Significance
For mechanism-based development of treatment for Alzheimer’s disease (AD), the precise molecular mechanism of γ-secretase modulators (GSMs), which have been extensively developed as possible therapeutic reagents, is required. Here, we analyzed the mode of actions of phenylimidazole-type GSMs using a chemical biology approach and systematic mutagenesis. We provide the first structural model, to our knowledge, that binding of the phenylimidazole-type GSMs at the luminal loop of presenilin induces a conformational change of the catalytic center to reduce toxic amyloid-β species selectively. Our results may facilitate the effective development of AD therapeutics.
Keywords: Alzheimer’s disease, intramembrane proteolysis, allosteric modulator, chemical biology, amyloid-β protein
Abstract
γ-Secretase is an intramembrane-cleaving protease responsible for the generation of amyloid-β (Aβ) peptides. Recently, a series of compounds called γ-secretase modulators (GSMs) has been shown to decrease the levels of long toxic Aβ species (i.e., Aβ42), with a concomitant elevation of the production of shorter Aβ species. In this study, we show that a phenylimidazole-type GSM allosterically induces conformational changes in the catalytic site of γ-secretase to augment the proteolytic activity. Analyses using the photoaffinity labeling technique and systematic mutational studies revealed that the phenylimidazole-type GSM targets a previously unidentified extracellular binding pocket within the N-terminal fragment of presenilin (PS). Collectively, we provide a model for the mechanism of action of the phenylimidazole-type GSM in which binding at the luminal side of PS induces a conformational change in the catalytic center of γ-secretase to modulate Aβ production.
Gamma-secretase is responsible for the production of amyloid-β (Aβ) peptide, which is thought to play a key role in the pathogenesis of Alzheimer’s disease (AD) (1, 2). γ-Secretase is a membrane protein complex comprising a catalytic subunit, presenilin (PS), and the other three membrane protein subunits: nicastrin, anterior pharynx-defective 1 (Aph-1), and presenilin enhancer 2 (Pen-2) (2, 3). PS is endoproteolyzed into N- and C-terminal fragments (NTF and CTF, respectively) through the maturation process of the γ-secretase complex. In the Aβ production pathway, amyloid-β precursor protein (APP) is first cleaved by β-secretase to generate a CTF of APP, C99. This stub is then cleaved by γ-secretase to release the APP intracellular domain at the cytoplasmic border of the membrane, which is called ε-cleavage. γ-Secretase then processively trims every three to four residues from the ε-site as γ-cleavage to generate Aβ fragments heterogeneous in their C termini, resulting in Aβ species ranging from 46 to 38 residues in length (4, 5). Previous immunohistological, genetic, and biochemical studies indicate that Aβ of 42 residues in length (Aβ42) is the most aggregation-prone and pathogenic species among the various Aβ peptides (6). To date, clinical trials of γ-secretase inhibitors (GSIs) have failed due to severe adverse effects, presumably attributable to the simultaneous inhibition of the cleavage of the other γ-secretase substrates, including the notch receptor (7). Recently, a series of compounds called γ-secretase modulators (GSMs) has emerged as promising therapeutic candidates, because GSMs selectively decrease Aβ42 production without affecting notch cleavage (8, 9). GSMs are chemically classified into two groups (10): acidic GSMs with carboxylic acid groups, which are derived from NSAIDs, and nonacidic or phenylimidazole-type GSMs, which are generally more potent than acidic GSMs. Phenylimidazole-type GSMs reduce the production of Aβ40 and Aβ42 while increasing the production of Aβ37, Aβ38, and Aβ39. Photoaffinity labeling experiments revealed that the phenylimidazole-type GSMs directly target the NTF of PS (11, 12). However, the precise mode of binding of phenylimidazole-type GSMs, as well as the molecular mechanism underlying the modulation of the proteolytic reaction of γ-secretase, still remains unclear. Here, we report that phenylimidazole-type GSMs facilitate the formation of the transition-state structure and augment the catalytic activity of γ-secretase by directly targeting the extracellular pocket formed by hydrophilic loop 1 (HL1) of PS. Together with the homology model derived from the crystal structure of presenilin homolog (PSH) from the archaea Methanoculleus marisnigri (13), we propose a mode of action of phenylimidazole-type GSMs, namely, the activation of the processive cleaving activity of γ-secretase, which reduces the number of long Aβ species.
Results
Phenylimidazole-Type GSM Activates Intrinsic γ-Secretase Activity.
