Significance
The kinesin family member 2C (KIF2C) that regulates microtubule depolymerization, is up-regulated in a number of human cancer cells in a Ras- and ERK1/2-dependent manner. In this study, we find that KIF2C expression positively regulates signaling pathways downstream of Ras, ERK1/2, and mechanistic target of rapamycin complex 1 in nontransformed cells to coordinate a cellular nutrient response. Transformation by oncogenic Ras overrides the need for this level of organization and repurposes ERK1/2 and KIF2C signaling events. Therefore, this study provides a link for how cytoskeletal transformation may interconnect with cancer metabolism.
Abstract
The kinesin family members (KIFs) KIF2A and KIF2C depolymerize microtubules, unlike the majority of other kinesins, which transport cargo along microtubules. KIF2A regulates the localization of lysosomes in the cytoplasm, which assists in activation of the mechanistic target of rapamycin complex 1 (mTORC1) on the lysosomal surface. We find that the closely related kinesin KIF2C also influences lysosomal organization in immortalized human bronchial epithelial cells (HBECs). Expression of KIF2C and, to a lesser extent, KIF2A in untransformed and mutant K-Ras–transformed cells is regulated by ERK1/2. Prolonged inhibition of ERK1/2 activation with PD0325901 mimics nutrient deprivation by disrupting lysosome organization and decreasing mTORC1 activity in HBEC, suggesting a long-term mechanism for optimization of mTORC1 activity by ERK1/2. We tested the hypothesis that up-regulation of KIF2C and KIF2A by ERK1/2 caused aberrant lysosomal positioning and mTORC1 activity in a mutant K-Ras–dependent cancer and cancer model. In Ras-transformed cells, however, mTORC1 activity and lysosome organization appear independent of ERK1/2 and these kinesins although ERK1/2 activity and the kinesins are required for Ras-dependent proliferation and migration. We conclude that mutant K-Ras repurposes these signaling and regulatory proteins to support the transformed phenotype.
Complex signaling pathways are engaged to coordinate cellular growth and proliferation as dictated by stimulatory signals, such as abundant nutrients and growth factors, or inhibitory signals, such as limiting nutrients or other cell stresses. Disturbances in coordination of signaling events are often observed in pathologies such as cancer (1). Some cancers use oncogenic mutations of the small GTPase K-Ras to promote their survival and proliferation (2–6). Mutant K-Ras activates central signaling cascades, among them the phosphatidylinositol 3-kinase (Ras-PI3K) and the extracellular signal-regulated kinase ERK1/2 (Ras-ERK) pathways, to facilitate continued proliferation under suboptimal conditions and bypass inhibitory signals from energy stress pathways.
Mechanistic target of rapamycin (mTOR), a serine/threonine protein kinase in a PI3K-related family, is a central regulator of events leading to cell growth and proliferation (7–12). It carries out this role primarily as part of a complex [mTOR complex 1 (mTORC1)], and, in this capacity, it is exquisitely responsive to nutrient availability. In certain cancers, positive regulatory signals to mTORC1 can originate from mutant K-Ras, even in nutrient-poor settings (13, 14). Because cancers often use mTORC1 to optimize cellular functions to promote growth and survival, we have compared how the Ras-ERK pathway influences nutrient sensing and mTORC1 in untransformed immortalized cells and in cancer cells.
Kinesins are motor proteins that transport cargo along microtubules. Currently, more than 40 kinesins have been found in mammals, most of which have primary functions as cargo carriers. Kinesin family member (KIF) 2A is one of a handful of kinesins that apparently do not transport cargo, but instead depolymerize microtubules. In HeLa cells, KIF2A was shown to regulate the location of lysosomes in the cytoplasm, which is thought to be essential for mTORC1 activation (15–18). We find that KIF2A and the closely related kinesin KIF2C (also known as MCAK) also control lysosomal organization in human bronchial epithelial cells.
We recently determined that expression of KIF2C and KIF2A is up-regulated by the Ras-ERK pathway (19). Therefore, we asked whether ERK1/2 and these kinesins are required to maintain lysosomal organization in immortalized human bronchial epithelial cells (HBECs) and also whether the elevated expression of KIF2C and KIF2A accounts for the nutrient insensitivity of lysosomal positioning and mTORC1 activity in a mutant K-Ras–dependent cancer and cancer model.
Results
KIF2A and KIF2C Are Required for the Amino Acid-Dependent Localization of mTORC1 to Lysosomes in Immortalized HBEC.
To explore the relationships among KIF2A and KIF2C expression, ERK1/2, and lysosomal organization, we used a model system to assess signaling differences between immortalized cells before and after transformation under relatively well-defined conditions. We examined the characteristics of a nontumor human bronchial epithelial cell line (HBEC) that was immortalized by expression of cyclin-dependent kinase 4 (CDK4) and telomerase (hTERT), yielding HBEC3KT (number distinguishes different cell donors) as described (20). These cells maintain epithelial morphology and express E-cadherin but not N-cadherin (19). We compared the behavior of this immortalized untransformed cell line to an isogenic non-small cell lung cancer (NSCLC) model that was generated by introducing mutant K-Ras and a stable knockdown of p53 to yield HBEC3KTRL53 (21, 22). We also examined the patient-derived colorectal cancer cell HCT116, which has an oncogenic K-Ras mutation.
