Significance
The emergence of highly pathogenic avian influenza viruses has received much attention due to the severe consequences of their occasional spread to humans, as well as the large toll they take on the poultry industry. Here we argue that the main barriers to the emergence of these viruses are imposed by immunity to related strains rather than the ability of the virus to acquire the necessary mutations. We show that, under these circumstances, patterns of influenza in different avian species are strongly dependent on their lifespan and that processes that alter the interactions between species of different lifespans—such as changes in farming practices—could promote the emergence of highly pathogenic types.
Keywords: antigenic evolution, virulence, multistrain model, population dynamics, hemagglutinin
Abstract
Outbreaks of highly pathogenic strains of avian influenza viruses (AIVs) cause considerable economic losses to the poultry industry and also pose a threat to human life. The possibility that one of these strains will evolve to become transmissible between humans, sparking a major influenza pandemic, is a matter of great concern. Most studies so far have focused on assessing these odds from the perspective of the intrinsic mutability of AIV rather than the ecological constraints to invasion faced by the virus population. Here we present an alternative multihost model for the evolution of AIV in which the mode and tempo of mutation play a limited role, with the emergence of strains being determined instead principally by the prevailing profile of population-level immunity. We show that (i) many of the observed differences in influenza virus dynamics among species can be captured by our model by simply varying host lifespan and (ii) increased contact between species of different lifespans can promote the emergence of potentially more virulent strains that were hitherto suppressed in one of the species.
Avian influenza viruses (AIVs) exist within a complex ecology that includes common interspecies transmission among birds (1, 2). Like all influenza viruses, AIV can be divided into subtypes (e.g., H5N1) on the basis of variation in the hemagglutinin (HA) and neuraminidase (NA) surface proteins and also exhibit extensive antigenic diversity within a particular subtype (e.g., refs. 3 and 4). Most AIV cocirculating between domestic and wild birds are classified as being of low pathogenicity (LPAIV). Occasionally, however, highly pathogenic (HPAIV) forms arise, causing high mortality in poultry. Severe illness and death also occur in humans infected by HPAIV: There have been around 650 human cases of subtype H5N1 HPAIV, with 384 deaths, since 2003. However, a recent epidemic of subtype H7N9 LPAIV, which has to date claimed more than 100 lives from ∼400 confirmed cases, illustrates that inducing severe disease in humans is not the sole preserve of HPAIV.
In avian species other than domestic poultry, AIV infection is largely asymptomatic (5, 6) with the notable exception of HP H5N1, which has caused recorded deaths in domestic and exotic waterfowl. Among ducks, the outcome of HP H5N1 infection is variable (7–11) and several factors suggest that domestic ducks may act as an asymptomatic reservoir (or “Trojan horse”) (12–14). Since 2002, HP H5N1 viruses have been regularly found among wild birds, including various species of migratory ducks and geese (15) in Asia, but other HPAIV have been isolated only sporadically from wildfowl (e.g., ref. 7).
HP H5N1 was first isolated in 1997 from chickens, ducks, and geese but the direction of transmission among these species remains unclear (16, 17). By contrast, genetic analyses of the virus responsible for the 2013 H7N9 outbreak show that virus to have moved first from wild birds to geese and ducks and then to chickens (18). Such studies offer insights into the dispersal history of AIV but they do not explain why new viral lineages or strains associated with phenotypes of interest (including, but not limited to, pathogenicity) arise and spread. The current understanding is that their emergence is limited by the occurrence of mutational events that are assumed to be rare (4, 19), but this is not wholly consistent with both the high observed rates of AIV nucleotide substitution (20) and the fact that many viral phenotypes are defined by a small number of genetic changes (21): Changes at as few as seven amino acid sites can explain recent patterns of antigenic evolution in humans (22). Here, we show many of these questions can be answered by recognizing that the virus population comprises a large pool of interchangeable gene segments (2) circulating within a variety of wild and domestic species of widely different lifespans. We demonstrate that host lifespan is a key determinant of the population dynamics of the virus and that the emergence of particular phenotypes may be driven by the immunodynamics of multispecies transmission rather than by the generation of those phenotypes by mutation.
A Multispecies Model for Avian Influenza
We combine three properties—pathogenicity, antigenicity, and transmissibility—of the influenza virus within a single framework to investigate its population dynamics within a multihost system.