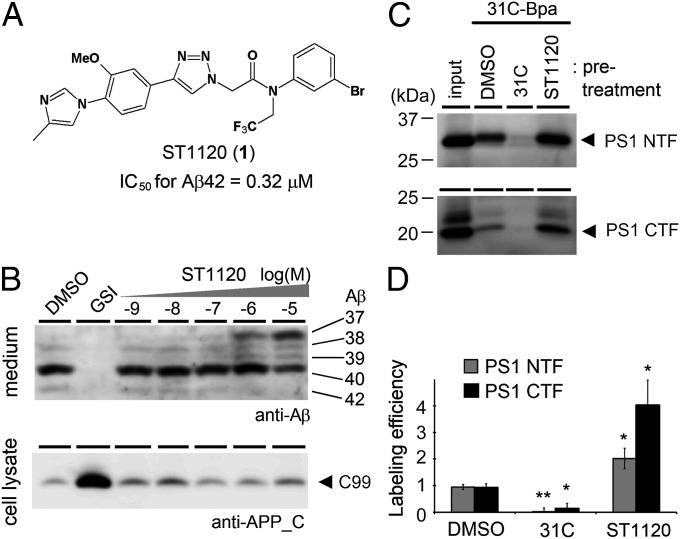
In this study, we used compound ST1120 (1) (Fig. 1A) as a representative nonacidic GSM, which contains a phenylimidazole connected to a triazole (14). In a cell-based assay, ST1120 decreased the levels of Aβ42 and Aβ40 in the conditioned medium, with a concomitant increase in the secretion of Aβ37 and Aβ39, in a concentration-dependent manner (Fig. 1B and SI Appendix, Fig. S1A). The IC50 for the production of Aβ42 [IC50 (Aβ42)] was 0.32 μM. In contrast, ST1120 showed no effect on the levels of APP CTFs or the intracellular domain of both APP and Notch (SI Appendix, Fig. S1B), as previously described for other nonacidic GSMs (9, 11, 12). To investigate the effect of GSMs on the conformation of γ-secretase, we next analyzed the effect of ST1120 on the photoaffinity labeling of PS1 proteins using two different GSI-based probes. The 31C-Bpa probe is a derivative of a transition-state analog-type GSI, and it directly targets the catalytic center and binds to the PS1 NTF and CTF (15, 16). The Pep.11-Bt probe is a derivative of the helical peptide-type GSI pep.11, and it binds to the initial substrate-binding site within the PS1 NTF (16, 17). Notably, pretreatment with ST1120 increased the labeling efficiency of PS1 by 31C-Bpa to 214% (NTF) and 437% (CTF) compared with DMSO (Fig. 1 C and D). On the other hand, the labeling of the PS1 NTF by pep.11-Bt was decreased to 65% of the original level by pretreatment with ST1120 (SI Appendix, Fig. S2). These results raised the possibility that ST1120 affects the conformation of the functional sites for γ-secretase cleavage, especially by facilitating the formation of the transition state of γ-secretase.
Fig. 1.
ST1120 increased the binding of a transition-state analog inhibitor. (A) Chemical structure of the phenylimidazole-type GSM ST1120 (1). IC50 for Aβ42 production was determined by a cell-based assay. (B, Upper) Conditioned media of HEK 293 cells expressing an APP-carrying Swedish mutation (APPNL) were separated by urea SDS/PAGE and analyzed by immunoblotting with the anti-Aβ antibody 82E1. (B, Lower) Cell lysates were analyzed by standard immunoblotting. GSI, 10 μM DAPT [(N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester)]. (C) Effect of ST1120 on the binding of the transition-state analog-based photoprobe 31C-Bpa (100 nM) in DKO cells expressing PS1, in which 50 μM 31C and ST1120 were used. (D) Labeling efficiencies in C were quantified by densitometric analysis (n = 3; data represent mean ± SE; *P < 0.05, **P < 0.01, Student t test).
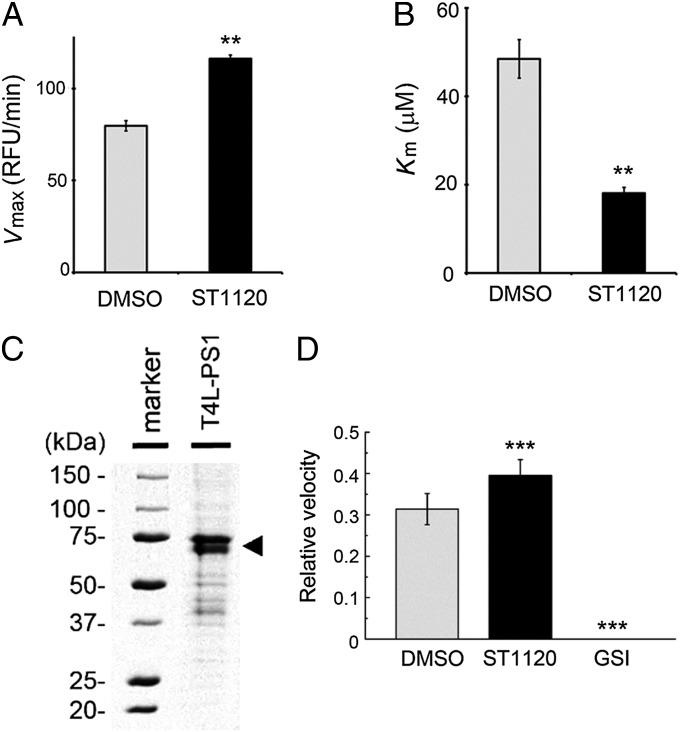
We next performed an in vitro assay to analyze the effect of the GSM on the enzymatic activity of the purified recombinant γ-secretase complex (18–20) (SI Appendix, Fig. S3 A and B). ST1120 modulated the production of Aβ from the recombinant C99 substrate in a fashion similar to that observed in cell-based assays (SI Appendix, Fig. S3C), suggesting that ST1120 acts directly on the processive cleavage activity of γ-secretase. The potency of ST1120 was lower than that in the cell-based assay, presumably due to the solubilized condition of the assay. To examine the catalytic activity in detail, we used a short fluorescent peptide-based γ-secretase substrate, Nma-APP-Dnp (21). Addition of Nma-APP-Dnp to the purified γ-secretase complex led to a linear increase in fluorescence (SI Appendix, Fig. S3D), which was completely abolished by a GSI (SI Appendix, Fig. S3F). The initial velocity of the cleavage activity under ST1120 treatment increased in a concentration-dependent manner, and was fourfold higher than that of the DMSO control with 100 μM ST1120 (SI Appendix, Fig. S3 E and F). We further analyzed the enzymatic parameters of this proteolytic reaction by the Michaelis–Menten plot (SI Appendix, Fig. S3E). Notably, the Vmax of the ST1120 treatment group was 1.5-fold higher than that of the DMSO control (Fig. 2A), and the Km of the ST1120 treatment group was 60% lower (Fig. 2B). These results are consistent with the results of the photoaffinity labeling experiment, demonstrating that ST1120 facilitates the formation of the proteolytically active conformation of the catalytic center of γ-secretase. Furthermore, we tested the effect of ST1120 on the intrinsic proteolytic activity of the recombinant PS protein produced by an Escherichia coli-based cell-free protein synthesis system (22) (Fig. 2C). As reported previously (23), the purified recombinant PS1 polypeptide possessed endoproteolytic activity for the Nma-APP-Dnp peptide (SI Appendix, Fig. S3G). In addition, we observed that 100 μM ST1120 increased the proteolytic activity of the PS1 polypeptide by 1.2-fold (Fig. 2D). These data indicate that the PS1 polypeptide is necessary, but not fully sufficient, to provide the full response to the phenylimidazole-type GSM. Taken together, these results support our notion that the GSM activates the intrinsic proteolytic activity of PS.
