Significance
There is considerable information about the spatial regulation of gene expression during pattern formation in animal development. Significantly less is known about temporal control, in part due to our inability to analyze gene activity in real time. Using a recently developed approach for the visualization of gene expression in living Drosophila embryos, we examined the well-known even-skipped stripe 2 expression pattern. Surprisingly, we observe that this classic pattern is quite transient and generated by discontinuous surges of transcriptional activity in individual cells. These results challenge a purely static framework for dissecting developmental programs and emphasize the importance of the dynamic features of pattern formation.
Abstract
We present the use of recently developed live imaging methods to examine the dynamic regulation of even-skipped (eve) stripe 2 expression in the precellular Drosophila embryo. Nascent transcripts were visualized via MS2 RNA stem loops. The eve stripe 2 transgene exhibits a highly dynamic pattern of de novo transcription, beginning with a broad domain of expression during nuclear cycle 12 (nc12), and progressive refinement during nc13 and nc14. The mature stripe 2 pattern is surprisingly transient, constituting just ∼15 min of the ∼90-min period of expression. Nonetheless, this dynamic transcription profile faithfully predicts the limits of the mature stripe visualized by conventional in situ detection methods. Analysis of individual transcription foci reveals intermittent bursts of de novo transcription, with duration cycles of 4–10 min. We discuss a multistate model of transcription regulation and speculate on its role in the dynamic repression of the eve stripe 2 expression pattern during development.
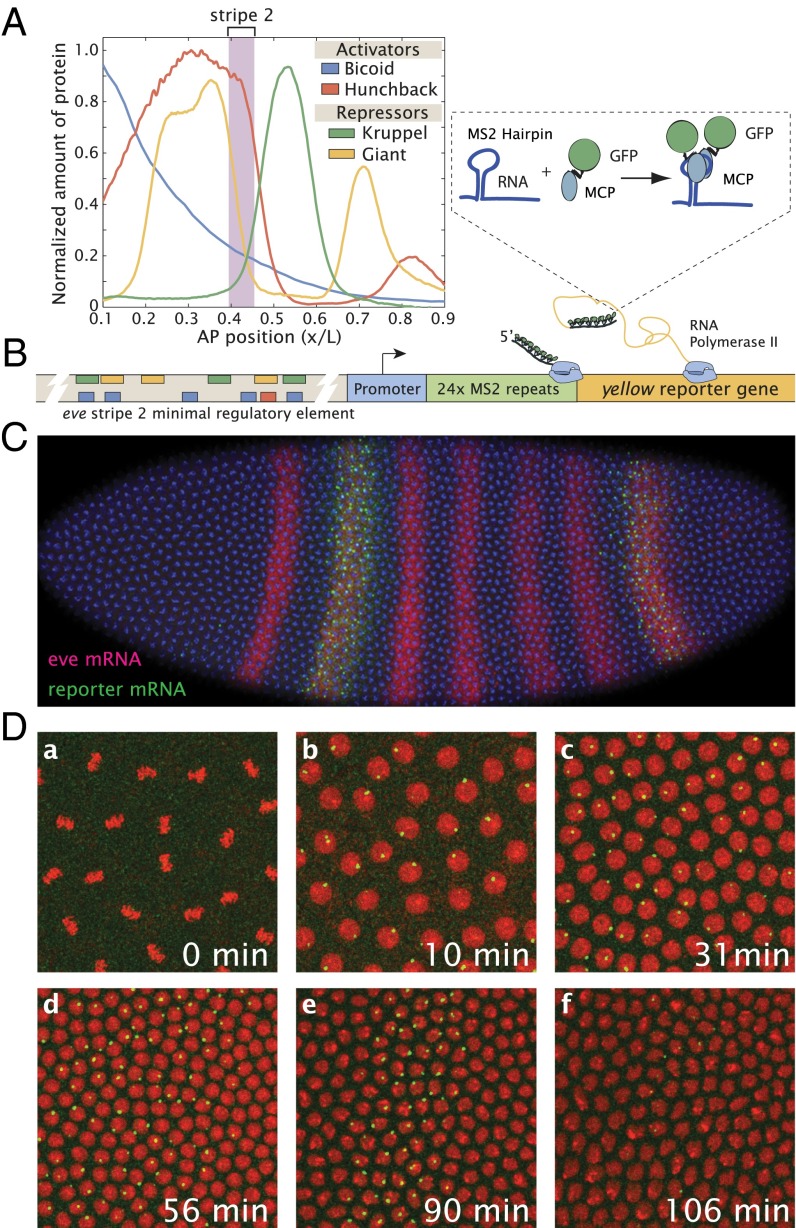
The Drosophila even-skipped (eve) stripe 2 enhancer is one of the best-characterized cis-regulatory DNAs in animal development (1). A combination of genetic analyses, DNA binding assays, and site-directed mutagenesis led to a detailed model for the regulation of stripe 2, whereby the maternal Bicoid gradient, in concert with zygotic Hunchback protein, defines a broad domain of activation in the anterior half of the embryo (2–5). Localized gap repressors, Giant in anterior regions and Kruppel in central regions, establish the anterior and posterior stripe borders, respectively (summarized in Fig. 1A).
