Significance
Sleep restriction and circadian clock disruption are associated with metabolic disorders including obesity and diabetes; this association can be studied by using the powerful tool of metabolomics. By using liquid chromatography/MS metabolomics, we have characterized plasma metabolites that were significantly affected by acute sleep deprivation (mainly lipids and acylcarnitines), all increasing during sleep deprivation. Observed increased levels of serotonin, tryptophan, and taurine may explain the antidepressive effect of sleep deprivation and deserve further study. Clear daily rhythms were observed in most metabolites, with 24 h wakefulness mainly reducing the amplitude of these rhythms. Our results further the understanding of sleep/wake regulation and the associated metabolic processes, and will be vital when using metabolic profiling to identify robust biomarkers for disease states and drug efficacy.
Keywords: circadian rhythms, total sleep deprivation, melatonin, depression, biomarker
Abstract
Sleep restriction and circadian clock disruption are associated with metabolic disorders such as obesity, insulin resistance, and diabetes. The metabolic pathways involved in human sleep, however, have yet to be investigated with the use of a metabolomics approach. Here we have used untargeted and targeted liquid chromatography (LC)/MS metabolomics to examine the effect of acute sleep deprivation on plasma metabolite rhythms. Twelve healthy young male subjects remained in controlled laboratory conditions with respect to environmental light, sleep, meals, and posture during a 24-h wake/sleep cycle, followed by 24 h of wakefulness. Two-hourly plasma samples collected over the 48 h period were analyzed by LC/MS. Principal component analysis revealed a clear time of day variation with a significant cosine fit during the wake/sleep cycle and during 24 h of wakefulness in untargeted and targeted analysis. Of 171 metabolites quantified, daily rhythms were observed in the majority (n = 109), with 78 of these maintaining their rhythmicity during 24 h of wakefulness, most with reduced amplitude (n = 66). During sleep deprivation, 27 metabolites (tryptophan, serotonin, taurine, 8 acylcarnitines, 13 glycerophospholipids, and 3 sphingolipids) exhibited significantly increased levels compared with during sleep. The increased levels of serotonin, tryptophan, and taurine may explain the antidepressive effect of acute sleep deprivation and deserve further study. This report, to our knowledge the first of metabolic profiling during sleep and sleep deprivation and characterization of 24 h rhythms under these conditions, offers a novel view of human sleep/wake regulation.
Circadian clocks control the timing of most daily biological processes, including cyclic changes in metabolism and the sleep/wake cycle (1). There is a clear link between the circadian timing system and metabolism (2–4), with disrupted circadian rhythms, sleep restriction, and sleep deprivation associated with metabolic disorders (obesity, insulin resistance, diabetes) and cardiovascular disease (5–8). The underlying mechanisms linking metabolic disease, circadian clock misalignment, and sleep restriction are the subject of current research, elucidation of which will require a global “systems” approach (9). Transcriptomic studies have shown that rhythmic gene expression may be affected by sleep restriction, sleep deprivation, and mistimed sleep (10–12), but, as yet, no studies have directly investigated the effect that sleep and sleep deprivation may have on the metabolic profile. Metabolic profiling, or “metabolomics,” is the profiling of small-molecule metabolites and offers the potential to characterize specific metabolic phenotypes associated with disrupted circadian timing and sleep loss. Metabolomics has an advantage over other “omics” techniques, in that it directly samples the metabolic changes in an organism and integrates information from changes at the gene, transcript, and protein levels, as well as posttranslational modification (13). Additionally, metabolomics can provide insight into the combination of genotype and environmental effects, leading to its use in predicting responses to drugs in a “pharmacometabolomic” approach (14, 15).
Daily rhythms have been identified in the metabolomes of mice (16–18) and humans (19). Recent studies conducted in constant routine conditions, in which the impact of exogenous factors such as light, food, posture, and sleep are minimized, have demonstrated endogenous circadian variation in the human metabolome (20, 21), with one study reporting that ∼15% of the metabolites quantified in human plasma and saliva showed circadian variation, particularly amino acids in saliva and fatty acids in plasma (20). These human study protocols aimed to specifically identify circadian variation in the metabolome. However, if metabolic profiling is to be applied in “real-life” clinical settings, and in the identification of robust biomarkers, time-of-day variation in the human metabolome also needs to be characterized, and the effect of sleep, wakefulness, light/dark conditions, and meals on these daily rhythms assessed. Previously, we have characterized significant time of day variation in ∼19% of the metabolite features detected (19). These metabolites included corticosteroids, bilirubin, amino acids, acylcarnitines, and lysophospholipids. Not all of these metabolites, however, have been found to show circadian variation in the constant routine studies (20), suggesting that some metabolites may be affected by light/dark, sleep/wake, or food.
The two-process model of sleep regulation proposes that sleep is driven by a homeostatic component (i.e., process S) and an endogenous circadian oscillator (i.e., process C) (22). The link between the homeostatic and circadian regulation of sleep and metabolic pathways remains ill-defined. Sleep restriction or total sleep deprivation has been shown to reduce the number and the amplitude of genes exhibiting circadian rhythmicity (10, 11, 23). The expression of genes affected by sleep restriction included circadian clock and sleep homeostasis genes, as well those associated with oxidative stress and metabolism. How these changes in the transcriptome translate into changes in the metabolome, however, remains unknown.