Fig. 2.
ST1120 increased the cleavage of a short fluorescent γ-secretase substrate peptide. (A and B) Effect of ST1120 on Vmax and Km of the recombinant γ-secretase complex derived from Sf9 cells calculated by fitting of the Hanes–Woolf plot (n = 4; mean ± SE; **P < 0.01). RFU, relative fluorescent units. (C) Coomassie Brilliant Blue staining of purified T4L-human PS1 protein (arrowhead) synthesized by an E. coli cell-free protein synthesis system. (D) Effect of 100 μM ST1120 on the cleavage of Nma-APP-Dnp by the purified PS1 protein. Relative velocities are shown (n = 4; data represent mean ± SE; **P < 0.01, ***P < 0.001, Student t test).
Identification of Residues Necessary for the Response to Phenylimidazole-Type GSM.
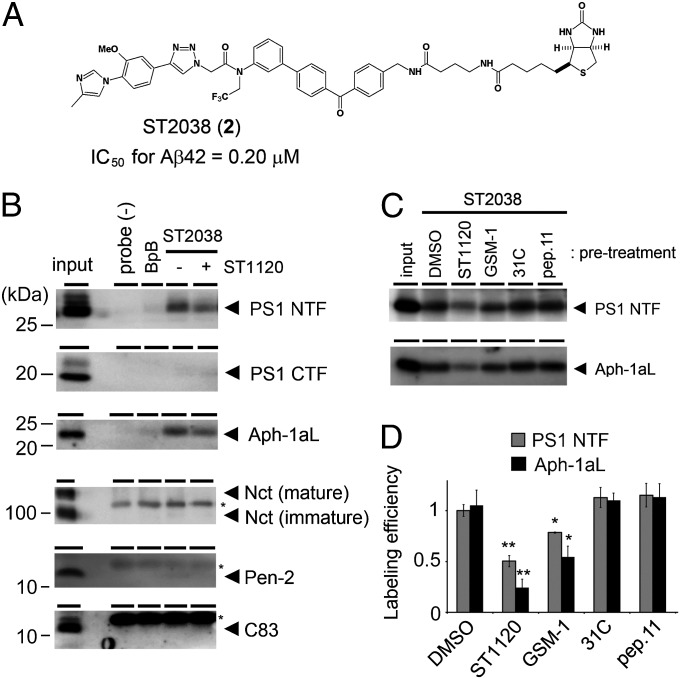
We next aimed to identify the target of the phenylimidazole-type GSM using a photoaffinity labeling experiment. We found that the probe ST2038 (2) (Fig. 3A) coupled with benzophenone and biotin moieties (3) (SI Appendix, Fig. S4A) retained the ability to exert a GSM effect (IC50 for Aβ42 secretion = 0.20 μM) (SI Appendix, Fig. S4 B and C). We performed photoaffinity labeling and biotin-based pull-down experiments using membrane fractions isolated from mouse brains (24) and found that the photoaffinity probe ST2038 specifically labeled the PS1 NTF and Aph-1aL (Fig. 3B). Next, we investigated the binding capabilities of ST2038 to PS2 or Aph-1b expressed on fibroblasts derived from Psen1−/−;Psen2−/− double-KO mice (DKO cells) (25) or Aph1a−/−;Aph1b−/− KO mice (AKO cells) (26). We found that the NTF of PS2 expressed in DKO cells was labeled by ST2038 (SI Appendix, Fig. S4D). We also observed that Flag-Aph-1aL-V5/His and Flag-Aph-1b-V5/His expressed in AKO cells were biotinylated by ST2038 (SI Appendix, Fig. S4 E and F). These results suggest that the phenylimidazole-type GSM-based probe ST2038 binds to both the PS NTF and Aph-1. Finally, we performed a cross-competition study to clarify whether ST2038 shares its binding site with known compounds. The labeling of the PS1 NTF and Aph-1aL by ST2038 was substantially decreased by pretreatment with GSM-1, an acidic GSM (24, 27). However, we observed no changes in the labeling of PS1 or Aph-1aL by pretreatment with either 31C or pep.11 (Fig. 3 C and D). These results indicate that the binding site of ST2038 is different from those of the GSIs and partially overlaps with that of GSM-1. Alternatively, GSM-1 prevents the conformation that allows binding of the phenylimidazole-type GSMs.
Fig. 3.
Photoprobe ST2038-labeled PS1 NTF and Aph-1aL. (A) Chemical structure of probe ST2038 (2). (B) Photoaffinity labeling of microsomes from mouse brains using ST2038. Endogenous proteins labeled by ST2038 were detected by immunoblotting using specific antibodies. ST2038, benzophenone, and biotin moieties (BpB) were used at 1 μM. Pretreatment with 50 μM ST1120 significantly reduced the labeling of the PS1 NTF and Aph-1aL. Input corresponds to 1% and 4% (Pen-2) of the pull-down fraction. Asterisks indicate nonspecific binding proteins. (C and D) Cross-competition experiment using ST2038 and 50 μM known inhibitors and modulators in DKO cells expressing human PS1. One micromolar ST2038 was used. Labeling efficiencies were quantified by densitometric analysis (n = 3–5; data represent mean ± SE; *P < 0.05, **P < 0.01, Student t test).