Fig. 1.
Live imaging of eve stripe 2 transcriptional activity. (A) Schematic representation of eve stripe 2 regulation (data from refs. 38 and 39). The stripe is the result of the combined activation of Bicoid and Hunchback, which define a broad activation domain in the anterior part of the embryo, and repressors Giant and Kruppel, which restrict expression anterior and posterior of this domain, respectively (4). (B) Structure of the reporter construct: the eve stripe 2 DNA region (−1.7 kbp, +50 bp) was placed upstream of 24 repeats of the MS2 stem loops and a yellow reporter gene. The MCP-GFP protein that binds to these stem loops is present in the unfertilized egg and in the early embryo. (C) Confocal image of a transgenic embryo carrying the eve>MS2 transgene labeled via in situ hybridization with full-length probes for the yellow reporter gene (plasmid transgene) and endogenous eve RNAs in the same embryo during nc14. (D) Projected confocal stack of a live Drosophila embryo at six time points (a–f) centered at ∼40% embryo length, expressing the eve>MS2 transgene, histone RFP (red), and MCP-GFP (green). Each image is 77 μm × 77 μm. (a) At metaphase of nc11, no foci of transcription are detectable. (b) Same embryo 10.3 min later than a during nc12 interphase. There are clear fluorescent foci indicating sites of nascent transcript formation. (c) Embryo in nc13 interphase showing broad expression of the transgene. (d) At the onset of nc14, the stripe pattern has started to refine. (e) Refined stripe by late nc14. (f) Embryo just before gastrulation when the transgene expression has diminished significantly.
Most of our information regarding the regulation of the stripe 2 expression pattern is derived from the analysis of fixed preparations of staged embryos (2, 5, 6). Here, we use a newly developed live-imaging technique (7, 8) to explore the detailed temporal dynamics of the eve stripe 2 expression pattern in living embryos. Multiple copies of an MS2 stem loop sequence were inserted into the 5′-UTR of a yellow reporter transgene (Fig. 1B). The loops form upon transcription by RNA polymerase II (Pol II) and are bound by a maternally provided MS2 coat protein fused to GFP (MCP-GFP) (9–14). As a result, fluorescence signals are detected at sites of Pol II elongation and de novo transcription, and the strength of the signals are proportional to the number of elongating Pol II complexes (7).
This method was recently used to examine the activation of the proximal hunchback enhancer by the Bicoid gradient in the anterior half of the precellular embryo (7, 8). Diminishing levels of Bicoid were shown to cause stochastic on/off transcription of the hunchback>MS2 transgene at the posterior limits of the Hunchback expression pattern. This observation suggests that the Bicoid activator not only augments the levels of transcription but also increases the probability that a given cell within a population will initiate expression (15).
The regulation of the hunchback>MS2 transgene is rather static. Once activated by Bicoid during nuclear division cycle 10 (nc10) (16), the spatial features of the pattern remain essentially constant for the next hour until transcription is lost at the midpoint of nc14 (7, 17). In contrast, the eve stripe 2 pattern is highly dynamic, with broad activation during nc11 and nc12, followed by progressive refinement during nc13 and nc14 (4). These regulatory dynamics are nicely captured by the MS2 detection system and reveal surprisingly transient expression of the mature stripe (Fig. 1D and Movie S1). We also present evidence for the occurrence of sporadic transcriptional bursts, with fluctuation cycles of 4–10 min. We discuss the possibility that these discontinuities in de novo transcription facilitate the dynamic regulation of eve stripe 2 expression by the localized Giant and Kruppel repressors.