Metabolomics can be used not only to study homeostatic regulation but also system perturbations. The metabolic pathways involved in human sleep and during sleep deprivation have not yet been systematically studied by using a metabolomics approach. Since 24 h sleep deprivation primarily affects the homeostatic regulator of sleep, acute total sleep deprivation permits assessment of the metabolic basis of this homeostatic process. The aim of the present study was thus to characterize plasma metabolite rhythms using untargeted and targeted liquid chromatography (LC)/MS metabolomics in healthy male participants during a 24 h wake/sleep cycle followed by 24 h of wakefulness.
Results
All participants had a melatonin onset time within a similar range (mean ± SEM, 21:47 ± 0:35 h, day 1, wake/sleep), and this did not change significantly between study days (21:58 ± 0:31 h, day 2, wake/wake). Levels of melatonin were significantly increased (27 ± 5%) during sleep deprivation [00:00–06:00 h, night 2 (N2)] compared with during sleep [00:00–06:00 h, night 1 (N1); P < 0.005; SI Appendix, Fig. S1].
By using the untargeted LC/MS metabolomics method described, we extracted 367 metabolite features for each sample. A principal component analysis (PCA) of all data showed there was clear time-of-day variation in the metabolome in principal component (PC) 1 [amount of variance in the x matrix explained by PC1 (R2) = 0.145, estimate of the predictive ability of the model (Q2) (cumulative) = 0.437; Fig. 1A], with the mean score on PC1 having a significant fit to a cosine curve on day 1 (wake/sleep) and day 2 (wake/wake). There was a 14% reduction in cosinor amplitude from day 1 (wake/sleep) to day 2 (wake/wake). Orthogonal partial least squares discriminant analysis (OPLS-DA) models, validated by permutation analysis, showed some separation between sleep (00:00–06:00 h day 1) and sleep deprivation periods [00:00–06:00 h day 2; Q2 (cumulative) = 0.0692, total amount of variance explained in the x matrix (R2X) (cumulative) = 0.188, total amount of variance explained in the y matrix (R2Y) (cumulative) = 0.546; Fig. 1B], and clear separation with time of day (14:00–18:00 h vs. 02:00–06:00 h) on day 1 [Q2 (cumulative) = 0.546, R2X (cumulative) = 0.262, R2Y (cumulative) = 0.886; Fig. 1C] and day 2 [Q2 (cumulative) = 0.604, R2X (cumulative) = 0.250, R2Y (cumulative) = 0.867; Fig. 1D].
Fig. 1.
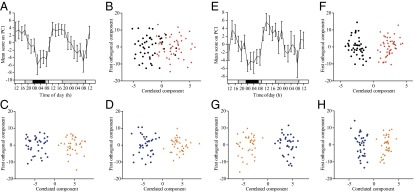
Multivariate analysis of untargeted (A–D) and targeted (E–H) metabolomics data. (A and E) PCA of all analysis data were carried out, and the change in mean score (±SEM) across all subjects on PC1 with time is shown (A, untargeted; E, targeted). White bars, awake, 90 lx, free to move; gray bars, awake, <5 lx, semirecumbent; black bars, sleeping with eye masks, 0 lx, supine. (B and F) OPLS-DA models separating selected time points according to sleep status (B, untargeted; F, targeted); black circles, sleep, day 1 00:00–06:00 h; red diamonds, sleep deprivation, day 2 00:00–06:00 h). (C, D, G, and H) OPLS-DA models (validated by permutation analysis) in which selected time periods were classed according to time of day (blue circles, 02:00–06:00 h; orange triangles, 14:00–18:00 h). This analysis was carried out for day 1 (C, untargeted; G, targeted) and day 2 (D, untargeted; H, targeted).
From the 367 features, we identified metabolites including acylcarnitines, lysophospholipids, phosphocholines, amino acids, cortisol, and bilirubin by using our previously published masses and fragments (19) [intraassay quality control (QC) coefficient of variation (CV), 25 ± 3%]. Eight acylcarnitines [propionylcarnitine (C3:0), octenoylcarnitine (C8:1), decanoylcarnitine (C10:0), dodecanoylcarnitine (C12:0), tetradecanoylcarnitine (C14:0), tetradenenoylcarnitine (C14:1), tetradecadienoylcarnitine (C14:2), hexadecanoylcarnitine (C16:0)] of the 14 identified were found to have significantly different levels between sleep (00:00–06:00 h, day 1) and sleep deprivation (00:00–06:00 h, day 2). All but octenoylcarnitine (C8:1) were increased during sleep deprivation compared with during sleep. As many of the metabolite classes identified using our established LC/MS methodology could also be measured using the Biocrates AbsoluteIDQ p180 targeted metabolomics kit, we elected to run duplicate samples using the AbsoluteIDQ p180 kit to increase the number of metabolites screened (n = 171) and to be able to measure absolute metabolite concentrations against validated standards. In a PCA of all targeted metabolomic analysis data, there was clear time-of-day variation in the metabolome in principal component 1 [R2 (PC1) = 0.364, Q2 (cumulative) = 0.768; Fig. 1E], with the mean score on PC1 having a significant fit to a cosine curve on day 1 (wake/sleep) and day 2 (wake/wake). There was a 24% reduction in cosinor amplitude from day 1 to day 2. OPLS-DA models, validated by permutation analysis, showed clear separation between sleep (00:00–06:00 h, day 1) and sleep deprivation [(00:00–06:00 h, day 2; Q2 (cumulative) = 0.746, R2X (cumulative) = 0.635, R2Y (cumulative) = 0.892; Fig. 1F], and clear separation with time of day (14:00–18:00 h vs. 02:00–06:00 h) on day 1 [Q2 (cumulative) = 0.830, R2X (cumulative) = 0.555, R2Y (cumulative) = 0.920; Fig. 1G] and day 2 [Q2 (cumulative) = 0.825, R2X (cumulative) = 0.530, R2Y (cumulative) = 0.913; Fig. 1H]. The p(corr) values for each OPLS-DA model are given in SI Appendix, Table S2. For completeness, OPLS-DA models of selected time windows across the 24 h period were generated. These OPLS-DA plots and the p(corr) values for each OPLS-DA model are presented in SI Appendix, Fig. S2 and Table S3, respectively.