Although the photoaffinity labeling experiments suggest that ST2038 binds to both the PS NTF and Aph-1, the enzymatic study indicates that ST1120 is capable of acting directly on PS. However, biochemical identification of the precise binding site of ST2038 in the PS NTF was difficult because of the low quantity of proteins bound to the probe. To annotate the binding site of ST1120, we focused on the difference between PS1 and PS2 in their response to ST1120. Ebke et al. (11) previously reported that the phenylimidazole-type GSM RO-02 potently modulated the activity of the PS2-containing γ-secretase in cultured cells and that the photoaffinity probe RO-57 preferentially bound to the PS2 NTF (11). We also confirmed that ST1120 preferentially targets the PS2-containing γ-secretase (IC50 for Aβ40/Aβ42 against PS1 or PS2 was 5,538/320 or 271/51 nM, respectively) (SI Appendix, Table S1). To narrow down the critical domain for pharmacological action of the phenylimidazole-type GSMs, we examined the response of PS1/PS2 chimeric molecules. Large differences in the primary sequences of PS1 and PS2 are found in the N terminus and the sixth loop, whereas the midportion of the PS NTFs encompassing transmembrane domain (TM) 1–6 is relatively conserved. Thus, we designed PS1-2-2, PS2-1-1, PS1-1-2, and PS2-2-1 chimeras [denoted as PS(N terminus) − (TM1 to TM6) − (sixth loop to CTF)] by swapping at the 71st residue in PS1 (77th residue in PS2) and at the 269th residue in PS1 (275th residue in PS2). We found that the IC50s for PS1-2-2, PS2-2-1, and PS1-2-1 were similar to that for PS2. In contrast, the IC50s for PS2-1-1, PS1-1-2, and PS2-1-2 were similar to that for PS1 (SI Appendix, Table S1). These results indicate the importance of the relatively conserved TM1 to TM6 of PS as a critical determinant of the potency of ST1120.
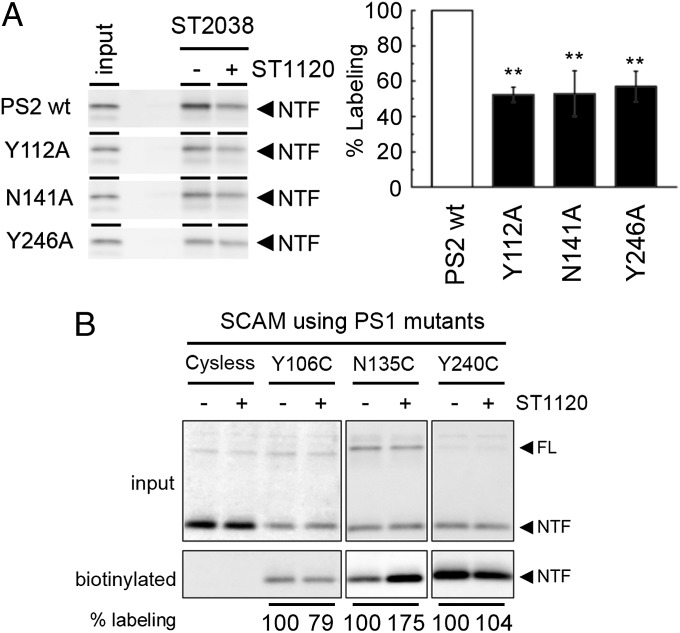
To identify the amino acid residues of PS2 important for the GSM response, we performed alanine scanning analyses. Phenylimidazole moieties typically bind via π–π interactions to the phenylalanine, tyrosine, histidine, and tryptophan residues of the target molecule, or form hydrogen bonds with the serine, threonine, aspartic acid, asparagine, glutamic acid, glutamine, lysine, and arginine residues in the target molecule (28). Given that both PS1- and PS2-containing γ-secretase activity is modulated by ST1120, we substituted these amino acid residues in PS2 that are conserved in PS1 to alanine. As a first screen, we compared the secretion of Aβ from DKO cells expressing PS2 mutants in the presence of DMSO or 1 μM ST1120 (SI Appendix, Tables S2–S5). Among these mutants, we selected 12 that showed a greater than 50% increase in the secretion of both Aβ40 and Aβ42 in the presence of 1 μM ST1120 compared with that in the presence of DMSO (SI Appendix, Fig. S5 A–D). We also tested the activities of similar mutants in Aph1-aL–overexpressing AKO cells. However, none of the mutants affected the GSM response in these cells (SI Appendix, Fig. S5E). We then analyzed the effect of ST1120 and IZ2083 (4) (SI Appendix, Fig. S6B), with the latter being a different phenylimidazole-type GSM, containing thiazole (patent WO2004110350) (9) on Aβ secretion from DKO cells expressing one of the 12 selected PS2 mutants. We found that the PS2 Y112A, N141A, and Y246A mutants were almost devoid of a response to ST1120 or IZ2083 treatment (SI Appendix, Fig. S6 A–C). The other mutants decreased, but did not abolish, the response to the GSMs (SI Appendix, Fig. S7). We then analyzed the alanine substitutions at the homologous residues of Y112, N141, and Y246 in PS1 (i.e., Y106, N135, and Y240, respectively). These PS1 mutants also showed a reduced response to ST1120 and IZ2038 (SI Appendix, Fig. S6D), suggesting that these residues are involved in the molecular machinery for the GSM response. We further validated the binding potency of the photoaffinity probe ST2038 to PS2 alanine substitution mutants. Consistent with the results of the cell-based assay, the labeling efficiencies of the photoaffinity probe ST2038 on PS2 Y112A, N141A, and Y246A were significantly decreased (Fig. 4A), raising two possibilities: These residues are either directly involved in the binding with ST2038, or these alanine substitutions allosterically affected the structural architecture of the binding site.
Fig. 4.