Results
The first 1.7 kb of the eve 5′ flanking region was attached to a yellow reporter gene containing 24 tandem repeats of the 60- to 70-bp MS2 stem loop motif [summarized in Fig. 1B (18)]. The eve>MS2 fusion gene contains the “full-length” 720-bp eve stripe 2 enhancer, located between –1.5 kb and –800 bp upstream of the eve transcription start (4, 19). It also contains dispersed regulatory sequences that mediate weak expression within the limits of stripe 7 (Fig. 1C). Conventional in situ hybridization assays identify authentic stripe 2 and stripe 7 expression patterns, as seen for similar eve reporter genes lacking MS2 stem loop sequences (e.g., refs. 20 and 21), confirming that the presence of the stem loops does not significantly affect the output pattern of expression.
Dynamics of Stripe Formation.
The eve>MS2 transgene was introduced into embryos containing a maternally expressed MCP-GFP fusion protein, as described in ref. 7. Sites of de novo transcription were imaged by sampling a series of confocal z sections through the entirety of cortical nuclei. The fluorescence intensities of these expression puncta is a proxy for the number of Pol II molecules actively transcribing the reporter gene and hence is an instantaneous measure of activity (7). A broad spatial domain of transcriptional activity is detected during nc11, nc12, and nc13 (e.g., Fig. 1 D, a–c and Movies S1–S3). The stripe is gradually refined during nc14 (Fig. 1 D, d and e), and ultimately disappears before the onset of gastrulation (Fig. 1 D, f; see below). The dynamics of the stripe 2 expression pattern, broad activation followed by localized repression, is consistent with previous studies of fixed embryos (e.g., ref. 4). As observed in classical studies (e.g., ref. 22), there is no evidence of transcription during mitosis (e.g., Fig. 1 D, a).
There is a marked restriction in the posterior limits of the reactivated expression pattern following the mitosis at nc13/nc14 (Movies S2 and S3). During nc13, de novo transcription is observed throughout most of the length of the embryo, spanning 10–70% along the anterior-posterior (AP) limits of the embryo. However, at the onset of nc14, the transgene is reactivated within tighter spatial limits, from 20% to 50% across the AP axis. This initial nc14 pattern is broader than the final limits of the mature stripe, but considerably more restricted than the pre-nc14 pattern (Movies S2 and S3).
The progressive refinement of the mature stripe 2 expression pattern (Fig. 2 A–C) is revealed by quantifying the instantaneous fraction of nuclei exhibiting fluorescent signals of de novo transcription. There is relatively uniform activation of eve>MS2 transcription within a broad domain centered at 40% embryo length of nc13 embryos, This pattern is refined into the mature stripe during a 20-min window of nc14 (Fig. 2D, dashed line), but quickly disappears before the onset of gastrulation (Fig. 2E). The mature stripe 2 pattern is far more transient than the picture formed from conventional in situ hybridization assays using fixed embryos (e.g., refs. 20 and 21).
Fig. 2.
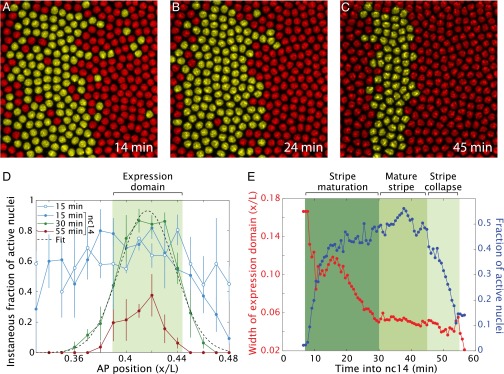
Formation and refinement of stripe 2 expression domain. (A–C) Snapshots of a Drosophila embryo expressing the eve>MS2 reporter at different times in nc14 centered at ∼37% embryo length. Nuclei that show foci of active transcription have been false-colored yellow. (D) Instantaneous fraction of active nuclei as a function of position in nc13 and at different times during nc14. The expression domain is defined as the area within the full width at half-maximum of a Gaussian fit to the profile at each time point. (E) Expression domain width and fraction of active nuclei within the domain as a function of time obtained from Gaussian fits as shown in D. After entry into nc14, the width of the domain refines and the fraction of active nuclei within it increases. The mature stripe is stable for 15 min and decays rapidly as gastrulation approaches. The temporal progression of the spatial profile of the fraction of active nuclei is also shown in Movie S5. (All data were obtained by averaging over four embryos; error bars correspond to SEMs.)
