 2, red), at four Ca2+ concentrations. Note that the probability not only includes the pathway with blue (red) color shown, but also includes all other pathways satisfying mechanism 1 (mechanism 2).
At the lowest Ca2+ concentration, [Ca2+] = 3.3 μM (Fig. 4A), the blue pathway is dominant. Along this pathway, CaM-C first folds to the closed state, which is followed by the first Ca2+ binding. This process can be viewed as conformational selection. Then, the first Ca2+ binding induces conformational change to the open state, which is the induced-fit process. The conformational change to the open state is followed by the second Ca2+ binding. This part can be viewed as conformational selection. Thus, one pathway can be viewed as sequential occurrence of conformational selection, induced fit, and conformational selection. The whole folding process may be roughly classified as the combined spontaneous folding and induced-fit mechanism (Fig. S3B).
By increasing the Ca2+ concentration, we see changes in the dominant pathways (Fig. 4 B and C). At [Ca2+] = 33.0 μM (Fig. 4B), the dominant pathway is the green one, in which Ca2+ binds to the unfolded state first, which is followed by folding to the open state. After folding, the second Ca2+ binds to CaM-C. At an even higher concentration (Fig. 4C and Fig. S3B), we see that two Ca2+ ions can bind to the unfolded state along the red pathway. Put simply, at a high Ca2+ concentration, the binding-induced folding mechanism dominates.
In Fig. 4D, we plot probabilities of the above-mentioned two overall folding/binding mechanisms. Clearly, as the Ca2+ concentration increases, mechanisms switch from combined spontaneous folding and induced fit to binding-induced folding. Undoubtedly, due to the reshaping of the two-basin energy landscape by Ca2+ binding, the competition of different folding pathways can be modulated by Ca2+ binding.
Other folding mechanisms can also be possible. For example, the CaM domain may first fold spontaneously to the open (or closed) state and then bind Ca2+ ions directly (or bind Ca2+ ions after converting to the open state), which can be termed as the “conformational selection” mechanism. We also note that similar results were observed for the CaM-N (Fig. S5). We propose that such cofactor-dependent coupling between the folding and allosteric motions can be a general mechanism for the folding of allosteric proteins.
Interplay Between Binding and Allostery at Lower Temperature.
At 300 K and with sufficient Ca2+, CaM-C is thermodynamically stable in the folded state and thus equilibrium fluctuation primarily involves only (un)binding and allosteric motions. Under this condition, we investigate interplay between binding and allosteric motions. For this purpose, we performed MD simulations at 300 K with different Ca2+ concentrations, starting from a folded conformation.
Fig. 5A shows the representative time courses of two Ca2+-binding energies to EF hands (Top and Middle) and Qopen (Bottom) at [Ca2+] = 3.3 μM. Two Ca2+ ions can bind and unbind frequently and the binding of two Ca2+ ions is primarily synchronized, showing binding cooperativity. In addition, the binding of Ca2+ ions is clearly correlated with conformational changes of CaM domains, as shown by the trajectory and the free energy landscape (Fig. 5 A and B). Without Ca2+ binding, the closed state dominates. With the binding of Ca2+ ions, the open state becomes more stable. Particularly, even for the one Ca2+ ion bound state, the open state is much more stable than the closed state. Therefore, binding of one Ca2+ ion can lead to the opening of the whole CaM domain, suggesting cooperativity between the two EF hands. Such cooperativity may be of biological importance because it can reduce the threshold of Ca2+ concentration for inducing the conformational changes of the whole protein (19). We note that even without Ca2+ binding, the open state can be accessed with a small probability (Fig. 5B, Left), which is the restraint from experimental data in the model parameterization (SI Text).
Fig. 5.
(A) Representative trajectory for the isolated CaM-C monitored by binding energies of the two binding sites (Top, EF-hand 1; Middle, EF-hand 2) and Qopen (Bottom) at 300 K with Ca2+ concentration of 3.3 μM. (B) Free energy profile of the reaction coordinates Qopen and total binding energy (Left) and the projection of the trajectory on the same reaction coordinates (Right). (C) The probabilities of the induced-fit (IF, open square) and conformational selection (CS, solid square) mechanisms for the allosteric transitions from closed to open state due to the binding of the first Ca2+ (black) and the second Ca2+ (red). The conformational transition events from the closed state with 0 Ca2+ bound (1 Ca2+ bound) to the open state with 1 Ca2+ bound (2 Ca2+ bound) were collected in calculating the allosteric mechanism due to the binding of the first (second) Ca2+.