To examine the effects of nutrient restriction, cells were placed in Earl’s balanced salt solution (EBSS), which contains glucose but lacks serum and amino acids. Because mTORC1 is, at least in part, activated through interaction with Rag small GTPases and the Ragulator complex on lysosomes (16), we examined how removal of amino acids and serum affected lysosomes and mTOR localization. After 60–90 min of starvation in EBSS, lysosomes in HBEC3KT lost the typical perinuclear concentrated pattern and instead gained a more diffuse and symmetric distribution, similar to what has been observed in HEK293 and HeLa cells following removal of amino acids (15, 16, 23) (Fig. 1 A and B). mTOR was also dispersed and less concentrated on lysosomes in HBEC3KT cells, and mTORC1 activity was also reduced (Fig. 1C), as measured by a decrease in phosphorylation of ribosomal protein S6. ERK1/2 were also less active after an hour in EBSS. Readdition of amino acids or complete medium with amino acids and serum reactivated both ERK1/2 and mTORC1 (Fig. 1C).
Fig. 1.
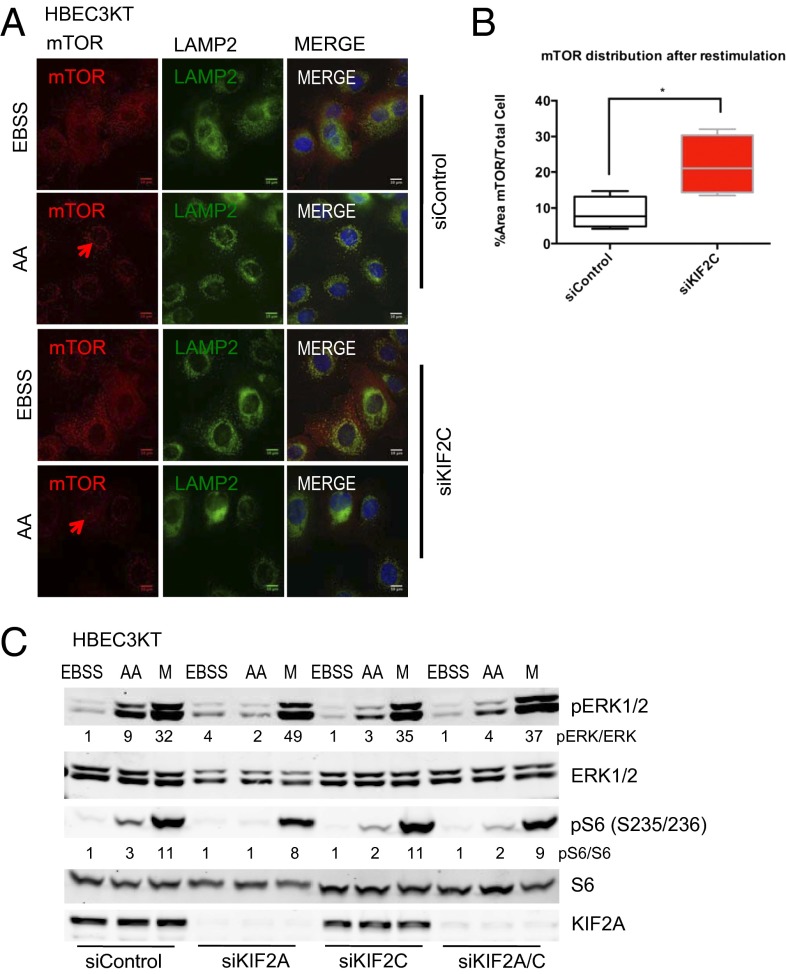
KIF2C influences lysosomal organization and mTORC1 activity in HBEC3KT cells. (A) HBEC3KT cells were treated with control or KIF2C siRNA twice 48 h apart for a total of 96 h. Cells were then starved for 90 min and restimulated with amino acids (combination of each amino acid contained in l-glutamine-free DMEM plus 0.5 mM l-glutamine) for 30 min. Cells were coimmunostained for endogenous mTOR (red) and endogenous LAMP2 (green). (B) The eight-bit grayscale images from Fig. 1A were analyzed after thresholding to measure the area of mTOR localization after restimulation with amino acids by obtaining the ratio of mTOR staining per whole-cell area. (C) Cells were starved treated as above with control, KIF2C, or KIF2A siRNA, starved, and then stimulated with amino acids (AA) or fresh medium with serum (M). Lysate proteins were resolved on gels, transferred to membranes, and immunoblotted with antibodies to the indicated proteins.
Knockdown studies revealed that the microtubule-depolymerizing kinesin KIF2A is required for lysosomal organization and mTORC1 positioning on lysosomes (15). We recently found that KIF2A and the closely related kinesin KIF2C are both up-regulated in many cancers and that their persistent expression is supported by oncogenic K-Ras and ERK1/2 (19). As previously found in HeLa and 293 cells, depletion of KIF2A from HBEC3KT prevented typical lysosomal organization, and mTOR also remained diffuse and less well-localized with lysosomes (Fig. S1 A, B, and F). Because KIF2C has many functions that overlap with KIF2A (19), we tested its effects as well and found that depletion of KIF2C also prevented restoration of lysosomal organization or mTOR activity by amino acids (Fig. 1 A–C and Fig. S1 D and G), suggesting that more than one kinesin participates in lysosomal positioning. Although KIF2A siRNA and KIF2C siRNA caused a reduction in mTORC1 activity in amino acid-replete cells, knockdown of either or both had little or no effect on mTORC1 activity in cells in serum-containing medium (Fig. 1C and Fig. S1 F and G). ERK1/2 activation in HBEC3KT was also reduced following KIF2A or KIF2C siRNA.