Mutations in residues of the HL1/TM2/TM5 of PS2 lowered the interactions with the phenylimidazole-type GSMs. (A) Photoaffinity labeling of PS2 mutants using probe ST2038. (Right) Labeling efficiencies are shown (n = 4; data represent mean ± SE; **P < 0.05, Dunnett’s test). (B) Competition assay of SCAM-based biotinylation of PS1 mutants by 50 μM ST1120. Changes in the labeling are indicated below the panel (n = 3; data represent mean). Note that ST1120 decreased the hydrophilicity around N106C (P < 0.001, Student t test) but augmented it at N135C (P < 0.05, Student t test). FL, full length.
We then examined the effect of ST1120 on the solvent accessibility of these residues in PS1 using the substituted cysteine accessibility method (SCAM). The SCAM has been repeatedly used to gain structural information about various multispanning membrane proteins in a functional state by covalently modifying the introduced cysteine residues using the sulfhydryl reagent N-biotinylaminoethyl methanethiosulfonate (MTSEA-biotin). We found that the Y106C, N135C, and Y240C mutants in PS1, which does not contain cysteine residues, were each labeled by MTSEA-biotin from the extracellular side of the cell (29). Notably, the biotinylation of the Y106C mutant showed statistically significant reduction by ST1120 treatment (79 ± 0.5% compared with DMSO treatment) (Fig. 4B), suggesting the possibility that ST1120 targets an area of PS1 around this residue. In contrast, the labeling of N135C was drastically increased (175 ± 23% compared with DMSO treatment), suggesting that ST1120 might allosterically affect the hydrophilic environment around N135 rather than directly binding to this residue. The labeling efficiency of Y240C was not significantly altered (104 ± 7.8% compared with DMSO treatment). To visualize these regions further, we generated a homology model of human PS1 based on the crystal structure of archaea PSH (13) and annotated the residues critical to the GSM response. Notably, among six critical residues for GSM action, two residues (PS2 Y112 and Y121 corresponding to PS1 Y106 and Y115, respectively) exist within the extracellular HL1 region, although this region was excluded in the crystal structure of PSH. Two other residues of TM2 and TM5 (PS2 N141 and Y246 corresponding to PS1 N135 and Y240, respectively) face to the extracellular side (SI Appendix, Fig. S8). Taken together, these results suggest that the extracellular pocket formed by HL1/TM2/TM5 is critical for the interaction of PS with the phenylimidazole-type GSM.
Allosteric Effect on the Catalytic Center of PS by the Binding of Phenylimidazole-Type GSM.
To clarify further the allosteric effects of the binding of phenylimidazole-type GSMs on the catalytic site of PS, we performed the SCAM assay and cross-linking of the residues around the catalytic residues located in the PS1 CTF, which is not a direct target region (Fig. 5A). We have shown previously that residues L381 and L383, with the latter being a part of the Gly-x-Gly-Asp catalytic motif at TM7 of the PS1 CTF, face the hydrophilic catalytic pore within the membrane (30). Intriguingly, pretreatment with ST1120 reduced the labeling of L381C, but the levels of the biotinylated CTF carrying L383C were increased (Fig. 5B). These results suggest that ST1120 induces allosteric changes in the hydrophilic environment around the catalytic site of PS1. We next analyzed residues I383 in TM7 and L435 in the Pro-Ala-Leu (PAL) motif (31), which are located in proximity to L250 in TM6 (32). The NTF-CTF heterodimer was then formed using cross-linking reagents in double-cysteine mutants of PS1 harboring either L250C/I387C or L250C/L435C (30, 32, 33). We found that preincubation with ST1120 decreased the formation of cross-linked heterodimers of the L250C/L435C mutant (Fig. 5C), whereas it had no effect on the L250C/I387C mutant, suggesting that the binding of ST1120 selectively affects the conformation of the catalytic site. Taken together, this study revealed that phenylimidazole-type GSMs, which bind to HL1/TM2/TM5 (Fig. 6), induce a conformational change in the catalytic site of PS to increase the processive cleavage activity of γ-secretase.
Fig. 5.
Allosteric changes around the catalytic site of PS1 induced by ST1120. (A) Positions of the mutated residues in the homology model of PS1, based on the crystal structure of archaea PSH. Catalytic aspartates are shown as red spheres in TM6 and TM7. (B) Competition assay in SCAM-based biotinylation of the PS1 L381C and L383C mutants by ST1120. (C) Effects of ST1120 on the cross-linking of double-cysteine mutants of PS1. We used 1,2-ethanedithiol dimethanesulphonate that harbors a 5.0-Å spacer as a cross-linking reagent. Note that the levels of cross-linked products and fragments of L250C/L435C were specifically decreased and increased, respectively, by ST1120.
Fig. 6.
Homology model of PS1 and molecular mechanism of the GSM. A human PS1 homology model was derived from the crystal structure of archaea PSH. PS11–71 and PS1274–376 are not shown. PS1105–130 as HL1 is shown by a dotted line. The catalytic cavity is shown as a cyan circle. The catalytic aspartates D257 and D385 are shown as cyan spheres. The red sticks denote Y106, N135, and Y240 (SI Appendix, Fig. S6A), and the orange sticks represent Y115, F177, and F386. The allosteric effect on the catalytic site structure by GSM (shown by stick model) is shown by red arrows.
Discussion
In this study, we aimed to clarify the binding site and molecular mechanism of action of phenylimidazole-type GSMs using enzymatic, biochemical, and chemical biological approaches. As a result, we found the following. First, phenylimidazole-type GSMs enhance the catalytic activity of γ-secretase by directly interacting with the PS protein. Second, phenylimidazole-type GSMs target the extracellular pocket formed by the HL1/TM2/TM5 of PS. Finally, GSM binding allosterically affects the structure of the catalytic center of PS. This study provides molecular insights into the mechanisms of action of phenylimidazole-type GSMs, which should aid in the development of structure-based drugs for AD.