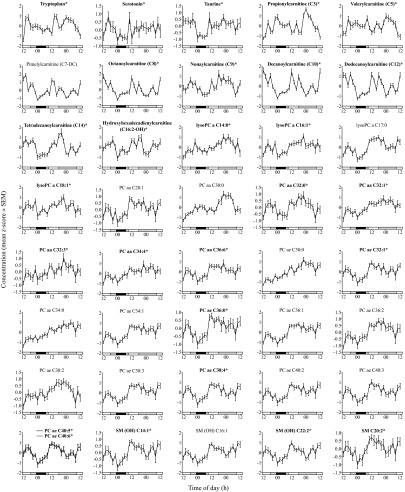
Metabolites showing significant differences (P < 0.05 and q < 0.05) across selected time periods, time of day, and selected time period-by-time of day interaction are shown in SI Appendix, Table S4. As expected, most significant differences in metabolite levels were observed in the time of day grouping (range, 59–159 of 171). Looking at selected time periods across the protocol, comparison between the sleep and sleep deprivation period revealed the most significant differences (n = 41) compared with the other time periods (range, 0–21). A Venn diagram showing the number of metabolites that were significantly different between the sleep and sleep-deprivation time periods (00:00–06:00 h), time of day, and interaction, as well as the overlap, is presented in SI Appendix, Fig. S3. Of the total 171 metabolites quantified, 41 metabolites exhibited significant changes (P < 0.05, q < 0.05) during sleep deprivation compared with during sleep. All these metabolites (n = 41) exhibited increased levels during sleep deprivation. The percent changes between sleep and sleep deprivation for these metabolites is presented in SI Appendix, Table S5, with serotonin showing the largest change (44 ± 20%) between the two conditions. Full 48-h profiles of these metabolites are presented in Fig. 2 and are annotated in bold in SI Appendix, Tables S6 and S7. The metabolites showing significantly increased levels during sleep deprivation come from a range of classes (25 glycerophospholipids, 9 acylcarnitines, 4 sphingolipids, 2 biogenic amines, 1 amino acid). Most of these metabolites (n = 27; 66%) only increased significantly (P < 0.05, q < 0.05) during sleep deprivation compared with during sleep (annotated in Fig. 2). These metabolites comprised the amino acid, tryptophan, biogenic amines, serotonin, and taurine, eight acylcarnitines [propionylcarnitine (C3), valerylcarnitine (C5), octanoylcarnitine (C8), nonaylcarnitine (C9), decanoylcarnitine (C10), dodecanoylcarnitine (C12), tetradecanoylcarnitine (C14), hydroxyhexadecadienylcarnitine (C16:2-OH)], 13 glycerophospholipids, and three sphingolipids. Overrepresentation analysis has been performed with the IMPaLA Web tool (http://impala.molgen.mpg.de) using these 27 metabolites significantly increased in sleep deprivation (dataset of interest) and all 171 metabolites identified by using targeted analysis (background dataset; SI Appendix, Results and Discussion). The remaining metabolites (n = 14; 34%), in addition to increasing between the sleep/sleep deprivation conditions, also changed significantly (P < 0.05, q < 0.05) between the wake conditions (14:00–22:00 h on day 1 and day 2). These were predominately glycerophospholipids (n = 12, 86%) as well as one acylcarnitine (pimelylcarnitine, C7-DC) and one sphingolipid [SM (OH) C16:1].
Fig. 2.
Concentrations of 41 individual metabolites (z-score mean ± SEM) found at significantly higher levels (P < 0.05, q < 0.05) during sleep deprivation compared with during sleep (00:00–06:00 h). Asterisks and bold labels denote metabolites only showing significant changes between the sleep and sleep-deprivation (00:00–06:00 h) conditions. The metabolites not annotated also changed significantly (P < 0.05, q < 0.05) between wake periods (14:00–22:00 h) on day 1 and day 2. White bars, awake, 90 lx, free to move; gray bars, awake, < 5 lx, semirecumbent; black bars, sleeping with eye masks, 0 lx, supine.