Inhibition of the ERK1/2 Pathway Alters Lysosome Positioning and Reduces mTORC1 Activity in Immortalized Cells.
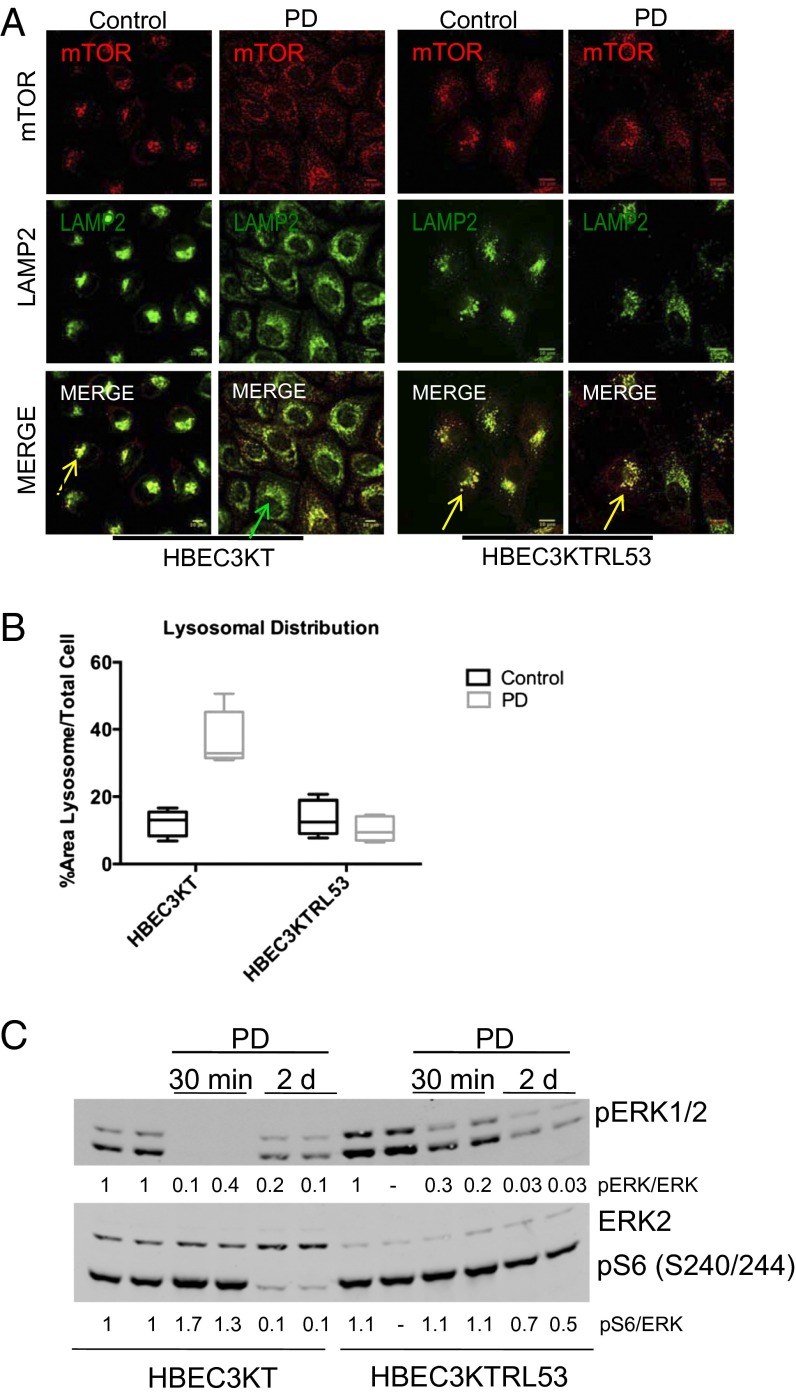
mRNAs encoding KIF2C and, to a lesser extent, KIF2A are decreased in both HBEC3KT and HBEC3KTRL53 by extended exposure to PD0325901, which inhibits the enzymes MEK1 and MEK2 that activate ERK1/2 (19). These and other studies indicated that ERK1/2 stimulate KIF2A/C expression. Therefore, we explored the possibility that ERK1/2 may also influence organelle organization in immortalized cells through their ability to control the expression of proteins that alter microtubule dynamics including these kinesins. We inhibited ERK1/2 activation by exposure of cells to the MEK inhibitor PD0325901 briefly or for up to 2 d. We showed previously that serum can stimulate ERK1/2 immediately after washing out PD0325901 even if cells have been treated with the inhibitor for 2 d (19). In HBEC3KT, prolonged blockade of the ERK pathway with the MEK inhibitor had significant effects on mTOR and LAMP2 localizations, causing dispersal of both (Fig. 2 A and B). mTORC1 activity was not affected by inhibition of ERK1/2 activation for 30 min but was almost completely inhibited in cells treated with the MEK inhibitor for 2 d (Fig. 2C), suggesting that inhibition of mTORC1 required changes in expression of protein. Similar observations were made in HBEC30KT, a lung cell line from a different donor (Fig. S2A) and in H358, another cancer cell line (Fig. S2B).
Fig. 2.