To characterize the mechanism of allosteric motions in depth, we replotted the same trajectory on the 2D conformational space (Fig. 5B, Right). One can see that at the current calcium concentration, the first Ca2+ preferentially binds to the closed state, inducing the conformational transition to the open state, which corresponds to the “induced-fit” mechanism. On the contrary, the second Ca2+ dominantly binds to the open state, possibly due to the tight coupling between the two EF hands, suggesting the “conformational selection” mechanism. To get a more quantitative understanding, we conducted 200 independent MD simulations at each calcium concentration, starting from the closed state without calcium binding (Fig. 5C). The results show that for the binding of the first Ca2+, the induced-fit mechanism dominates for a wide range of calcium concentrations. In comparison, for the binding of the second Ca2+, at relatively low [Ca2+], the conformational selection mechanism dominates. With increasing [Ca2+], the conformational selection mechanism becomes less and less probable. Particularly, at very high [Ca2+], i.e., 3.3 mM, the induced-fit mechanism dominates. All these results demonstrate that the mechanism of allosteric motions can be modulated by the calcium concentrations. We note that modulation of the allostery mechanism by ligand concentration has also been demonstrated by flux analysis of NMR data for other two allosteric proteins (36).
Interplay Between Folding and Allostery with No Calcium.
Next, focusing on the interplay between folding and allosteric motions, we performed folding simulations at 300 K with no calcium. Without calcium binding, the closed state is the most stable state. Then, we investigated folding pathways from the unfolded state to the closed state; the results are summarized in Fig. 6. We see that even without Ca2+ binding, the pathway via the transient open state has significant populations. Interestingly, for the isolated CaM-N, a “U” → “O” → “C” pathway even becomes dominant. Because the metastable open state represents a trap along the folding pathway to the most stable closed state, these results may indicate that allostery introduces frustrations to folding processes. The above observation is consistent with a recent suggestion by Wolynes and coworkers that the allostery requirement may introduce frustrations to the energy landscape (37). Such frustrations can inevitably be reflected by the folding pathways.
Fig. 6.
(A and B) Folding pathways on the conformational space formed by three states, i.e., unfolded state (U), closed state (C), and open state (O), with the Ca2+ concentration of 0 for the isolated CaM-N (A) and CaM-C (B) at 300 K.
The above results show that the isolated CaM-N demonstrates more significant frustrations than the isolated CaM-C. Such asymmetry is not surprising because the two domains have some differences in both sequence and structure. Equilibrium MD simulations with single-basin energy functions constructed based on the experimental structures at closed and open states, respectively, showed that the open structure is kinetically more easily accessible (lower folding barrier) than the closed structure for the CaM-N (Fig. S6). Consequently, the populations of folding pathways in the CaM-N are more resistant to the change of relative stability between the two states. In comparison, the folding barriers to two end structures are comparable for the CaM-C. Such differences can lead to the observed difference in folding behaviors of the two domains. We conducted the same type of folding simulations at 300 K as well, finding that both domains fold directly to the closed state without passing through the metastable open state.
Full-Length Calmodulin Under Constant Mechanical Extension.
Finally, we also conducted simulations for the full-length CaM with a wide range of mechanical extensions. Our results show that, as the separation increases, the population of the open (closed) state increases (deceases) for both domains (Fig. S7), which suggests that the tension between terminal residues can modulate the relative populations of the functional states. A possible reason for the tension-induced population shift is that the terminal residues of the open state have larger separation than that of the closed state. Consequently, tension shifts the populations to the open state. We also observed that once the CaM-C becomes unfolded, the CaM-N shifts its population to the closed state to some extent due to the release of tension between the terminal residues by CaM-C unfolding. This demonstrates the tension-induced coupling between the two domains. The effect of tension on the populations of the conformational states, and therefore the Ca2+-binding affinity, can also be observed for the isolated domains (Fig. S7). In addition, our results show that the intrinsic folding mechanism of the CaM domain can be modulated by applying tensions (Fig. S7).