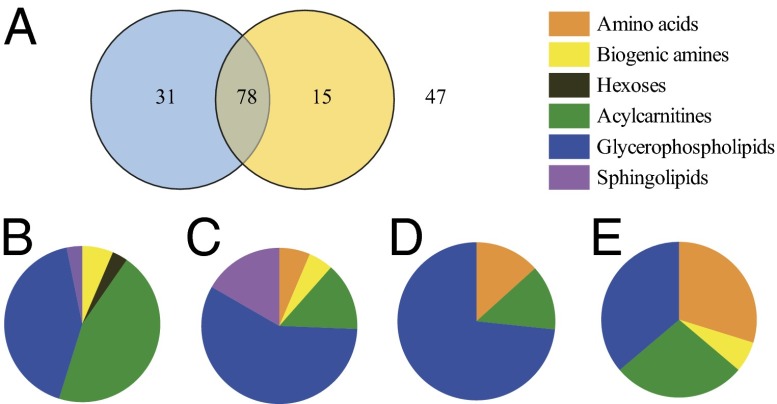
The mean (±SEM) concentration and minimum and maximum concentrations measured for each metabolite is presented in SI Appendix, Table S7. To assess rhythmic variation of the quantified metabolites, cosinor analysis was performed. Fig. 3 presents the number of rhythmic metabolites in the sleep/wake and wake/wake conditions and the classes to which these metabolites belong. Of the 171 metabolites, 109 (64%) exhibited a diurnal rhythm that had a significant fit to a cosine curve on day 1 (wake/sleep), the levels of most (n = 95; 87%) peaking during the day (06:00–18:00 h). Of those with daily rhythms, most (n = 78; 72%) maintained their rhythmicity on day 2 during the 24 h of wakefulness. These included four of the nine biogenic amines measured (α-aminoadipic acid, symmetrical dimethylarginine, kynurenine, sarcosine), 11 of the 40 acetylcarnitines, 45 of the 86 glycerophospholipids, and 13 of the 14 sphingolipids. Amino acids with significant 24 h rhythms irrespective of the presence or absence of sleep included the branched chain amino acids (BCAAs) isoleucine and valine, glutamate, ornithine, and proline, all peaking at night between 21.28 and 01.45 decimal h on day 1. The metabolites that lost their rhythmicity on day 2 (wake/wake; n = 31, 28%) were 14 acylcarnitines, 13 glycerophospholipids, one sphingolipid [SM (OH) C14:1], hexose, total dimethylarginine, and taurine. A few metabolites (n = 15, 9%) did not exhibit a significant cosine rhythm on day 1 but did on day 2. These included 11 glycerophospholipids, 2 acylcarnitines, tyrosine, and aspartate. A heat map of the metabolites that showed significant cosinor rhythms on day 1 (n = 31), day 2 (n = 15), or both day 1 and day 2 (n = 78) is presented in SI Appendix, Fig. S4. The 47 remaining metabolites (28%) that had no significant cosinor rhythm on either day comprised 17 glycerophospholipids, 14 amino acids, 13 acylcarnitines, and 3 biogenic amines (Fig. 3). The results of the cosinor analysis for all 171 metabolites (amplitude, acrophase, P value) are given in SI Appendix, Table S6. A change in acrophase and amplitude between day 1 and day 2 was calculated for the 78 metabolites with significant fits to a cosine curve on day 1 and day 2. The mean phase shift (±SEM) between day 1 and day 2 was an advance of 0.67 ± 0.14 h. The majority of these metabolites (n = 66; 85%) exhibited reduced amplitude during 24 h of wakefulness. For the metabolites that peaked during the day (n = 67; 86%), most (n = 63; 94%) exhibited a reduction in amplitude during the 24 h of wakefulness. On the contrary, for those metabolites that peaked during the night (n = 11; 14%), 24 h of wakefulness increased the amplitude of the cosinor rhythm in most cases (n = 8; 73%; SI Appendix, Table S6).
Fig. 3.
(A) Venn diagram showing the number of metabolites exhibiting a significant fit to a cosine curve on day 1 (left circle, blue), day 2 (right circle, yellow), both days (n = 78), or neither (n = 47). (B–E) Pie charts showing the proportion of metabolites from each metabolite class [exhibiting a significant fit to a cosine curve on day 1 only (B), on days 1 and day 2 (C), on day 2 only (D), and on neither day (E)].
Discussion
To our knowledge, this is the first report of metabolic profiling during sleep and acute total sleep deprivation conditions in humans. Characterizing the 24-h rhythms in plasma metabolites under these conditions by using LC/MS metabolomics offers a unique view of sleep processing and sleep/wake regulation.
A consistent finding in the untargeted and targeted analysis was that, in comparison with the number of ions and metabolites showing time-of-day rhythms, fewer ions/metabolites were significantly changed during sleep deprivation. In the targeted analysis, only 27 of the measured metabolites (16%) were significantly different between the sleep and sleep-deprivation periods without exhibiting differences during the two wake conditions. All these metabolites (serotonin, taurine, tryptophan, 8 acylcarnitines, 13 glycerophospholipids, 3 sphingolipids) increased during the sleep-deprivation period. It may be that sleep has an inhibitory effect on their synthesis or a stimulatory effect on their degradation. A single metabolomics study attempting to identify the pathways activated by waking in mice also showed that relatively few metabolite pathways were affected by 6 h of sleep deprivation, including glycolysis and lipid metabolism (24). Both in vivo after sleep deprivation and in vitro following stimulation of cortical cultures, increased levels of lysolipids were measured, suggesting that sleep may be involved in neuronal membrane homeostasis.