Inhibition of ERK1/2 inhibits mTORC1 in immortalized bronchial epithelial cells but not in Raf-transformed cells. (A) HBEC3KT and HBEC3KTRL53 cells were treated for 2 d with 100 nM PD0325901 and immunostained as in Fig. 1A. (B) Images were analyzed as in Fig. 1 to measure the area of lysosomal distribution (LAMP2 staining) by obtaining the ratio of the organelle area per whole-cell area. (C) HBEC3KT and HBEC3KTRL53 cells were treated for 30 min or 2 d with 100 nM PD0325901. Lysate proteins were immunoblotted with the indicated antibodies.
To confirm that these effects on mTORC1 and lysosomes were comparable with those triggered by pathways that respond to limiting nutrients, we activated the AMP-activated protein kinase AMPK with the AMP-like molecule AICAR (5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside). The activities of AMPK and mTORC1 are intertwined on multiple levels (11, 24–29). AMPK can inhibit mTORC1 and can induce autophagy to aid in replenishing nutrients. Inhibition of mTORC1 as a consequence of AMPK activation occurred in HBEC3KT as early as 6 h after treatment with AICAR (Fig. S3 A and B). Metformin, which activates AMPK by inhibition of complex I of the respiratory chain and perhaps other mechanisms (30), also inhibited mTORC1 in HBEC3KT, but to a lesser extent in HBEC3KTRL53 (Fig. S3 A–C). Exposure to metformin also appeared to disperse mTOR and lysosomes in HBEC3KT, but not in HBEC3KTRL53 (Fig. S3C). These data are consistent with the idea that oncogene-induced transformation prevents a comprehensive shutdown of signaling pathways coordinated in normal cells in response to nutrient limitation.
Cancer-Cell Transformation Unlinks the Nutrient Sensitivity of mTOR Localization with Lysosomes.
Fast growth and limited vascularization are just some of the environmental hurdles that provide a challenge to cancer-cell survival. Signaling in cancer cells must be altered compared with normal cells to adjust to the high demand for nutrients and other stresses. In HCT116 or HBEC3KTRL53 in EBSS, mTOR remained concentrated in the proximity of the lysosomal marker LAMP2 (Fig. 3A). In nutrient-rich medium, these cells exhibited a more dispersed lysosomal phenotype than untransformed cells. Lysosomes in both cell types showed a range of changes in organization but did not acquire the morphology typical of starved HBEC3KT. ERK1/2 were inactivated as quickly as 15 min following transfer of HBEC3KT to EBSS (Fig. 3B); mTORC1 was inactivated within 45 min in EBSS. On the other hand, in HBEC3KTRL53 or HCT116 cells, ERK1/2 phosphorylation was not reduced after an hour in EBSS, and relatively small decreases in mTORC1 were observed (Fig. 3 B and C).
Fig. 3.
Cancer cells with K-Ras mutations are less sensitive to starvation than normal cells. (A) HBEC3KTRL53 and HCT116 cells were starved in EBSS for 30 min or 120 min, as indicated, and coimmunostained as in Fig. 1A. (B) HBEC3KT and HBEC3KTRL53 cells were starved in EBSS for 0 min, 30 min, or 60 min. Lysate proteins were resolved on gels followed by immunoblotting with the indicated antibodies. (C) HBEC3KT and HCT116 cells were starved in EBSS for the specified times and immunoblotted as above.
The mTOR localization pattern in HBEC3KTRL53 was also not substantially altered by prolonged inhibition with the MEK inhibitor PD0325901, nor was mTORC1 activity greatly reduced by the drug, suggesting that oncogenic transformation overrides the changes in organelle compartments required for signaling in normal cells after growth pathway inhibition (Fig. 2 A–C and Fig. S1 C and E). Also noted, ERK2 expression relative to ERK1 is decreased in these cells, most clearly observed using an ERK2-specific antibody. These findings are consistent with the idea that association of mTOR with lysosomes permits mTOR to maintain an activated state in cancer cells even in nutrient-poor and growth pathway-inhibited conditions (Fig. S2C). Inhibition of MEK/ERK also had no effect on mTORC1 in H358 lung-cancer cells that have oncogenic K-Ras mutation as well as loss of p53 (Fig. S3B).
In HBEC3KT, mTORC1 could be inhibited by various means, including starvation, inhibition of growth pathways, energy stress, and loss of the kinesins KIF2A or KIF2C. Because HBECKTRL53 and HCT116 cells were resistant to mTORC1 inhibition by any of these interventions, we hypothesized that overriding mechanisms mediated by oncogenic transformation contributed to the capacity of these cells to withstand these challenges. Activation of growth-factor pathways may provide more options to overcome amino acid depletion to maintain mTORC1 activity. To test this idea, we asked whether normal cells would be resistant to mTORC1 down-regulation due to loss of these kinesins in the presence of abundant nutrients and growth factors. Following KIF2A or KIF2C siRNA in HBEC3KT, starved cells were restimulated with medium containing amino acids and serum. In contrast to the results observed with amino acids alone, restimulation with amino acids plus serum activated mTORC1 even in cells with reduced KIF2A and KIF2C expression (M lanes in Fig. 1C). Consistent with these results, mTOR was more localized to lysosomes in the presence of serum plus amino acids even following loss of KIF2A or KIF2C in both HBEC3KT and HBEC3KTRL53 (Fig. S1H). Loss of KIF2A or KIF2C was dispensable for restimulation of ERK1/2 and mTORC1 in HBEC3KT or HBEC3KTRL53 by EGF, suggesting that KIF2A/C may be important for signaling through amino acid-regulated pathways but not in the presence of high concentrations of growth factors (Fig. S4 A–C).
Elevated Expression of KIF2A and KIF2C Increased mTORC1 and ERK1/2 Activity in HBEC and HeLa Cells.