Dynamic Regulation Predicts the Mature Expression Pattern.
Image analysis methods were used to quantify the signal intensities of individual transcription foci in an effort to understand how the dynamic de novo transcription profile produces a steady-state stripe of expression (7). Signal intensities were tracked for individual nuclei during the entirety of nc14 development. Some of these nuclei reside within the limits of the mature stripe, whereas others are located just beyond the anterior and posterior borders of the stripe (Fig. 3A). Interestingly, there is an overall reduction of signal intensities within the stripe 2 domain at the midpoint of nc14 (arrow, Fig. 3A). This transient reduction might reflect a change in the regulatory landscape, for example, due to the accumulation of Giant and Kruppel repressors as the mature stripe is formed. A similar reduction in expression was observed for the hunchback>MS2 transgene at ∼12 min after onset of nc14 (7).
Fig. 3.
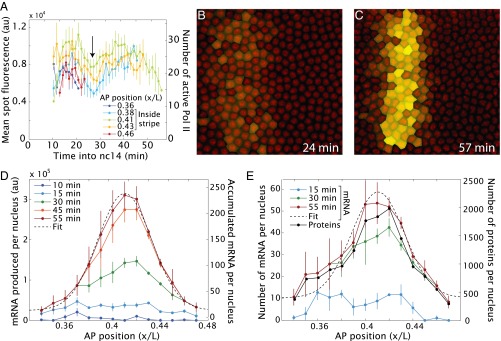
Dynamics of eve stripe 2 mRNA distribution. (A) Mean spot fluorescence, indicating transcriptional activity, as a function of time for different positions along the AP axis of the embryo. The arrow indicates a reduction in the average fluorescence that consistently occurs at about 28 min into nc14 in all embryos observed. (B and C) By integrating the total fluorescence as a function of time and assuming no mRNA degradation, it is possible to predict the amount of accumulated mRNA (SI Text). The intensity of the yellow false-color label is proportional to the amount of mRNA produced in each nucleus. (D) Total amount of mRNA produced per nucleus, assuming no degradation as a function of position along the AP axis at different time points during nc14. The absolute number of mRNA molecules should be seen as an estimate (SI Text). (E) Number of mRNA and protein molecules per nucleus assuming an eve mRNA half-life of 7 min (24), a protein translation rate of one protein per mRNA per min (25) and a protein half-life of 6–40 min (26). The exact half-life of eve mRNA that is used for this model has little influence on the qualitative appearance of the stripe (Fig. S1 and Movie S4). The temporal progression of all parameters is shown in Movie S5. (All data were obtained by averaging over four embryos; error bars correspond to SEMs.)
By integrating fluorescent traces over time, we calculated the spatial profile of mRNAs produced per nucleus at each time point during nc14 (Fig. 3 B–D, SI Text, and Movie S4) (7). The mature stripe of steady-state mRNAs becomes apparent at ∼30 min into nc14, when each nucleus produces ∼130 mRNAs (SI Text). To systematically quantify the width of the stripe, a Gaussian curve was fitted to the mRNA accumulation profiles at the end of nc14 (Fig. 3D). This analysis reveals a mature stripe that is centered at 41.0 ± 0.1% egg length with half-maximum at full-width limits of 5.3 ± 0.1% egg length. The stripe position is entirely consistent with previous reports based on in situ hybridization assays and antibody staining of fixed embryos (located between 39% and 43% egg length; see ref. 23).