Serotonin is one of several neurotransmitters involved in sleep/wake regulation, functioning primarily to promote wakefulness (25). The increased plasma concentrations observed in the present study support previous studies in which higher serotonin levels have been measured in the hippocampus, dorsal raphe, and suprachiasmatic nuclei (SCN) of sleep-deprived rats (26, 27). The antidepressive effect of one night of total sleep deprivation is well established (28, 29), although the mechanism of action of this intervention remains undefined. Given that low levels of serotonin and reduced serotoninergic neurotransmission have long been associated with major depressive disorder (30), and the related efficacy of selective serotonin reuptake inhibitors in this illness (30, 31), the raised levels of serotonin we observed during sleep deprivation may provide a possible antidepressive mechanism for this intervention in humans. It also supports recent human in vivo evidence showing increased cerebral serotonin 2A receptor binding during 24 h of wakefulness (32).
Of the 21 amino acids quantified in the targeted analysis, only tryptophan was found to vary significantly with sleep status, with increased levels during acute sleep deprivation. Tryptophan is vital for the formation of serotonin and melatonin via the indoleamine pathway. Tryptophan itself, as well as 5-hydroxytryptophan, an intermediate in the indoleamine pathway, has also been used to treat depressive disorder (33, 34). The increased levels of tryptophan measured during sleep deprivation may contribute to the antidepressive effect of sleep deprivation, directly or indirectly via serotonin synthesis. The biogenic amine taurine also exhibited significantly increased levels during sleep deprivation compared with during sleep. Increased levels of taurine have been reported in the rat brain cortex following paradoxical sleep deprivation (35) and have been shown to be a activator of extrasynaptic GABA(A) receptors in the mouse ventrobasal thalamus (36), an area involved in regulating the transitions between sleep and wakefulness (37). Taurine has also been found at altered levels in the plasma of depressed patients, at increased concentrations in some cases (38), and at lower concentrations compared with controls in others (39).
It remains intriguing that the only amino acid and biogenic amines shown here to increase during acute sleep deprivation (serotonin, tryptophan, taurine) have been implicated in the etiology of depression (30, 33, 34, 38, 39). In addition, all three metabolites have relatively high concentrations in the pineal gland (40, 41). The pineal hormone melatonin was also measured in the present and previous study (11), with significantly increased plasma concentrations being observed during sleep deprivation. Increased tryptophan and taurine could explain the increased melatonin levels, as taurine has been shown to increase pineal melatonin by stimulating the activity of its rate limiting biosynthetic enzyme, N-acetyltransferase (42). Whether the antidepressive effect of acute sleep deprivation is linked to the increased circulating levels of tryptophan, serotonin, taurine, and melatonin deserves further study. In addition, whether acute sleep deprivation activates the SCN–pineal pathway requires clarification.
Levels of glycerophospholipids (25 of 86, 29%) and sphingolipids (4 of 14, 29%) also significantly increased during sleep deprivation compared with sleep. However, some of these glycerophospholipids (12 of 25) and sphingolipids (1 of 4) also increased significantly during the wake period on day 2 compared with day 1. This steady increase in circulating glycerophospholipids and sphingolipids across the study period confounds any sleep/sleep deprivation analysis. The food content at each set meal was identical, and thus diet is unlikely to explain the findings. Although the participants were free to move during some of the wake period, inactivity and lack of exercise during the laboratory study protocol may explain this accumulation. A buildup of some metabolites has previously been observed during a constant routine protocol (20). Despite the accumulation of glycerophospholipids and sphingolipids during the study protocol, most of the glycerophospholipids and sphingolipids quantified here exhibited a significant fit to a cosine curve regardless of sleep status and meal pulses. This finding is consistent with the idea that the endogenous circadian system controls lipid metabolism (43) and with previous human metabolomics and transcriptomic studies examining circadian variation (20, 21, 23) (SI Appendix, Results and Discussion). Transcriptomic data (12) also support the metabolomics data showing that transcripts associated with lipid metabolism are robustly rhythmic, and are not affected by mistimed sleep.