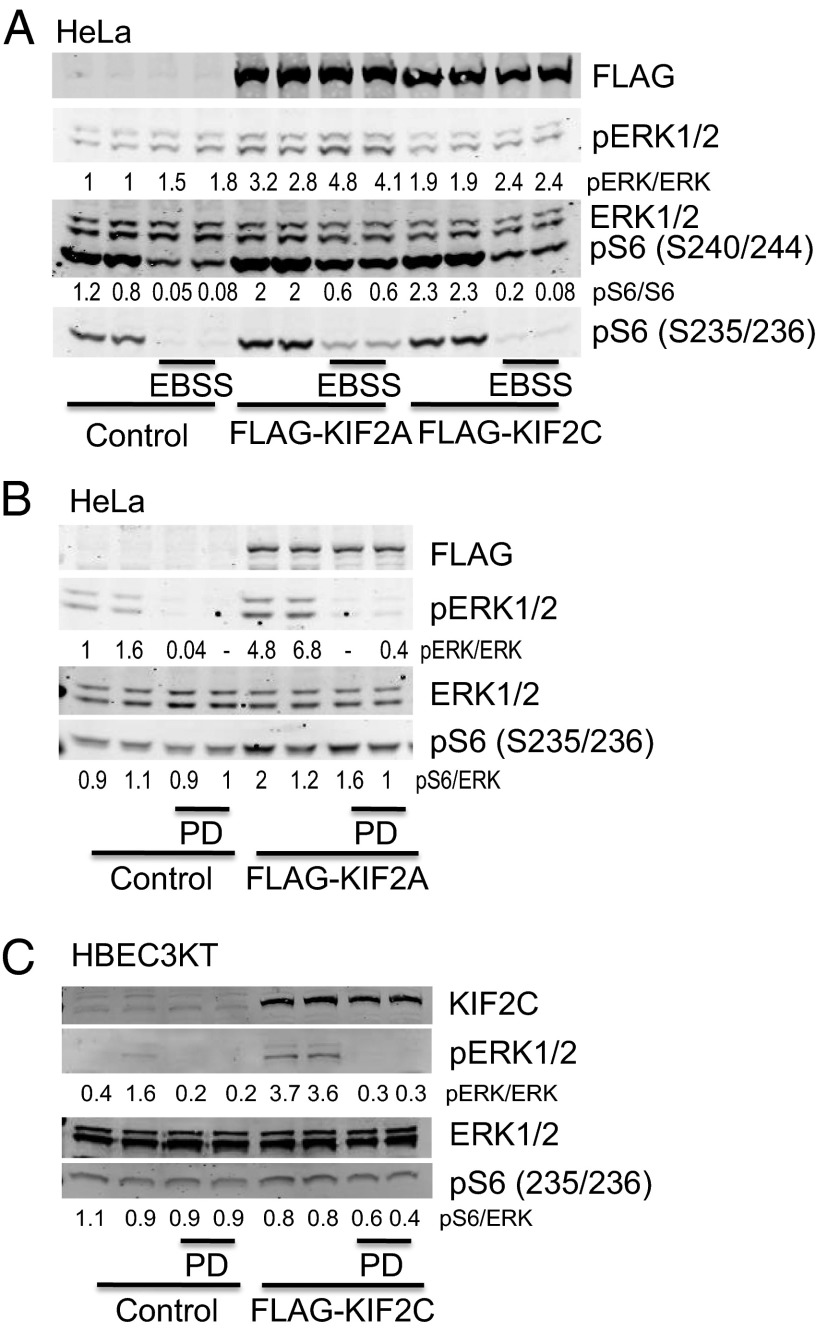
In HBEC3KTRL53, loss of KF2A or KIF2C had little effect on the localization of lysosomes or mTOR whether or not amino acids were present. We examined heterologous expression of KIF2C in HBEC3KT. Because KIF2A expresses poorly in HBEC cells, we also examined the effects of forced KIF2A/C expression in HeLa cells. Expression of KIF2A and KIF2C increased mTORC1 and ERK1/2 activities in cells in medium or EBSS (Fig. 4 A–C). These results support the idea that changes in KIF2A/C expression affect the lysosomal compartment and mTOR activity.
Fig. 4.
KIF2A and KIF2C overexpression in HeLa and HBEC3KT activates pERK and pS6. (A) FLAG-KIF2A were expressed in HeLa cells that were starved in EBSS or for 90 min. (B) Cells expressing FLAG-KIF2A were treated with PD0325901 for 90 min. (C) FLAG-KIF2C was expressed in HBEC3KT cells that were treated with PD0325901 for 90 min. Lysate proteins were resolved on gels, followed by immunoblotting with the indicated antibodies.
Effects of Depletion of K-Ras and KIF2C on Autophagy.