The increased and decreased production of short and long Aβ peptides, respectively, by GSM can be explained by two possible mechanisms: the decreased probability of release of longer Aβ from the enzyme–substrate complex or increased processive cleavage activity of the γ-secretase. γ-Secretase performs multiple turnovers; therefore, product release and proteolytic reaction are not mutually exclusive. However, our study supports the latter based on two lines of evidence. We observed that ST1120 causes a significant increase in the binding of 31C-Bpa (Fig. 1C) in a manner similar to that by piperidine-type GSM (34), as well as a 1.5-fold increase in the Vmax of γ-secretase against short peptide substrates (Fig. 2). Recently, another nonacidic type of GSM was reported to increase the Vmax of γ-secretase by 2.3-fold when Aβ42 was used as a substrate (35), which correlates well with our study. However, an obvious increase in total Aβ levels that would reflect an increased efficiency of ε-cleavage has not been observed by GSMs (Fig. 1B and SI Appendix, Fig. S1B). Biochemical analysis of proteolytic reaction of γ-secretase revealed that ε-cleavage is the first process in Aβ generation (4), suggesting that these compounds selectively augment the carboxypeptidase-like γ-cleavage activity. A similar mode of action of GSMs was reported by Chávez-Gutiérrez et al. (36) using a comprehensive in vitro γ-secretase assay. A different possibility still remains that GSM promotes all of the hydrolytic processes by γ-secretase, whereas another mechanism, such as a substrate docking, breaking the α-helix, or transferring the substrate to the catalytic site, would limit the ε-cleavage and total Aβ production. In fact, we observed a decrease in biotinylation by pep.11-Bt, which targets the initial substrate-binding site of PS, suggesting the possibility that ST1120 also affects the binding mode of the PS with the substrate. Nevertheless, our findings indicate that phenylimidazole-type GSMs activate the catalytic mechanism(s) of PS to accelerate the processive cleavage of longer forms of Aβ.
Our photoaffinity probe labeled both the PS NTF and Aph-1 (Fig. 3B). However, other photoprobes based on the nonacidic GSM labeled only the PS NTF (11, 12), and ST1120 directly activated the catalytic activity of recombinant PS without Aph-1 (SI Appendix, Fig. 3H). In addition, no mutants were found to reduce the enzymatic activity in response to ST1120 in the alanine scanning mutagenesis of Aph-1aL (SI Appendix, Fig. S5E and Table S6). These results strongly indicate that the PS NTF is the molecular target of the phenylimidazole-type GSM. However, the molecular function of Aph-1 still remains unclear. Notably, a high concentration of ST1120 was required to augment the proteolytic activity of the purified recombinant PS1 protein, suggesting that a full response to GSM would require Aph-1. In addition, Aph-1 was implicated in the allosteric regulation for γ-cleavage (37, 38). Further structural analyses of the γ-secretase complex should provide information regarding the molecular function of Aph-1 in the GSM response.
Based on the structure of PSH (13), we have generated a model of the binding of ST1120 to PS1 (SI Appendix, Fig. S6B). In this model, ST1120 targets the extracellular pocket formed by HL1/TM2/TM5, which is located far from the catalytic cavity formed by TM6 and TM7 (30). In addition, the initial substrate entry site, in which TM2, TM6, and TM9 are involved (39), is also located on a different side of the binding pocket from ST1120. Supporting this model, pretreatment with 31C and pep.11 did not affect the binding of the photoaffinity probe ST2038 (Fig. 3D). In contrast, helical peptide enabled ibuprofen and fenofibrate to alter the conformation of PS1 (40). Notably, we have reported that acidic-type GSMs target TM1 (24, 41). These results suggest that occupation of the substrate binding site differently affected the conformation of these acidic and nonacidic GSM binding sites (TM1 and HL1, respectively) that are directly connected. Furthermore, we found that the allosteric structural changes in the catalytic center were induced upon the binding of ST1120 (Fig. 5). In addition, alanine substitutions around the catalytic aspartate in the CTF (i.e., PS2 F367A and F369A mutants) significantly reduced the GSM response. Moreover, the distance between L250 in TM6 and L435 in the PAL motif/TM9, both of which are located near the cytoplasmic side of the pore, was affected by ST1120. This result suggests the functional structural connections between different PS TMs that comprise the catalytic pocket. Moreover, this conformational change is reminiscent of that of transporters in which ligand binding induces conformational changes at both the extracellular and intracellular sides to relocate the substrates to the opposite side (42). HL1 is the biggest extracellular loop region in PS, containing ∼40 residues, and it is implicated in the γ-secretase activity and substrate selectivity (29, 43). Interestingly, the location of the residues critical for GSM activity is similar to that of the residues mutated in familial AD (SI Appendix, Fig. S8), supporting the notion that these residues in HL1 play an important role in the processive cleavage activity of γ-secretase. The dynamic motion of the HL in rhomboid protease, the other intramembrane cleaving enzyme, influences the whole conformation and activity of the rhomboid (44). In addition, allosteric activation of the Vibrio cholerae repeats-in-toxin cysteine protease by inositol hexakisphosphate has been reported (45). In this protease, the binding of inositol hexakisphosphate to the flexible β-flap structure contributes to enzyme activation by properly ordering the P1 pocket and active site. Thus, it is possible that the HL of intramembrane cleaving enzymes harbors a critical function in the allosteric regulation of the proteolytic reaction. Taken together, we identified the extracellular pocket formed by PS HL1/TM2/TM5 as a critical binding site for phenylimidazole-type GSMs. Further fine structural analyses to clarify the molecular connection between this pocket and the catalytic site would contribute to the development of novel AD therapeutics.
Materials and Methods
Materials.
IZ2038 was prepared according to the WO2004110350 patent (9). Maintenance of cultured cells, transfection, retroviral infection, and selection of stably expressing cells were performed as described previously (30, 33, 39, 46). Full descriptions of experiments are detailed in SI Appendix, SI Materials and Methods.
γ-Secretase Assay.