A critical reason that avian influenza viruses differ in their pathogenicity is that the HA surface molecule, which mediates entry into host cells, has to be cleaved into HA1 and HA2 by cellular proteases. In LPAIV, the cleavage site contains a single basic amino acid; in HPAIV, this is replaced by a string of basic amino acids. The “polybasic” cleavage site of HA in HPAIV is susceptible to a greater number of proteases than the “monobasic” cleavage sites of other AIVs and HPAIV is therefore able to disseminate to a wider variety of organs, causing severe disease.
The HA molecule also contains a number of variable antigenic sites that are the target of protective antibodies; this makes it amenable to representation by a set of multilocus genotypes, with each locus corresponding to a variable antigenic site or epitope (23). We have previously used a similar framework to explain the dynamics of human influenza (24, 25) and it has also recently been applied to avian influenza (26). An important feature of this model is that antigenic variability at each locus is restricted; viral antigenic diversity arises from the large number of allele combinations (e.g., mn for an n-locus system with m alleles at each locus) and the success of an antigenic type is strongly constrained by cross-immunity with other antigenic types with which it shares alleles.
We assume that it is possible for each antigenic type within our framework to exist in either HP or LP form but that these may differ in their transmission potential (R0) due to biochemical constraints and epistatic interactions between antigenic sites and the cleavage site. Also, although wider dissemination of HPAIV in host tissues may provide a mechanism for enhanced transmissibility, the attendant pathology could have a negative effect on its transmission potential (for example, by killing the host) and the balance between these factors will dictate overall transmission success. To avoid any dependence on mutation rates, we ensure that all LP and HP forms of each antigenic types are continuously generated.
Results
We examine the dynamics of this system under the assumption that the LP forms of most antigenic types are more transmissible (i.e., have a higher R0) than their HP counterparts. Under our framework, these antigenic types will exist only in LP form because, although all HP forms may be continuously generated within every species, they will be outcompeted by the more transmissible LP variant of the same antigenic type. However, if the converse is true (as is the case for the antigenic types shown in red and black in Fig. 1A), the LP form will be outcompeted by the HP form: Thus each antigenic type will uniquely associate with either HP or LP cleavage sites.
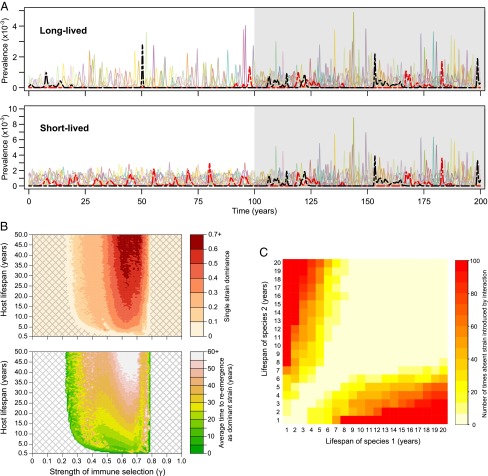
Fig. 1.
Effects of lifespan on antigenic dynamics and emergence of highly pathogenic forms. (A) Antigenic dynamics within long-lived (Upper, life expectancy = 15 y) and short-lived (Lower, life expectancy = 2 y) species. The shaded area (right-hand side) corresponds to the period that the two populations are linked (i.e., transmit to each other). Each colored line corresponds to an individual antigenic type within a {2, 3, 5} antigenic system, with solid/dashed lines referring, respectively, to LP/HP versions of each antigenic type (βi′ < βi = 146 for all strains, except for the red and black strains where βi < βi′ = 146; 1/σ = 5 d, p = 0.5, γ = 0.68). (B) Impact of life expectancy on the likelihood of single-strain dominance (Upper) and average time to reemergence as a dominant strain (Lower) (precise definitions in Materials and Methods). Within the cross-hatched areas, the model exhibits antigenic stasis (βi′ < βi = 146 for all strains; 1/σ = 5 d). (C) Effect of contact between two species on appearance of a strain that was previously absent in both. For each combination of lifespans, we selected 100 simulations where the same strain was absent from both species for a period of at least 800 y (βi′ < βi = 146 for all strains; 1/σ = 5 d, p = 0.5, γ = 0.72). We then reran these exact simulations but now allowed contact between the two species after 300 y and recorded whether the previously absent strain now appeared in the following 100 y.