Reduced mTORC1 activity often results in autophagy to replenish nutrients needed for survival. mTORC1 usually inhibits autophagy under conditions in which nutrients such as glucose and amino acids or growth factors are abundant by phosphorylating the kinase ULK1, which is involved in autophagosome membrane formation (31). One indicator of accelerated autophagy is the conversion of the ubiquitin-like protein LC3-I to its lipidated form, LC3-II, which accumulates on autophagosome membranes (32, 33). LC3-II was increased in HBEC3KTRL53 (Fig. 5A). Because a number of Ras functions are executed by PI3K, we tested the extent to which this pathway contributes to sustained autophagy. Accumulation of LC3-II was decreased by LY294002, which inhibits the PI3K pathway, and more strongly by the MEK inhibitor PD0325901, suggesting that oncogenic Ras can promote autophagy through both of these effectors (Fig. S5A). We hypothesized that depleting mutant K-Ras would reverse the autophagy observed in K-Ras–transformed cells. Instead, autophagy was further enhanced by K-Ras knockdown, as assessed by loss of p62, a marker of autophagic flux, which is sequestered by autophagosomes and degraded in lysosomes when autophagy is induced (Fig. 5B and Fig. S5B) (34, 35). Because loss of KIF2A was suggested to induce autophagy, as a consequence of decreased mTORC1 activity (15), we tested whether loss of KIF2C also promotes autophagy. Even in HBEC3KTRL53, where loss of KIF2C had little effect on mTORC1 activity, we observed an enhancement of autophagy, as assessed by a decrease in p62 (Fig. 5C, lanes 1 and 2; compare lanes 7 and 8).
Fig. 5.
Loss of KIF2C induces autophagy. (A) LC3-II was immunoblotted in equal amounts of lysates from HBEC3KT and HBEC3KTRL53. (B) Ras was depleted from HBEC3KTRL53 with siRNA. Cells were then treated for 3 d with 100 nM PD0325901, followed by immunoblotting of lysates as indicated. (C) KIF2C was depleted with siRNA from HBEC3KTRL53. Cells were starved for 12 h in EBSS or treated with 10 nM Baflomycin A and immunoblotted with antibodies to p62. ERK1/2 was used as the loading control.
Discussion
We demonstrate that prolonged inhibition of the ERK1/2 pathway alters lysosomal distribution and suppresses mTORC1 in a manner that mimics nutrient deprivation. ERK1/2 are well known to have regulatory inputs via TSC1/2 and Raptor to growth factor-stimulated mTOR activities both directly and through the downstream effector Rsk (7–12, 36). ERKs have also been shown to be targeted to endosomes through a MEK partner 1 (MP1, gene LAMTOR3) complex (37), a component of the Ragulator complex, which is required for mTORC1 activity at the lysosome (16). Here, we define a long-term requirement for ERK1/2-dependent gene regulation in control of lysosomal organization in normal cells that is necessary for optimum mTORC1 performance.
Among ERK1/2-regulated genes in both untransformed and mutant K-Ras–transformed cells are the kinesins KIF2A and KIF2C (19). These KIFs are overrepresented in lung tumors, are induced by K-Ras in an ERK1/2-dependent manner, and enhance migration of Ras-transformed cancer cells. Because KIF2A was previously shown to be important for mTORC1 accumulation at lysosomes, we hypothesized that the insensitivity of mTORC1 and lysosome organization to growth factor and amino acid deprivation in Ras-transformed cells resulted from high expression of KIF2A/C. In agreement with published work (15), we established that KIF2A is important for lysosomal positioning in the immortalized lung-cell model and further showed that the related kinesin KIF2C also performs this function. Knocking down either of these kinesins inhibited mTORC1 stimulation by amino acids and altered lysosomal organization in HBEC.
In contrast, neither ERK1/2 activation nor these kinesins were required for lysosomal organization and mTORC1 activity in Ras-transformed cells. The kinesin requirement could also be bypassed in untransformed cells under nutrient-replete conditions in the presence of high concentrations of growth factors found in serum. Despite the fact that these proteins have the capacity to induce a functional organization of lysosomes in untransformed cells, in Ras-transformed cells, these kinesins are disconnected from this function. Wild-type and mutant K-Ras appear to induce morphological changes and perpetuate mTORC1 activity by distinct mechanisms in untransformed and cancer cells. In addition, a signaling hierarchy is apparent in untransformed cells that leads to mTORC1 activation with a requirement for KIF2A and KIF2C that is detected only in growth factor-poor conditions. Because two kinesins share this function, we considered the possibility that a third kinesin, closely related to KIF2A and -C, may also collaborate to position organelles. Expression of this third microtubule depolymerizing family member, KIF2B, is low to undetectable in these cells, and its expression is increased, not decreased, by the MEK inhibitor (19), suggesting that it is not a candidate for such a function.
Microtubule instability, as is observed in K-Ras–transformed cells, can be regulated by many proteins in addition to depolymerizing kinesins. Other factors proposed to account for K-Ras–induced microtubule instability include stathmin, a destabilizing factor, and reduced expression of the stabilizing factors discs large 1 (Dlg1), RASSF1A, and adenomatous polyposis coli (APC) (38–41). ERK1/2 MAPKs themselves were linked to changes in microtubule dynamics many years ago (42–44). Lysosomes are localized at the microtubule organizing center (MTOC), require microtubule dynamics to maintain proper organization (45), and would be expected to have different distributions in cells depending on microtubule dynamics. Ras-dependent changes in microtubule dynamics affected by these other molecules may bypass KIF2A/C and account for distinct lysosomal distributions in HBEC3KT and HBEC3KTRL53.
We also noted that manipulating KIF2A/C expression affected ERK1/2 activity. Activity differences may also be a consequence of the effects of these kinesins on microtubule dynamics. ERK activity was originally identified using a microtubule-associated protein as substrate (46), and, in early studies, ERK activity was shown to be increased by microtubule depolymerization (47). From 30% to 50% of the cytosolic ERK1/2 was estimated to be microtubule bound (48). Decreases in amounts of microtubule-depolymerizing kinesins should increase the average extent of microtubule polymerization and thereby increase the number of microtubule binding sites present at any given time to scaffold or sequester the kinases. We have found that KIF2A knockdown impaired ERK1/2 activation. Likewise, overexpression of KIF2A/C in HeLa cells decreased microtubule polymerization and increased ERK1/2 activity. For these reasons, we suggest that Ras-ERK induction of KIF2A/C may form a positive loop to perpetuate some level of ERK1/2 activity by increasing the unbound fraction of the proteins.