Aβ levels in the conditioned media from cultured cells were analyzed by ELISA and immunoblotting (23, 47). Intrinsic γ-secretase activity was measured by in vitro assay using recombinant C99 (18, 19, 24) or the fluorescent short peptide, Nma-APP-Dnp (no. 3217-v; Peptide institute) (21). Recombinant γ-secretase complex by Sf9 cells and T4 lysozyme-human PS1 chimeric protein by an E. coli cell-free protein synthesis system were prepared as described previously (19, 22). Full descriptions of experiments are detailed in SI Appendix, SI Materials and Methods.
Photoaffinity Labeling.
Photoaffinity labeling was performed as described (24, 46). Briefly, membrane fractions were incubated with indicated compounds and collected by centrifugation after UV irradiation. The pellets were solubilized by 1% SDS buffer, and Streptavidin Sepharose (GE Healthcare) was added to the supernatants to pull down the biotinylated proteins.
Supplementary Material
Acknowledgments
We thank Drs. Gopal Thinakaran (The University of Chicago), Bart De Strooper (Vlaams Instituut vor Biotechnologie Leuven), Yigong Shi (Tsinghua University), Tong Li and Philip C. Wong (Johns Hopkins University), Naoki Umezawa and Tsunehiko Higuchi (Nagoya City University), and Toshio Kitamura (The University of Tokyo) for valuable reagents; Ms. Mio Inoue for generating plasmids; Dr. Jeanne Hardy (University of Massachusetts, Amherst) for critical reading; Takeda Pharmaceutical Company for Aβ ELISA; and our current and previous laboratory members for helpful discussions and technical assistance. K.T. and S.T.-N. are research fellows of the Japan Society for the Promotion of Science. This work was supported, in part, by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (to T.T.); by grants on Scientific Research on Innovative Areas for Foundation of Synapses and Neurocircuit Pathology Grant (to T.I.) and Brain Environment (to T.T.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan; by the Ministry of Health, Labor, and Welfare, Japan (Comprehensive Research on Aging and Health) (T.T.); by the Targeted Proteins Research Program of the MEXT, Japan (T.S., T.K.-S., M.S., S. Yokoyama, T.T., and T.I.); by Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency (T.T. and T.I.); and by a donation from Mr. Chuichi Imai (to T.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402171111/-/DCSupplemental.
References
- 1.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: The challenge of the second century. Sci Transl Med. 2011;3(77):sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomita T, Iwatsubo T. Structural biology of presenilins and signal peptide peptidases. J Biol Chem. 2013;288(21):14673–14680. doi: 10.1074/jbc.R113.463281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takasugi N, et al. The role of presenilin cofactors in the γ-secretase complex. Nature. 2003;422(6930):438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 4.Qi-Takahara Y, et al. Longer forms of amyloid β protein: Implications for the mechanism of intramembrane cleavage by γ-secretase. J Neurosci. 2005;25(2):436–445. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takami M, et al. γ-Secretase: Successive tripeptide and tetrapeptide release from the transmembrane domain of β-carboxyl terminal fragment. J Neurosci. 2009;29(41):13042–13052. doi: 10.1523/JNEUROSCI.2362-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwatsubo T, et al. Visualization of A β 42(43) and A β 40 in senile plaques with end-specific A β monoclonals: Evidence that an initially deposited species is A β 42(43) Neuron. 1994;13(1):45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 7.Doody RS, et al. Alzheimer’s Disease Cooperative Study Steering Committee Semagacestat Study Group A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N Engl J Med. 2013;369(4):341–350. doi: 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- 8.Weggen S, et al. A subset of NSAIDs lower amyloidogenic Aβ42 independently of cyclooxygenase activity. Nature. 2001;414(6860):212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 9.Kounnas MZ, et al. Modulation of γ-secretase reduces β-amyloid deposition in a transgenic mouse model of Alzheimer’s disease. Neuron. 2010;67(5):769–780. doi: 10.1016/j.neuron.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehlrich D, Berthelot DJ, Gijsen HJ. γ-Secretase modulators as potential disease modifying anti-Alzheimer’s drugs. J Med Chem. 2011;54(3):669–698. doi: 10.1021/jm101168r. [DOI] [PubMed] [Google Scholar]
- 11.Ebke A, et al. Novel γ-secretase enzyme modulators directly target presenilin protein. J Biol Chem. 2011;286(43):37181–37186. doi: 10.1074/jbc.C111.276972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pozdnyakov N, et al. γ-Secretase modulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin. J Biol Chem. 2013;288(14):9710–9720. doi: 10.1074/jbc.M112.398602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493(7430):56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 14.Fischer C, et al. Triazole derivatives for treating alzheimer's disease and related conditions. International patent. 2008: WO2008156580. [Google Scholar]
- 15.Micchelli CA, et al. γ-secretase/presenilin inhibitors for Alzheimer’s disease phenocopy Notch mutations in Drosophila. FASEB J. 2003;17(1):79–81. doi: 10.1096/fj.02-0394fje. [DOI] [PubMed] [Google Scholar]
- 16.Imamura Y, et al. Inhibition of γ-secretase activity by helical β-peptide foldamers. J Am Chem Soc. 2009;131(21):7353–7359. doi: 10.1021/ja9001458. [DOI] [PubMed] [Google Scholar]
- 17.Kornilova AY, Bihel F, Das C, Wolfe MS. The initial substrate-binding site of γ-secretase is located on presenilin near the active site. Proc Natl Acad Sci USA. 2005;102(9):3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi Y, et al. Sulindac sulfide is a noncompetitive γ-secretase inhibitor that preferentially reduces Aβ 42 generation. J Biol Chem. 2003;278(20):18664–18670. doi: 10.1074/jbc.M301619200. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi I, et al. Selective reconstitution and recovery of functional γ-secretase complex on budded baculovirus particles. J Biol Chem. 2004;279(36):38040–38046. doi: 10.1074/jbc.M405597200. [DOI] [PubMed] [Google Scholar]