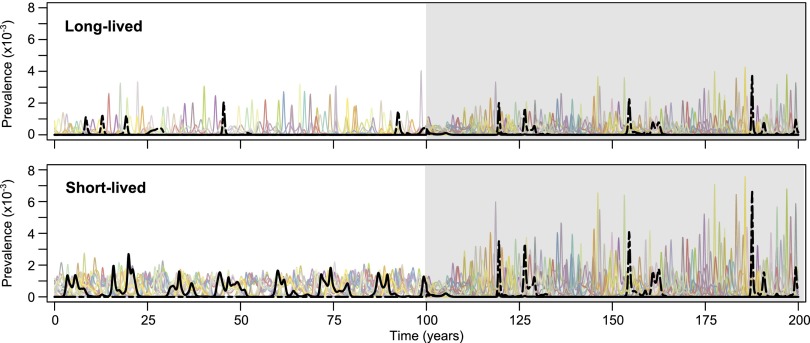
The unshaded areas of Fig. 1A contrast the dynamics of this model in species with lifespans of 15 y (Fig. 1A, Upper) and 2 y (Fig. 1A, Lower). We note that that there is greater cocirculation and more frequent reemergence of antigenic types (e.g., the red strain), among short-lived species, leading to higher overall influenza prevalence (Fig. S1). Fig. 1B illustrates more generally how the qualitative behavior of the system is affected by lifespan as well as the strength of immune selection against the relevant epitopes. At low levels of immune selection, all possible antigenic types coexist stably in either LP or HP form, whereas at very high levels of immune selection we observe stable coexistence of a smaller subset of antigenic types. Between these regions of antigenic stasis the strain dynamics are unstable (Fig. 1A) with both strain diversity within an epidemic (Fig. 1B, Upper) and the rate of strain reemergence (Fig. 1B, Lower) declining with host lifespan, as a consequence of lower rates of replenishment of the totally susceptible population in longer-lived species. High levels of immune selection can also cause certain antigenic types (e.g., the black strain in the short-lived population in Fig. 1A) to be suppressed permanently or, at least, over extended periods of time (Fig. S2), particularly if they are less transmissible (Fig. S3), although this is dependent on the precise representation of the multilocus structure of HA (Fig. S4).
The shaded area in Fig. 1A shows how the viral population dynamics are altered when the two species are able to transmit to each other due to some change, for example, in farming practice that brings shorter-lived birds such as ducks into closer contact with longer-lived wildfowl. Very small amounts of cross-species transmission can significantly alter the antigenic dynamics of influenza in longer-lived species without affecting the short-lived species (Fig. S5). Higher levels of connectivity result in a dynamic where strains reappear more frequently in long-lived species and less frequently in short-lived species than before they were connected (e.g., the red HP strain in Fig. 1A), and the difference in overall prevalence between species is reduced. Contact between species of different lifespans can also promote the emergence of an antigenic type (such as the black HP strain in Fig. 1A) that was previously suppressed in one or both species (Fig. 1C). Interestingly, the withdrawal of connections between the species does not necessarily return the dynamics to their previous state: Thus a strain that has emerged due to contact between the species cannot be guaranteed to disappear once contact is disrupted (Fig. S6).
Finally, subtle differences in the R0 values of HP and LP forms of an antigenic type in different species can cause the HP form to dominate in one and the LP form to dominate in another (Fig. 2). In these circumstances, contact between species will cause one of these forms to be displaced as coexistence between them is not possible within the same transmission system. Whether the HP or the LP form “wins” will be dictated by their relative transmissibilities in each species as well as the host lifespans (Fig. S7). In the example shown in Fig. 2, the HP form outcompetes the LP form when the species are connected (within the shaded area) and therefore a transition from LP to HP occurs within the same antigenic type in the short-lived population.
Fig. 2.
Effects of unequal transmissibility of highly pathogenic forms in different species. Here, the HP form (dashed line) of a particular antigenic type (shown in black) is more transmissible than the LP form (solid line) in the long-lived species and vice versa in the short-lived species. The shaded area (right-hand side) corresponds to the period that the two populations are linked (i.e., transmit to each other). (Parameters are identical to those used in Fig. 1A, other than for the black antigenic type where now βi′ = 153 in the long-lived species and βi′ = 143 in the short-lived species and the red antigenic type that now has βi′ < βi = 146.)