As mentioned above, the de novo transcription profiles predict the final limits of the mature stripe 2 pattern (Figs. 2C and 3C) as visualized by conventional methods (Fig. 1C). Thus, posttranscriptional mechanisms, such as differential mRNA stability, are unnecessary to account for the dynamics of the stripe 2 expression pattern. However, mRNA half-life does affect the total number of steady-state mRNAs and proteins present within the stripe. When we assume a half-life of ∼7 min, as measured for ftz mRNAs (24), the position and width of the stripe remains unchanged, but there is a reduction in the total number of steady-state mRNAs (Fig. 3E and Movie S5). Furthermore, assuming a protein translation rate of one protein per mRNA per min (25) and a protein half-life of 6–40 min (26), we predict an average of ∼1,200 Eve proteins per nucleus within the mature stripe 2 domain. It should be possible to test these predictions using quantitative in situ detection methods.
Bursts of Transcriptional Activity in Individual Nuclei.
The preceding analysis reveals a highly dynamic pattern of de novo transcription. In an effort to gain insights into the underlying mechanisms, we measured the fluorescence intensities of individual foci during the entirety of nc14. There are significant oscillatory fluctuations in fluorescence signal intensities during nc14 (Fig. 4A and Fig. S2). A typical nucleus within the definitive stripe 2 domain displays reactivation of de novo transcription within ∼10 min after the onset of nc14. There are variable reductions in the levels of transcription followed by surges or bursts of expression. These fluctuations are evocative of transcriptional bursts reported in a number of other systems subject to live-image analysis, including bacteria, yeast, Dictyostelium, and cultured mammalian cells (18). Moreover, previous analysis based on fixed Drosophila embryos found evidence of transcriptional bursting at the Hox gene Scr (27) and in gap gene expression (17).
Fig. 4.
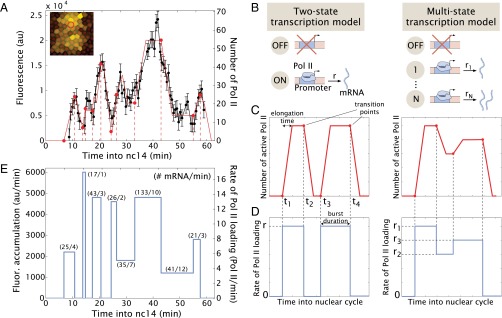
Transcriptional bursting in eve stripe 2 activity. (A) Fluorescence intensity of an individual spot within the stripe (black) and manual fits consistent with the simple model put forth in B–D (red); error bars are imaging errors as in Garcia et al. (7). Inset (51 μm × 51 μm) shows the nuclear location corresponding to the spot using false coloring as in Fig. 3C. (B) The widespread two-state model of transcription posits that promoters can be in an OFF or ON state. Transcription factors can then regulate the rates of interconversion between these two states or the rate of transcriptional initiation in the ON state. In a more general multistate model of transcription, the promoter can be found in the OFF state as well as several ON states, each one of which has a characteristic rate of transcription initiation, i.e., polymerase loading rate. (C) The strength of eve>MS2 fluorescent foci are proportionate to the number of elongating Pol II complexes across the gene template. (D) The rate of Pol II loading is related to the spot fluorescence intensity through the time Pol II molecules spend bound to the gene during transcript elongation. Two example time traces of the rate of Pol II loading and their corresponding fluorescence dynamics are shown. In the example for the two-state model, Pol II molecules are loaded onto the gene at a rate r starting at a time t1 after mitosis resulting in a linear increase of fluorescence. Once the first Pol II molecule reaches the end of the gene and falls off [∼4.2 ± 0.4 min; see Garcia et al. (7)], the number of Pol II molecules on the gene will reach steady state, resulting in a constant fluorescence value. At time t2, the promoter is switched OFF and the fluorescence intensity will decline as Pol II molecules terminate transcription. (E) Estimated rate of Pol II loading resulting from the manual fits in A. The estimated number of mRNA molecules produced per state and their duration are shown.