Comparison with Recent Single-Molecule Experiments.
The results presented in previous subsections are highly compatible with available experimental data. For example, the multiple-state fluctuations observed in MD simulations can be supported by recent single-molecule force spectroscopy experiments. In ref. 23, the authors identified two folded conformations with significantly different lifetimes at low Ca2+ concentration for the isolated CaM-C, although the force trajectories show typical two-state folding/unfolding transitions. The conformation with a long lifetime was assigned as the holo state, and that with a short lifetime was assigned as the apo state, suggesting the coexistence of the unfolded, apo and holo conformations. Our simulation results also support the experimental assignment (Fig. S8).
In addition, the Ca2+-regulated multiroute folding/binding pathways revealed in this work are consistent with the Ca2+-dependent folding kinetics of CaM domains measured by a recent single-molecule experiment. In ref. 23, by fitting the transition-state model of protein folding, the authors proposed that at low Ca2+ concentration, the folding of CaM domains precedes the Ca2+ binding. At high Ca2+concentration, the pathway with Ca2+ bound to the denatured state becomes dominant. The results of our MD simulations not only accord with the experimental data, but also provide direct time-resolved structural evidence for the experimentally proposed model. In addition, we provide previously unidentified information on the Ca2+-dependent coupling between folding and allosteric motions.
Although it is a big challenge, it will be very interesting to experimentally examine the coupling mechanisms among folding, cofactor binding, and allostery revealed in this work. Especially, with the currently available single-molecule force spectroscopy technique, it is possible to measure the force-modulated folding/allostery mechanisms and the tension-induced interdomain coupling observed in the MD simulations.
Conclusion
In summary, we have developed a computational model that can integrate folding, cofactor binding, and allosteric motions. Using this model, we revealed the tight coupling among the above three processes for the CaM domains. Particularly, our results showed that the allosteric feature of the energy landscape tends to introduce new folding pathways, therefore adding complexity to folding processes. Moreover, the folding/allostery can be further modulated by Ca2+ binding. As the Ca2+ concentration increases, the folding mechanism switches from combined spontaneous folding and induced fit to binding-induced folding. Finally, the intrinsic folding/allosteric mechanism can be modulated by applying mechanical force, which leads to interdomain coupling of the full-length CaM.
Materials and Methods
Integrated Modeling of Folding, Binding, and Allosteric Motions.
To study the folding of allosteric proteins, a model that can simultaneously describe the folding, ligand binding, and allosteric motions is necessary. In our previous work, within the framework of perfect-funnel approximation (27, 38), we developed an atomic interaction-based coarse-grained model with flexible local potential (AICG2) by using a multiscale strategy (25, 26, 39). The AICG2 model can reasonably describe the physiochemical features and chain flexibility of proteins and was proved to be successful in modeling the folding of topologically complex proteins (25). After that work, we noticed that folding simulations with the AICG2 model could lead to mirror-imaged misfolds for some proteins with a small probability. To solve this issue, in this work, we improved the AICG2 model (termed as AICG2+) and used local dihedral angles rather than the distance in the local potential to better discriminate the mirror images (SI Text). Then, we constructed a double-basin Hamiltonian by interpolating the two AICG2+ energy functions built from the closed and open structures of isolated CaM domains, respectively, using the framework developed in ref. 28. Similar models with a double-basin Hamiltonian have also been developed and successfully used in studying the allosteric motions of a number of proteins (12, 15, 40–43). The Ca2+ ions were considered implicitly (29, 41), and the effects of Ca2+ binding were modeled by strengthening the contacting interactions formed by the liganding residues of Ca2+ ions at the open state. Although the precise description of energetic consequences of the coordination events of ligand binding needs more sophisticated quantum mechanics methods and an atomically detailed structural model, the available experimentally measured binding affinities and populations of conformational states provide a reasonable parameterization of the energies (Fig. S9) (7, 16, 23, 44–46). With such an integrated model, we can concisely describe the folding, ligand binding, and allosteric motions in an integrated way. More details on the model are given in SI Text.
Reaction Coordinates.