During sleep deprivation, nine acylcarnitines had significantly increased levels compared with during the sleep period. Most of these acylcarnitines (eight of nine) were significantly increased only during sleep deprivation compared with sleep, not between the wake conditions, suggesting an effect of high sleep pressure on acylcarnitine levels. These affected acylcarnitines were medium- and long-chain saturated acylcarnitines (C3, C5, C8, C9, C10, C12, C14, and C16:2-OH), pointing to possible changes in β-oxidation of fatty acids during sleep deprivation. The increased plasma 3-hydroxydecanoate, an intermediate in the β-oxidation of fatty acids, observed by Dallmann et al. (20) during 40 h of continual wakefulness, supports this hypothesis. Our findings support reports indicating a role for the carnitine system and fatty acid oxidation in sleep/wake regulation. Acylcarnitine levels have been reported to be low, and carnitine palmitoyltransferase 1B expression, the rate-limiting enzyme in the β-oxidation of long-chain fatty acids, has been reported to be significantly higher in patients with narcolepsy than in control subjects (44). Oral l-carnitine was recently tested in patients with narcolepsy and shown to reduce their excessive daytime sleepiness (45). Similarly low serum acylcarnitine levels have been measured in patients with chronic fatigue syndrome (46), another condition with reported sleep problems (47), and l-carnitine administration has also shown some benefit in reducing fatigue (48). These studies support the growing evidence from mutant mice studies that altered fatty acid metabolism affects sleep signaling (49). The mechanism underlying these observed changes (e.g., sympathetic tone and lipolysis or mitochondrial function and fatty acid oxidation) and their source (liver, skeletal muscle, fat) require further study.
The present study protocol was also designed to be able to assess the effect of 24 h wakefulness on the daily metabolite rhythms observed in a regular sleep/wake cycle. When all metabolites were considered simultaneously, PCA revealed a clear time-of-day variation with a significant fit to a cosine curve during a wake/sleep cycle and during 24 h of wakefulness in untargeted and targeted analysis. Cosinor analysis of the individual plasma metabolites identified by the targeted screens (n = 171) demonstrated robust daily rhythms in the majority of metabolites, confirming and extending the findings from previous human diurnal and circadian metabolomics studies (19–21) (SI Appendix, Results and Discussion). The overall effect of 24 h wakefulness on the PCA-derived daily wake/sleep rhythm was a reduction in amplitude in the untargeted and targeted analysis, which is consistent with reported transcriptomic sleep deprivation data (11, 23). In the targeted screens (n = 171), of the 109 metabolites that exhibited significant fits to a cosine curve during wake/sleep, the majority (72%) maintained their rhythmicity during the 24 h of wakefulness. Metabolites that peaked during the day predominately showed reduced amplitude during the 24 h wake period, whereas metabolites that peaked during the night showed increased amplitude. These time-specific changes in amplitude are consistent with increased levels of metabolites during sleep deprivation.
Of the 40 acylcarnitines quantified with targeted analysis, 63% were shown to vary significantly with time of day on day 1 (wake/sleep), and 56% retained this rhythmicity during sleep deprivation. The daily rhythms in acylcarnitines support previous studies (19, 20) (SI Appendix, Results and Discussion) and gene expression data, with key transporters of long-chain acylcarnitines exhibiting clear oscillation with time of day in mice (50).
Most of the amino acids (76%) did not vary significantly with time of day, possibly reflecting the effect of the timed meals on these amino acid profiles (51). However, glutamate, ornithine, proline, and two of the BCAAs, isoleucine and valine, had 24-h rhythms with significant fits to a cosine curve, irrespective of the presence or absence of sleep or meals, with levels peaking at night (between 21:00 and 01:30 h). Knowledge of this daily variation in isoleucine and valine may aid in optimizing their therapeutic efficacy, for example, to improve protein metabolism in patients with liver cirrhosis, in whom administration of BCAAs at night is known to be more effective than during the day (52). Further characterization of the endogenous fluctuations of metabolites such as BCAAs under different conditions (e.g., in type 2 diabetes) may help to generate testable hypotheses relating to the best time of day to administer treatments most effectively.
The present study assessed metabolite profiles during a single night of total sleep deprivation with the aim of characterizing the effect of increased sleep pressure on metabolite levels as well as investigating the masking effect of sleep on daily metabolite rhythms. Acute total sleep deprivation, in addition to permitting investigation of the homeostatic sleep drive, also mimics the first night of night shift work and one methodology of antidepressive therapy used in psychiatry (28, 29). The protocol, however, does not allow identification of metabolites that might only be affected by prolonged, repeated sleep deprivation; the metabolic changes observed under chronic partial sleep deprivation remain to be determined.
In conclusion, we have identified plasma metabolites that were significantly altered during acute sleep deprivation (mainly lipids and acylcarnitines, serotonin, tryptophan, and taurine), all increasing during sleep deprivation. The 24-h variation in several metabolite classes (amino acids, biogenic amines, acylcarnitines, glycerophospholipids, sphingolipids) has also been characterized in the presence and absence of a night’s sleep. Determining the full impact of exogenous factors such as sleep on the metabolome will be crucial for the future metabolic profiling-based identification of biomarkers of disease and drug effects. In addition, this metabolomics approach is a step toward understanding sleep/wake regulation and the associated metabolic pathways involved in these processes.
Materials and Methods
Clinical Study.
The study was approved by the University of Surrey Ethics Committee and conducted according to the Declaration of Helsinki. Subjects had to meet defined inclusion/exclusion criteria to be deemed eligible for the study (SI Appendix, Materials and Methods).
In-Laboratory Session.
The 4-d in-laboratory session was conducted at the Surrey Clinical Research Centre and consisted of an adaptation night followed by a 48-h sampling period beginning at 12:00 h, which comprised a 24-h period (day 1) incorporating an 8-h sleep opportunity (23:00–07:00 h; N1), followed by 24 h (beginning at 12:00 h, day 2) during which subjects remained continually awake (day 2/N2; SI Appendix, Fig. S5). Blood samples were collected for 48 h at hourly and 2-hourly intervals from 12:00 h on day 1 for melatonin and LC/MS analysis, respectively (SI Appendix, Materials and Methods).