Discussion
We have adapted a multilocus model of influenza antigens to consider the effects of host lifespan, variable transmissibility, and cross-species transmission on the antigenic evolution of the virus. A number of empirical observations endorse this discrete representation of the antigenic properties of HA (25), including recent work showing that (i) antigenic cluster transitions in H3N2 are governed by a small number of critical amino acid positions in HA, with restricted amino acid use therein (22), and (ii) HA epitopes of H1N1 strains appear to be periodically recycled (27, 28), suggesting that there are fundamental restrictions on available antigenic space. Extended overlap between the antigenic repertoires of influenza viruses is also supported by the observation that mixtures of sera raised in ferrets against historic strains exhibit neutralizing activity toward HA of the 2009 pH1N1 virus (29). Within our framework, epitope regions with high variability may exhibit a linear change over an extended time period [in line with data obtained using techniques such as the hemagglutination inhibition (HI) assay (22)] whereas epitope regions of low variability will exhibit continuous cycling. Thus, there is no fundamental disagreement between this mechanistic model and other theoretical representations of the antigenic evolution of influenza (30–34).
The general trends of cross-sectional antigenic diversity and emergence shown by our model (Fig. 1B) fit well with our understanding of the qualitative features of influenza in major mammalian and avian host species. Human influenza A epidemics exhibit limited antigenic diversity and reemergence of antigenic types occurs only after several decades [of which the 2009 pH1N1 and 1918 H1N1 strains serve as examples (28)], in agreement with the behavior of our model under long lifespans. Our model predicts that the antigenic dynamics of AIV in longer-lived avian populations are more likely to resemble influenza A in humans, albeit with slightly higher levels of cocirculating antigenic diversity. Although there is now an abundance of genetic data from global surveillance studies, the serological studies required to test this hypothesis have yet to be performed. Patterns of influenza A among poultry agree with model behavior within short-lived species: At least two distinct antigenic types of H9N2 have cocirculated in South Korean poultry over the last 10 y (35–37) and circulating H5N1 strains in Southeast Asian poultry are also antigenically diverse (3, 4). The rapid reemergence of antigenic variants in short-lived species is also in agreement with patterns of cross-reactivity among AIVs isolated over extended periods of time in poultry (38–40) and provides an alternative explanation to the virus population being in antigenic stasis.
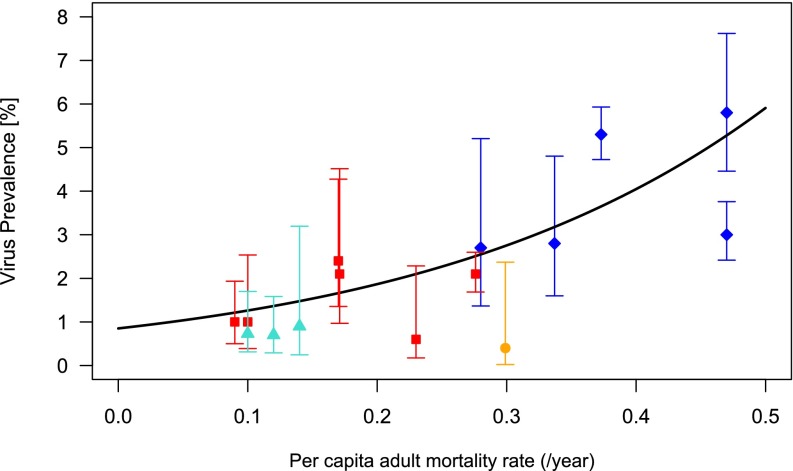
A critical assumption of our model is that avian immunity to influenza involves epitope-specific antibody responses against HA. There is both observational and experimental evidence that a variety of domestic and wild avian species produce specific hemagglutination-inhibiting and neutralizing antibodies against HA, although opinion is divided as to their efficacy and duration, and there are some mechanistic reasons (such as the existence of truncated variants of IgY among ducks) that could prevent certain species from mounting an effective immune response (41–43). Nonetheless, AIV prevalence tends to decline with age whereas seroprevalence increases (9, 44–46), suggesting that some form of functional, long-term immunity is acquired upon exposure. We also present here an analysis of published data from a large, multiannual survey of influenza prevalence in wild birds in The Netherlands (47) showing that virus prevalence decreases with lifespan (Fig. 3 and Table S1), which suggests that longer-lived populations contain a higher proportion of immune individuals (i.e., immunity is not short lived). The same pattern of higher prevalence in wild ducks compared with other, longer-lived bird species occurs in a number of other studies (1, 45, 48, 49). Naturally, numerous ecological and behavioral factors may also contribute to this trend, but it has been hard to find firm correlations. For example, the original analysis of the data in Fig. 3 yielded no relationship between flock size and prevalence (47). A link has been demonstrated between surface water feeding and prevalence (50) but, because dabbling ducks are themselves generally shorter lived than the diving ducks included in this study, their higher AIV prevalence may result from quicker population turnover rather than higher exposure. Other mechanisms that increase the supply of susceptibles, such as weak species-specific immunity or migration from AIV-free populations, could also lead to similar qualitative outcomes.