The reaction coordinates Qopen and Qclosed, which were defined as the fractions of native contacts of the CaM domains with the open and closed states used as the native structures, respectively, were used to describe the folding of the CaM domain. In describing the binding of Ca2+ ions, the binding energy of the two binding sites was used as the reaction coordinates. More details are given in SI Text.
Simulation Details.
We used CafeMol 2.0 (47) for the CG molecular simulations. The all-atom MD simulations were conducted by AMBER11 (48). The protein structures were visualized by PyMOL (49). Details are in SI Text.
Supplementary Material
Acknowledgments
This work was based on a joint project between the Japan Society for the Promotion of Science and the Natural Science Foundation of China, partly supported by the Natural Science Foundation of China (Grants 11174134, 11334004, and 81121062) and Jiangsu Province (BK2011546). W.L. also thanks the support by Program for New Century Excellent Talents in University and High Performance Computing Center of Nanjing University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402768111/-/DCSupplemental.
References
- 1.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Zhuravlev PI, Papoian GA. Protein functional landscapes, dynamics, allostery: A tortuous path towards a universal theoretical framework. Q Rev Biophys. 2010;43(3):295–332. doi: 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]
- 3.Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: Shifts in energy landscapes. Proc Natl Acad Sci USA. 1999;96(18):9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy Y, Cho SS, Onuchic JN, Wolynes PG. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J Mol Biol. 2005;346(4):1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 5.Chung HS, McHale K, Louis JM, Eaton WA. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335(6071):981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Eldik LJ, Watterson DM. Calmodulin and Signal Transduction. 1st Ed. New York: Academic; 1998. [Google Scholar]
- 7.Kuboniwa H, et al. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2(9):768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992;228(4):1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol. 1995;2(9):758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 10.Stigler J, Ziegler F, Gieseke A, Gebhardt JC, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334(6055):512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 11.Slaughter BD, et al. Sampling unfolding intermediates in calmodulin by single-molecule spectroscopy. J Am Chem Soc. 2005;127(34):12107–12114. doi: 10.1021/ja0526315. [DOI] [PubMed] [Google Scholar]
- 12.Zuckerman DM. Simulation of an ensemble of conformational transitions in a united-residue model of calmodulin. J Phys Chem B. 2004;108:5127–5137. [Google Scholar]
- 13.Park HY, et al. Conformational changes of calmodulin upon Ca2+ binding studied with a microfluidic mixer. Proc Natl Acad Sci USA. 2008;105(2):542–547. doi: 10.1073/pnas.0710810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Liang KC, Czader A, Waxham MN, Cheung MS. The effect of macromolecular crowding, ionic strength and calcium binding on calmodulin dynamics. PLoS Comput Biol. 2011;7(7):e1002114. doi: 10.1371/journal.pcbi.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YG, Hummer G. Slow conformational dynamics and unfolding of the calmodulin C-terminal domain. J Am Chem Soc. 2007;129(9):2414–2415. doi: 10.1021/ja067791a. [DOI] [PubMed] [Google Scholar]
- 16.Malmendal A, Evenäs J, Forsén S, Akke M. Structural dynamics in the C-terminal domain of calmodulin at low calcium levels. J Mol Biol. 1999;293(4):883–899. doi: 10.1006/jmbi.1999.3188. [DOI] [PubMed] [Google Scholar]
- 17.Rabl CR, Martin SR, Neumann E, Bayley PM. Temperature jump kinetic study of the stability of apo-calmodulin. Biophys Chem. 2002;101–102:553–564. doi: 10.1016/s0301-4622(02)00150-3. [DOI] [PubMed] [Google Scholar]
- 18.Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur J Biochem. 1995;230(3):1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 19.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405(2):199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi C, Takada S. Protein grabs a ligand by extending anchor residues: Molecular simulation for Ca2+ binding to calmodulin loop. Biophys J. 2006;90(9):3043–3051. doi: 10.1529/biophysj.105.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi S, Portman JJ. Inherent flexibility and protein function: The open/closed conformational transition in the N-terminal domain of calmodulin. J Chem Phys. 2008;128(20):205104. doi: 10.1063/1.2928634. [DOI] [PubMed] [Google Scholar]