Transcriptional “bursts” have been associated with promoters that switch between ON and OFF states (28, 29). In this simple “two-state model,” transcription occurs only when the promoter is in the ON state and no transcription is permitted when the promoter is in the OFF state (summarized in Fig. 4B). To determine whether this simple model can describe the fluctuations of eve transcription, we calculated the dynamics of Pol II loading based on the fluorescence signal intensities at individual sites of de novo transcription. These signals are a proxy for the number of Pol II molecules actively transcribing the gene (Fig. 4 A and C). Changes in signal intensities can be directly related to the rate of Pol II loading at the promoter (Fig. 4 D and E) using a previously described model (7) (SI Text). We observed highly variable burst cycles of 4–10 min, and the production of 20–100 mRNAs per typical burst (Fig. 4 A and E, and Fig. S2). Both the time of persistence and the number of mRNAs produced per burst are comparable to those observed in other systems using similar live-imaging methods as well as fixed tissue techniques (10, 11, 28, 30). Surprisingly, however, these bursts do not present a single characteristic rate of Pol II loading but correspond to discrete values ranging from a peak of 14 elongating Pol II complexes per min to a minimum of 4 Pol II per min.
The occurrence of multiple rates of Pol II loading argues against a simple two-state model of transcription (Fig. 4B). Instead, the data are consistent with a “multistate model,” with promoter switching between several discrete transcriptional states (Fig. 4B and Fig. S2). For both the multistate model and the simpler two-state model, the molecular mechanisms underlying these multiple transcriptional states are uncertain (see below).
Discussion
During the past 30 years, we have obtained a comprehensive picture of the spatial patterning processes underlying the segmentation of the Drosophila embryo [e.g., reviewed by Levine (1)]. However, considerably less is known about the temporal dynamics of this process. Here, we applied recently developed live-imaging methods to monitor the transcriptional activity of an eve stripe 2 fusion gene in living Drosophila embryos. We found that the mature eve stipe 2 expression pattern is surprisingly short-lived and persists for only ∼15 min after it is fully formed (Fig. 2). Nonetheless, the temporal dynamics of de novo transcription accurately account for the steady-state expression of eve stripe 2 seen with conventional in situ detection methods (Fig. 3). A critical observation of this study is the occurrence of transcriptional bursts underlying the dynamic eve expression pattern. These bursts are highly variable in duration and in mRNA output, and a simple two-state model cannot explain them (Fig. 4).
The ephemeral nature of the stripe 2 expression pattern highlights our ignorance of the temporal dynamics of the segmentation gene network, despite extensive insights into the spatial control of expression (e.g., ref. 1). Timing is just as important for developmental fate decisions as the control of the spatial limits, and it is now possible to measure the temporal control of gene expression using newly developed live-imaging methods (7, 8). Indeed, recent evidence suggests that promoters with poised Pol II exhibit more rapid activation dynamics than those lacking poised Pol II, and subtle differences in the timing of expression can influence the coordination of cell invagination events during gastrulation (31).
Previous live-imaging studies have reported transcription bursts, whereby promoters switch between ON and OFF states (10, 11, 28, 30) (Fig. 4B). Such bursts have also been inferred from fixed embryo data (17, 27). However, our analysis of eve stripe 2 regulation suggests a more nuanced picture of the transcription dynamics. We find evidence for multiple ON states, with each state exhibiting a distinct rate of Pol II loading and release from the eve promoter. Because the nc14 interphase occurs after DNA replication, there are two copies of each allele on adjoining sister chromatids. Independent burst cycles from each copy could contribute to the observed multistate complexity. Additional molecular mechanisms underlying promoter switching include occupancy of transcription factor binding sites, nucleosome remodeling, disassembly of the preinitiation complex, and stochastic enhancer-promoter looping events (32–35).
It is possible that transcriptional bursting could contribute to the dynamic regulation of the eve stripe 2 expression pattern. In the critical region of refinement, there are overlapping distributions of activators (Bicoid and Hunchback) and repressors (Giant and Kruppel). At the beginning of nc14, the activators have the upper hand and it is only as the concentration of Giant and Kruppel increase that the pattern becomes refined (36). Perhaps the “OFF phase” of the eve bursts is particularly susceptible to repression during this increase in the levels of Giant and Kruppel. The OFF phase might reflect the uncoupling of the stripe 2 enhancer and transcriptional machinery, thereby rendering the enhancer DNA more accessible to the newly synthesized repressors (e.g., ref. 37). The hunchback>MS2 transgene exhibits a relatively static pattern of expression (7, 8), and it is currently unclear whether individual nuclei exhibit transcriptional bursting behaviors.