The best-studied functions of KIF2A and KIF2C are in regulation of the mitotic spindle during mitosis (49–51). Despite the large number of kinesins in cells, KIF2A/C also have multiple functions in interphase cells, including effects on migration and autophagy (19, 52). As noted for loss of KIF2A (15), loss of KIF2C also induces autophagy even in HBEC3KTRL53 in which mTORC1 remains activated. This finding was unexpected because of the minimal effects of KIF2A/C on lysosomal organization in Ras-transformed cells and also because drugs that inhibit microtubule dynamics, such as nocodazole and taxol, prevent starvation-induced autophagy (53). It will be important to assess further whether or not the induction of autophagy by depletion of KIF2A/C is related to their ability to increase microtubule dynamics, their impact on ATP concentration, or other as yet unidentified actions.
Methods
Antibodies.
The following antibodies were used: KIF2A (cat. no. ab37005, Abcam; cat. no. NB500, Novus Biologicals), a-tubulin (cat. no. T6199; Sigma), Actin (cat. no. MAB1501MI; Millipore), FLAG (cat. no. F3165; Sigma), pS6 (cat. no. 5364; Cell Signaling Technology), pERK (cat. no. M8159, Sigma; cat. no. 4377; Cell Signaling), LC3 (cat. no. PM036; MBL), p62 (cat. no. SC28359; Santa Cruz Biotechnology), LAMP2 (cat. no. ab25631; Abcam), pAKT (cat. no. 4060; Cell Signaling), KIF2C (cat. no. A300-807A; Bethyl Labs), mTOR (cat. no. 2983; Cell Signaling), Ras (sc-166691; Santa Cruz), and ERK1/2 (Y691) and ERK2 (C357) as previously described) (54).
Cell Culture.
Immortalized HBEC3KT, HBEC30KT, and HBEC3KT53 cells were cultured in keratinocyte serum-free medium (KSFM) (Invitrogen) supplemented with 5 ng/mL epidermal growth factor and 50 μg/mL bovine pituitary extract according to the manufacturer’s recommendations. HBEC3KTRL53, HCT116, and H358 were cultured in RPMI-1640 medium supplemented with 5% (vol/vol) heat-inactivated FBS and 2 mM l-glutamine. Cells were grown at 37 °C in a humidified atmosphere of 5% CO2.
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde (vol/vol) in Tris-buffered saline TBS for 10 min and permeabilized with 0.1% Triton X-100 for 5 min. After blocking with 10% normal goat serum (vol/vol) at room temperature for 1 h, cells were incubated with the indicated antibodies at 4 °C overnight. Cells were incubated with Alexa Fluor-conjugated secondary antibody at room temperature for 1 h, mounted, and imaged. Fluorescent Z-stacks (0.2 mm) were acquired and deconvolved using the Deltavision RT deconvolution microscope. Images were processed using ImageJ software. The eight-bit grayscale images were analyzed after thresholding to measure the area of lysosomal distribution (LAMP2 staining, Fig. 2A) by obtaining the ratio of the organelle area per whole-cell area (Fig. 2B), or mTOR localization after restimulation with amino acids (Fig. 1A) by obtaining the ratio of mTOR staining per whole-cell area (Fig. 1B).
siRNA.
Cells were transfected for from 48 h to 96 h as indicated with dsRNA oligonucleotides using Lipofectamine RNAiMax according to the manufacturer’s protocol (Invitrogen). The following target sequences for KIF2A were used: GAAAACGACCACUCAAUAA (Thermo Scientific) and GACCCTCCTTCAAGAGATA (Thermo Scientific). For KIF2C, the following were used: GCAAGCAACAGGUGCAAGU (Thermo Scientific) and GGCAUAAGCUCCUGUGAAU (Thermo Scientific). For Ras, the following was used: GGAGGGCUUUCUUUGUGUA (Thermo Scientific).
Cell Harvest.
Cells were lysed on ice in 50 mM Hepes (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 0.2 mM Na3VO4, 100 mM NaF, 50 mM β-glycerophosphate, 10% glycerol, 0.1% Triton X-100, 1.6 μg/mL aprotinin, 0.1 mM phenylmethylsulfonyl fluoride, and 10 μg/mL each of Nα-p-tosyl-l-lysine chloromethyl ketone, Nα-p-tosyl-l-arginine methyl ester, pepstatin A, and leupeptin. Lysates were frozen in N2 (liquid) and thawed on ice, followed by centrifugation for 15 min at 16,000 × g in a microcentrifuge at 4 °C. Lysates were subsequently boiled in Laemmli sample buffer (2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue, 50 mM Tris⋅HCl) and subjected to polyacrylamide gel electrophoresis in SDS.
Immunoblotting and Li-Cor Imaging.
Gels were transferred to nitrocellulose and incubated with the indicated antibodies. Primary antibodies were detected by fluorescently labeled secondary antibodies (Fluor 800-labeled IgG or Fluor 680-labeled IgG secondary antibody), using the LI-COR Odyssey dual-color system. To quantify the relative intensity of bands, we subtracted the background and measured the ratio of band of interest to total loading control. Ratios were set equal to 1. If duplicates are shown, their average was set equal to 1.