Metabolomic Analysis.
Details of the untargeted and targeted metabolomics analysis, as well as all statistical analyses, are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Daniel Barrett, Cheryl Isherwood, and the Surrey Clinical Research Centre medical, clinical, and research teams for their help with the clinical study. This work was supported in part by the Netherlands Forensic Institute, Netherlands Genomics Initiative/Netherlands Organization for Scientific Research within the framework of the Forensic Genomics Consortium Netherlands, the 6th Framework project EUCLOCK (018741), and UK Biotechnology and Biological Sciences Research Council Grant BB/I019405/1. D.J.S. is a Royal Society Wolfson Research Merit Award holder.
Footnotes
Conflict of interest statement: D.J.S. has received research support from Philips Lighting. D.J.S. and B.M. are codirectors of Stockgrand. V.L.R. is a scientific advisor to Lumie and has received research support from Philips Lighting.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402663111/-/DCSupplemental.
References
- 1.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckel-Mahan K, Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev. 2013;93(1):107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass J, Turek FW. Sleepless in America: A pathway to obesity and the metabolic syndrome? Arch Intern Med. 2005;165(1):15–16. doi: 10.1001/archinte.165.1.15. [DOI] [PubMed] [Google Scholar]
- 6.Spiegel K, Knutson K, Leproult R, Tasali E, Van Cauter E. Sleep loss: A novel risk factor for insulin resistance and type 2 diabetes. J Appl Physiol (1985) 2005;99(5):2008–2019. doi: 10.1152/japplphysiol.00660.2005. [DOI] [PubMed] [Google Scholar]
- 7.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappuccio FP, Cooper D, D’Elia L, Strazzullo P, Miller MA. Sleep duration predicts cardiovascular outcomes: A systematic review and meta-analysis of prospective studies. Eur Heart J. 2011;32(12):1484–1492. doi: 10.1093/eurheartj/ehr007. [DOI] [PubMed] [Google Scholar]
- 9.Rey G, Reddy AB. Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 2013;23(5):234–241. doi: 10.1016/j.tcb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci USA. 2007;104(50):20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackermann K, et al. Effect of sleep deprivation on rhythms of clock gene expression and melatonin in humans. Chronobiol Int. 2013;30(7):901–909. doi: 10.3109/07420528.2013.784773. [DOI] [PubMed] [Google Scholar]
- 12.Archer SN, et al. Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci USA. 2014;111(6):E682–E691. doi: 10.1073/pnas.1316335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raamsdonk LM, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19(1):45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 14.Clayton TA, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440(7087):1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 15.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134(5):714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106(24):9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fustin JM, et al. Rhythmic nucleotide synthesis in the liver: Temporal segregation of metabolites. Cell Reports. 2012;1(4):341–349. doi: 10.1016/j.celrep.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ang JE, et al. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int. 2012;29(7):868–881. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasukawa T, et al. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci USA. 2012;109(37):15036–15041. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1(3):195–204. [PubMed] [Google Scholar]
- 23.Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110(12):E1132–E1141. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinard V, et al. Key electrophysiological, molecular, and metabolic signatures of sleep and wakefulness revealed in primary cortical cultures. J Neurosci. 2012;32(36):12506–12517. doi: 10.1523/JNEUROSCI.2306-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monti JM. Serotonin control of sleep-wake behavior. Sleep Med Rev. 2011;15(4):269–281. doi: 10.1016/j.smrv.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Peñalva RG, et al. Effect of sleep and sleep deprivation on serotonergic neurotransmission in the hippocampus: A combined in vivo microdialysis/EEG study in rats. Eur J Neurosci. 2003;17(9):1896–1906. doi: 10.1046/j.1460-9568.2003.02612.x. [DOI] [PubMed] [Google Scholar]
- 27.Alfaro-Rodríguez A, González-Piña R, González-Maciel A, Arch-Tirado E. Serotonin and 5-hydroxy-indole-acetic acid contents in dorsal raphe and suprachiasmatic nuclei in normal, malnourished and rehabilitated rats under 24 h of sleep deprivation. Brain Res. 2006;1110(1):95–101. doi: 10.1016/j.brainres.2006.06.069. [DOI] [PubMed] [Google Scholar]
- 28.Wu JC, Bunney WE. The biological basis of an antidepressant response to sleep deprivation and relapse: review and hypothesis. Am J Psychiatry. 1990;147(1):14–21. doi: 10.1176/ajp.147.1.14. [DOI] [PubMed] [Google Scholar]