- 22.Tripathi S, Portman JJ. Inherent flexibility determines the transition mechanisms of the EF-hands of calmodulin. Proc Natl Acad Sci USA. 2009;106(7):2104–2109. doi: 10.1073/pnas.0806872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stigler J, Rief M. Calcium-dependent folding of single calmodulin molecules. Proc Natl Acad Sci USA. 2012;109(44):17814–17819. doi: 10.1073/pnas.1201801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323(5914):633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Terakawa T, Wang W, Takada S. Energy landscape and multiroute folding of topologically complex proteins adenylate kinase and 2ouf-knot. Proc Natl Acad Sci USA. 2012;109(44):17789–17794. doi: 10.1073/pnas.1201807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Wolynes PG, Takada S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc Natl Acad Sci USA. 2011;108(9):3504–3509. doi: 10.1073/pnas.1018983108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21(3):167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 28.Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103(32):11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki K, Takada S. Dynamic energy landscape view of coupled binding and protein conformational change: Induced-fit versus population-shift mechanisms. Proc Natl Acad Sci USA. 2008;105(32):11182–11187. doi: 10.1073/pnas.0802524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoldák G, Rief M. Force as a single molecule probe of multidimensional protein energy landscapes. Curr Opin Struct Biol. 2013;23(1):48–57. doi: 10.1016/j.sbi.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Balamurali MM, Sharma D, Li H. A functional single-molecule binding assay via force spectroscopy. Proc Natl Acad Sci USA. 2007;104(40):15677–15681. doi: 10.1073/pnas.0705367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan HS, Shimizu S, Kaya H. Cooperativity principles in protein folding. Methods Enzymol. 2004;380:350–379. doi: 10.1016/S0076-6879(04)80016-8. [DOI] [PubMed] [Google Scholar]
- 33.Klimov DK, Thirumalai D. Symmetric connectivity of secondary structure elements enhances the diversity of folding pathways. J Mol Biol. 2005;353(5):1171–1186. doi: 10.1016/j.jmb.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 34.Noé F, Schütte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci USA. 2009;106(45):19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirchi M, et al. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat Commun. 2011;2:493. doi: 10.1038/ncomms1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: A flux description of reaction mechanism. Proc Natl Acad Sci USA. 2009;106(33):13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. On the role of frustration in the energy landscapes of allosteric proteins. Proc Natl Acad Sci USA. 2011;108(9):3499–3503. doi: 10.1073/pnas.1018980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298(5):937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Yoshii H, Hori N, Kameda T, Takada S. Multiscale methods for protein folding simulations. Methods. 2010;52(1):106–114. doi: 10.1016/j.ymeth.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Lu Q, Wang J. Single molecule conformational dynamics of adenylate kinase: Energy landscape, structural correlations, and transition state ensembles. J Am Chem Soc. 2008;130(14):4772–4783. doi: 10.1021/ja0780481. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Tang C, Wang E, Wang J. Exploration of multi-state conformational dynamics and underlying global functional landscape of maltose binding protein. PLoS Comput Biol. 2012;8(4):e1002471. doi: 10.1371/journal.pcbi.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maragakis P, Karplus M. Large amplitude conformational change in proteins explored with a plastic network model: Adenylate kinase. J Mol Biol. 2005;352(4):807–822. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 43.Whitford PC, Miyashita O, Levy Y, Onuchic JN. Conformational transitions of adenylate kinase: Switching by cracking. J Mol Biol. 2007;366(5):1661–1671. doi: 10.1016/j.jmb.2006.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca2+-calmodulin reveals flexible hand-like properties of its domains. Nat Struct Biol. 2001;8(11):990–997. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 45.Bayley PM, Findlay WA, Martin SR. Target recognition by calmodulin: Dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5(7):1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linse S, Helmersson A, Forsén S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266(13):8050–8054. [PubMed] [Google Scholar]
- 47.Kenzaki H, et al. CafeMol: A coarse-grained biomolecular simulator for simulating proteins at work. J Chem Theory Comput. 2011;7(6):1979–1989. doi: 10.1021/ct2001045. [DOI] [PubMed] [Google Scholar]
- 48.Case DA, et al. AMBER11. San Francisco: Univ of California; 2010. [Google Scholar]
- 49.DeLano WL. The PyMOL Molecular Graphics System. 2002 (DeLano Scientific, San Carlos, CA) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.