- 20.Ogura T, et al. Three-dimensional structure of the γ-secretase complex. Biochem Biophys Res Commun. 2006;343(2):525–534. doi: 10.1016/j.bbrc.2006.02.158. and correction (2006) 345(1):543. [DOI] [PubMed] [Google Scholar]
- 21.Farmery MR, et al. Partial purification and characterization of γ-secretase from post-mortem human brain. J Biol Chem. 2003;278(27):24277–24284. doi: 10.1074/jbc.M211992200. [DOI] [PubMed] [Google Scholar]
- 22.Shimono K, et al. Production of functional bacteriorhodopsin by an Escherichia coli cell-free protein synthesis system supplemented with steroid detergent and lipid. Protein Sci. 2009;18(10):2160–2171. doi: 10.1002/pro.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn K, et al. Activation and intrinsic γ-secretase activity of presenilin 1. Proc Natl Acad Sci USA. 2010;107(50):21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohki Y, et al. Phenylpiperidine-type γ-secretase modulators target the transmembrane domain 1 of presenilin 1. EMBO J. 2011;30(23):4815–4824. doi: 10.1038/emboj.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herreman A, et al. Total inactivation of γ-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol. 2000;2(7):461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- 26.Ma G, Li T, Price DL, Wong PC. APH-1a is the principal mammalian APH-1 isoform present in γ-secretase complexes during embryonic development. J Neurosci. 2005;25(1):192–198. doi: 10.1523/JNEUROSCI.3814-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page RM, et al. Generation of Aβ38 and Aβ42 is independently and differentially affected by familial Alzheimer disease-associated presenilin mutations and γ-secretase modulation. J Biol Chem. 2008;283(2):677–683. doi: 10.1074/jbc.M708754200. [DOI] [PubMed] [Google Scholar]
- 28.de Graaf C, Rein C, Piwnica D, Giordanetto F, Rognan D. Structure-based discovery of allosteric modulators of two related class B G-protein-coupled receptors. ChemMedChem. 2011;6(12):2159–2169. doi: 10.1002/cmdc.201100317. [DOI] [PubMed] [Google Scholar]
- 29.Takagi S, Tominaga A, Sato C, Tomita T, Iwatsubo T. Participation of transmembrane domain 1 of presenilin 1 in the catalytic pore structure of the γ-secretase. J Neurosci. 2010;30(47):15943–15950. doi: 10.1523/JNEUROSCI.3318-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato C, Morohashi Y, Tomita T, Iwatsubo T. Structure of the catalytic pore of γ-secretase probed by the accessibility of substituted cysteines. J Neurosci. 2006;26(46):12081–12088. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomita T, et al. The first proline of PALP motif at the C terminus of presenilins is obligatory for stabilization, complex formation, and γ-secretase activities of presenilins. J Biol Chem. 2001;276(35):33273–33281. doi: 10.1074/jbc.M011152200. [DOI] [PubMed] [Google Scholar]
- 32.Sato C, Takagi S, Tomita T, Iwatsubo T. The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the γ-secretase. J Neurosci. 2008;28(24):6264–6271. doi: 10.1523/JNEUROSCI.1163-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeo K, Watanabe N, Tomita T, Iwatsubo T. Contribution of the γ-secretase subunits to the formation of catalytic pore of presenilin 1 protein. J Biol Chem. 2012;287(31):25834–25843. doi: 10.1074/jbc.M111.336347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crump CJ, et al. Piperidine acetic acid based γ-secretase modulators directly bind to Presenilin-1. ACS Chem Neurosci. 2011;2(12):705–710. doi: 10.1021/cn200098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okochi M, et al. γ-secretase modulators and presenilin 1 mutants act differently on presenilin/γ-secretase function to cleave Aβ42 and Aβ43. Cell Reports. 2013;3(1):42–51. doi: 10.1016/j.celrep.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 36.Chávez-Gutiérrez L, et al. The mechanism of γ-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31(10):2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acx H, et al. Signature amyloid β profiles are produced by different γ-secretase complexes. J Biol Chem. 2014;289(7):4346–4355. doi: 10.1074/jbc.M113.530907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serneels L, et al. γ-Secretase heterogeneity in the Aph1 subunit: Relevance for Alzheimer’s disease. Science. 2009;324(5927):639–642. doi: 10.1126/science.1171176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe N, et al. Functional analysis of the transmembrane domains of presenilin 1: Participation of transmembrane domains 2 and 6 in the formation of initial substrate-binding site of γ-secretase. J Biol Chem. 2010;285(26):19738–19746. doi: 10.1074/jbc.M110.101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uemura K, et al. Substrate docking to γ-secretase allows access of γ-secretase modulators to an allosteric site. Nat Commun. 2010;1:130. doi: 10.1038/ncomms1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohki Y, et al. Binding of longer Aβ to transmembrane domain 1 of presenilin 1 impacts on Aβ42 generation. Mol Neurodegener. 2014;9:7. doi: 10.1186/1750-1326-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Focke PJ, Wang X, Larsson HP. Neurotransmitter transporters: Structure meets function. Structure. 2013;21(5):694–705. doi: 10.1016/j.str.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong P, et al. Mutation analysis of the presenilin 1 N-terminal domain reveals a broad spectrum of γ-secretase activity toward amyloid precursor protein and other substrates. J Biol Chem. 2010;285(49):38042–38052. doi: 10.1074/jbc.M110.132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17(3):395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322(5899):265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morohashi Y, et al. C-terminal fragment of presenilin is the molecular target of a dipeptidic γ-secretase-specific inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester) J Biol Chem. 2006;281(21):14670–14676. doi: 10.1074/jbc.M513012200. [DOI] [PubMed] [Google Scholar]
- 47.Tomita T, et al. The presenilin 2 mutation (N141I) linked to familial Alzheimer disease (Volga German families) increases the secretion of amyloid β protein ending at the 42nd (or 43rd) residue. Proc Natl Acad Sci USA. 1997;94(5):2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.