It remains to be seen whether the transcriptional bursts or surges identified in this study are a general property of gene expression in the Drosophila embryo, or a property of dynamically regulated genes such as eve. It is striking that eve transcription is stochastic and discontinuous because the Drosophila syncytium exhibits the most rapid regulatory dynamics known in animal development. Future studies will explore the possibility that previously described mechanisms of transcriptional precision, e.g., paused Pol II and shadow enhancers (e.g., ref. 37), somehow suppress transcriptional bursts to produce more uniform rates of mRNA synthesis.
Methods
Female virgins maternally expressing MCP-GFP and Histone-RFP from ref. 7 were crossed with males of the eve>MS2-yellow reporter line. Collected embryos were imaged using either two-photon or confocal microscopy. At each time point, a stack of at least 10 images separated by 0.5 µm (confocal) or 1 µm (two-photon) was acquired. MCP-GFP spots are detected, their fluorescence is quantified in 3D (7), and they are assigned to the closest segmented nucleus. See SI Methods for details on transgenic fly construction, sample preparation, and data acquisition and analysis.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM34431 (to M.L.), P50 GM071508 (to T.G.), and R01 GM097275 (to T.G.), and by Searle Scholar Award 10-SSP-274 (to T.G.). H.G.G. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and a Princeton Dicke Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410022111/-/DCSupplemental.
References
- 1.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20(17):R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5(5):827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 3.Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254(5036):1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 4.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11(11):4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122(1):205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 6.Surkova S, et al. Characterization of the Drosophila segment determination morphome. Dev Biol. 2008;313(2):844–862. doi: 10.1016/j.ydbio.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23(21):2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas T, et al. Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr Biol. 2013;23(21):2135–2139. doi: 10.1016/j.cub.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 10.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7(8):631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- 12.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332(6028):475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13(14):1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 15.Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281(5373):60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 16.Pisarev A, Poustelnikova E, Samsonova M, Reinitz J. FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution. Nucleic Acids Res. 2009;37(Database issue):D560–D566. doi: 10.1093/nar/gkn717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154(4):789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lionnet T, Singer RH. Transcription goes digital. EMBO Rep. 2012;13(4):313–321. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 2011;7(11):e1002364. doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 1989;8(4):1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57(3):413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 22.Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67(2):303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frasch M, Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987;1(9):981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- 24.Edgar BA, Weir MP, Schubiger G, Kornberg T. Repression and turnover pattern fushi tarazu RNA in the early Drosophila embryo. Cell. 1986;47(5):747–754. doi: 10.1016/0092-8674(86)90517-9. [DOI] [PubMed] [Google Scholar]
- 25.Petkova MD, Little SC, Liu F, Gregor T. Maternal origins of developmental reproducibility. Curr Biol. 2014;24(11):1283–1288. doi: 10.1016/j.cub.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellerman KA, Mattson DM, Duncan I. Mutations affecting the stability of the fushi tarazu protein of Drosophila. Genes Dev. 1990;4(11):1936–1950. doi: 10.1101/gad.4.11.1936. [DOI] [PubMed] [Google Scholar]
- 27.Paré A, et al. Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Curr Biol. 2009;19(23):2037–2042. doi: 10.1016/j.cub.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342(6163):1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336(6078):183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16(10):1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagha M, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153(5):976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322(5900):442–446. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buecker C, Wysocka J. Enhancers as information integration hubs in development: Lessons from genomics. Trends Genet. 2012;28(6):276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev. 2012;22(2):79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petesch SJ, Lis JT. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012;28(6):285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinitz J, Sharp DH. Mechanism of eve stripe formation. Mech Dev. 1995;49(1-2):133–158. doi: 10.1016/0925-4773(94)00310-j. [DOI] [PubMed] [Google Scholar]
- 37.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: Transcription enters a new era. Cell. 2014;157(1):13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F, Morrison AH, Gregor T. Dynamic interpretation of maternal inputs by the Drosophila segmentation gene network. Proc Natl Acad Sci USA. 2013;110(17):6724–6729. doi: 10.1073/pnas.1220912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubuis JO, Samanta R, Gregor T. Accurate measurements of dynamics and reproducibility in small genetic networks. Mol Syst Biol. 2013;9:639. doi: 10.1038/msb.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.