- 29.Wirz-Justice A, Van den Hoofdakker RH. Sleep deprivation in depression: What do we know, where do we go? Biol Psychiatry. 1999;46(4):445–453. doi: 10.1016/s0006-3223(99)00125-0. [DOI] [PubMed] [Google Scholar]
- 30.Ressler KJ, Nemeroff CB. Role of serotonergic and noradrenergic systems in the pathophysiology of depression and anxiety disorders. Depress Anxiety. 2000;12(suppl 1):2–19. doi: 10.1002/1520-6394(2000)12:1+<2::AID-DA2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Vaswani M, Linda FK, Ramesh S. Role of selective serotonin reuptake inhibitors in psychiatric disorders: a comprehensive review. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27(1):85–102. doi: 10.1016/s0278-5846(02)00338-x. [DOI] [PubMed] [Google Scholar]
- 32.Elmenhorst D, Kroll T, Matusch A, Bauer A. Sleep deprivation increases cerebral serotonin 2A receptor binding in humans. Sleep. 2012;35(12):1615–1623. doi: 10.5665/sleep.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coppen A, Noguera R. L-tryptophan in depression. Lancet. 1970;1(7656):1111. [PubMed] [Google Scholar]
- 34.Turner EH, Loftis JM, Blackwell AD. Serotonin a la carte: Supplementation with the serotonin precursor 5-hydroxytryptophan. Pharmacol Ther. 2006;109(3):325–338. doi: 10.1016/j.pharmthera.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Mohammed HS, Aboul Ezz HS, Khadrawy YA, Noor NA. Neurochemical and electrophysiological changes induced by paradoxical sleep deprivation in rats. Behav Brain Res. 2011;225(1):39–46. doi: 10.1016/j.bbr.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Jia F, et al. Taurine is a potent activator of extrasynaptic GABA(A) receptors in the thalamus. J Neurosci. 2008;28(1):106–115. doi: 10.1523/JNEUROSCI.3996-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coulon P, Budde T, Pape H-C. The sleep relay—the role of the thalamus in central and decentral sleep regulation. Pflugers Arch. 2012;463(1):53–71. doi: 10.1007/s00424-011-1014-6. [DOI] [PubMed] [Google Scholar]
- 38.Altamura C, Maes M, Dai J, Meltzer HY. Plasma concentrations of excitatory amino acids, serine, glycine, taurine and histidine in major depression. Eur Neuropsychopharmacol. 1995;5(suppl):71–75. doi: 10.1016/0924-977x(95)00033-l. [DOI] [PubMed] [Google Scholar]
- 39.Perry TL, et al. Hereditary mental depression and Parkinsonism with taurine deficiency. Arch Neurol. 1975;32(2):108–113. doi: 10.1001/archneur.1975.00490440058009. [DOI] [PubMed] [Google Scholar]
- 40.Quay WB. Circadian rhythm in rat pineal serotonin and its modifications by estrous cycle and photoperiod. Gen Comp Endocrinol. 1963;14:473–479. doi: 10.1016/0016-6480(63)90079-0. [DOI] [PubMed] [Google Scholar]
- 41.Vellan EJ, Gjessing LR, Stalsberg H. Free amino acids in the pineal and pituitary glands of human brain. J Neurochem. 1970;17(5):699–701. doi: 10.1111/j.1471-4159.1970.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 42.Wheler GH, Weller JL, Klein DC. Taurine: stimulation of pineal N-acetyltransferase activity and melatonin production via a beta-adrenergic mechanism. Brain Res. 1979;166(1):65–74. doi: 10.1016/0006-8993(79)90650-4. [DOI] [PubMed] [Google Scholar]
- 43.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121(6):2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyagawa T, et al. Abnormally low serum acylcarnitine levels in narcolepsy patients. Sleep. 2011;34(3):349–53A. doi: 10.1093/sleep/34.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyagawa T, et al. Effects of oral L-carnitine administration in narcolepsy patients: A randomized, double-blind, cross-over and placebo-controlled trial. PLoS ONE. 2013;8(1):e53707. doi: 10.1371/journal.pone.0053707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuratsune H, et al. Low levels of serum acylcarnitine in chronic fatigue syndrome and chronic hepatitis type C, but not seen in other diseases. Int J Mol Med. 1998;2(1):51–56. doi: 10.3892/ijmm.2.1.51. [DOI] [PubMed] [Google Scholar]
- 47.Fukuda K, et al. International Chronic Fatigue Syndrome Study Group The chronic fatigue syndrome: A comprehensive approach to its definition and study. Ann Intern Med. 1994;121(12):953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 48.Plioplys AV, Plioplys S. Amantadine and l-carnitine treatment of chronic fatigue syndrome. Neuropsychobiology. 1997;35(1):16–23. doi: 10.1159/000119325. [DOI] [PubMed] [Google Scholar]
- 49.Tafti M, et al. Deficiency in short-chain fatty acid β-oxidation affects theta oscillations during sleep. Nat Genet. 2003;34(3):320–325. doi: 10.1038/ng1174. [DOI] [PubMed] [Google Scholar]
- 50.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashley DV, Barclay DV, Chauffard FA, Moennoz D, Leathwood PD. Plasma amino acid responses in humans to evening meals of differing nutritional composition. Am J Clin Nutr. 1982;36(1):143–153. doi: 10.1093/ajcn/36.1.143. [DOI] [PubMed] [Google Scholar]
- 52.Fukushima H, et al. Nocturnal branched-chain amino acid administration improves protein metabolism in patients with liver cirrhosis: comparison with daytime administration. JPEN J Parenter Enteral Nutr. 2003;27(5):315–322. doi: 10.1177/0148607103027005315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.