Supplementary Material
Acknowledgments
We thank Stina Singel (Department of Cell Biology, University of Texas Southwestern Medical Center) and members of the M.H.C. laboratory for helpful discussions and Dionne Ware for administrative assistance. This work was supported by National Institutes of Health Grants R37DK34128 (to M.H.C.) and P50CA70907 (to J.D.M.), a grant from the Cancer Prevention and Research Institute of Texas (to J.D.M.), Welch Foundation Grant 1243 (to M.H.C.), and a grant from the Longenbaugh Foundation (to J.D.M.). For E.Z. and L.M.W., this article represents partial fulfillment for the PhD degree.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411016111/-/DCSupplemental.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karnoub AE, Weinberg RA. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White MA, et al. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80(4):533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 4.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 6.Pollock CB, Shirasawa S, Sasazuki T, Kolch W, Dhillon AS. Oncogenic K-RAS is required to maintain changes in cytoskeletal organization, adhesion, and motility in colon cancer cells. Cancer Res. 2005;65(4):1244–1250. doi: 10.1158/0008-5472.CAN-04-1911. [DOI] [PubMed] [Google Scholar]
- 7.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40(2):310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 2011;89(3):221–228. doi: 10.1007/s00109-011-0726-6. [DOI] [PubMed] [Google Scholar]
- 9.Avruch J, et al. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296(4):E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23(6):744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 12.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 13.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JY, White E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy. 2013;9(10):1636–1638. doi: 10.4161/auto.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13(4):453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21(5):833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narita M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332(6032):966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaganjor E, et al. Ras regulates kinesin 13 family members to control cell migration pathways in transformed human bronchial epithelial cells. Oncogene. 2013 doi: 10.1038/onc.2013.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66(4):2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 22.Sato M, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11(6):638–650. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wauson EM, et al. The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol Cell. 2012;47(6):851–862. doi: 10.1016/j.molcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: Evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18(13):1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn-Windgassen A, et al. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem. 2005;280(37):32081–32089. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- 26.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider A, Younis RH, Gutkind JS. Hypoxia-induced energy stress inhibits the mTOR pathway by activating an AMPK/REDD1 signaling axis in head and neck squamous cell carcinoma. Neoplasia. 2008;10(11):1295–1302. doi: 10.1593/neo.08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7(6):643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17(1):113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: A target for drugs both ancient and modern. Chem Biol. 2012;19(10):1222–1236. doi: 10.1016/j.chembiol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb CA, Dooley HC, Tooze SA. Endocytosis and autophagy: Shared machinery for degradation. BioEssays. 2013;35(1):34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- 34.Wooten MW, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283(11):6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 35.Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4(8):1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 36.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA. 2004;101(37):13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nada S, et al. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009;28(5):477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humbert PO, et al. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27(55):6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 39.van Es JH, Giles RH, Clevers HC. The many faces of the tumor suppressor gene APC. Exp Cell Res. 2001;264(1):126–134. doi: 10.1006/excr.2000.5142. [DOI] [PubMed] [Google Scholar]
- 40.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120(Pt 18):3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 41.Fotiadou PP, Takahashi C, Rajabi HN, Ewen ME. Wild-type NRas and KRas perform distinct functions during transformation. Mol Cell Biol. 2007;27(19):6742–6755. doi: 10.1128/MCB.00234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gotoh Y, et al. In vitro effects on microtubule dynamics of purified Xenopus M phase-activated MAP kinase. Nature. 1991;349(6306):251–254. doi: 10.1038/349251a0. [DOI] [PubMed] [Google Scholar]
- 43.Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell. 1992;3(10):1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison RE, Turley EA. Active erk regulates microtubule stability in H-ras-transformed cells. Neoplasia. 2001;3(5):385–394. doi: 10.1038/sj.neo.7900180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matteoni R, Kreis TE. Translocation and clustering of endosomes and lysosomes depends on microtubules. J Cell Biol. 1987;105(3):1253–1265. doi: 10.1083/jcb.105.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray LB, Sturgill TW. Rapid stimulation by insulin of a serine/threonine kinase in 3T3-L1 adipocytes that phosphorylates microtubule-associated protein 2 in vitro. Proc Natl Acad Sci USA. 1987;84(6):1502–1506. doi: 10.1073/pnas.84.6.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinohara-Gotoh Y, Nishida E, Hoshi M, Sakai H. Activation of microtubule-associated protein kinase by microtubule disruption in quiescent rat 3Y1 cells. Exp Cell Res. 1991;193(1):161–166. doi: 10.1016/0014-4827(91)90551-5. [DOI] [PubMed] [Google Scholar]
- 48.Reszka AA, Seger R, Diltz CD, Krebs EG, Fischer EH. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc Natl Acad Sci USA. 1995;92(19):8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166(4):473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manning AL, et al. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18(8):2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16(7):3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanhaji M, Friel CT, Wordeman L, Louwen F, Yuan J. Mitotic centromere-associated kinesin (MCAK): A potential cancer drug target. Oncotarget. 2011;2(12):935–947. doi: 10.18632/oncotarget.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA., Jr Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol. 1992;152(3):458–466. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- 54.Boulton TG, Cobb MH. Identification of multiple extracellular signal-regulated kinases (ERKs) with antipeptide antibodies. Cell Regul. 1991;2(5):357–371. doi: 10.1091/mbc.2.5.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.