Fig. 3.
Relationship between AIV prevalence and adult host mortality rate [Table S1; data reported for a single site in The Netherlands (47)]. Species were categorized into dabbling ducks [blue diamonds: mallard (adult mortality rate 0.373; prevalence 5.3%), Eurasian wigeon (0.470; 3.0%), common teal (0.470; 5.8%), northern pintail (0.337; 2.8%), gadwall (0.280; 2.7%)], geese [red squares: white fronted goose (0.276; 2.1%), barnacle goose (0.090; 1.0%)], greylag goose (0.170; 2.4%), brent goose (0.100; 1.0%), bean goose (0.230; 0.6%), pink-footed goose (0.171; 2.1%)], gulls [turquoise triangles: black-headed gull (0.100; 0.7%), common gull (0.140; 0.9%), herring gull (0.120; 0.7%)], and rails [orange circle: common coot (0.299, 0.4%)]. We plot the 95% confidence interval on viral prevalence, calculated via the Wilson method, for each species and the predicted relationship between prevalence and mortality based on a logistic regression (Materials and Methods). The effect of host mortality rate on prevalence offered a significantly better fit than that in the null model: χ2(df = 1, n = 15) = 101.26, P < 0.001.
With regard to the emergence of HPAIV, our model suggests that such variants may be continuously generated but excluded by competition from less pathogenic variants of similar antigenic types on account of having a lower transmission potential (R0). Recent cloning experiments (51) show that monobasic cleavage sites can be stably replaced in the majority of HA subtypes, leading to increased pathogenicity on at least H2, H4, H8, and H14 backgrounds. Our model provides one potential explanation of why such HP variants have never been seen to emerge: They cannot compete successfully with their LP counterparts, whereas certain HP variants on H5 and H7 backgrounds are able to do so. This, rather than a unique predisposition of H5 and H7 to acquire polybasic cleavage sites, may account for the restriction of highly pathogenic phenotypes to H5 and H7.
An important conclusion of our model is that HPAIV strains may be expected to emerge regularly in short-lived birds but only rarely in longer-lived species. Contact between species of different lifespans can reduce the incidence of any such strain in the short-lived species, but it can also provoke the emergence of a previously suppressed HPAIV strain. It is possible that changes in farming practices bringing domestic ducks into more prolonged contact with wildfowl have contributed to the increase in HPAI outbreaks in the last 20 y (4, 52, 53) and, indeed, duck flocks kept in open, rather than closed, systems are more susceptible to HPAIV infection (8).
Our model indicates that the main threat from ecological change in a multihost system may arise from a potential disturbance to the established immunodynamics of influenza rather than from the potential evolution of new virulent strains. Our studies underscore the importance of more “hypothesis-driven” surveillance (54) of AIV that routinely incorporates antigenic analyses [which is noted as “lagging behind” viral genetic analyses (26, 55)] and of further experimental work to establish the competitiveness of potentially pathogenic forms and their antigenic relationship with nonpathogenic forms. Such studies are required to understand the ecological interactions between circulating strains and for the design of effective preemptive measures against epizootics and potential human pandemics.
Materials and Methods
Model.
We extend a multilocus model of influenza (23, 24) to include two interacting populations, here named A and B. Within this model, antigenic types are described by an n-locus system, with varying numbers of alleles at each locus. We use the notation {2, 3, 5}, for example, to represent a multilocus structure in which there are three loci with two, three, and five alleles, respectively. We refer to each allelic combination as a “strain” of the pathogen. We also consider two types of each strain: one that has low pathogenicity (strain i) and one that has high pathogenicity (strain i′). These two types are immunologically identical, but may have different fitness.
The dynamics of the proportion immune to strains i and i′ in population A, , are given by
| [1] |
where is the per-capita risk of infection associated with strain i, is the per-capita risk of infection associated with strain i′, and is the per-capita death rate of population A (and hence is the average life expectancy).
The dynamics of the proportion of the population that has been exposed to any strain j that shares alleles with i (including i itself), , are given by
| [2] |
where j ∼ i denotes the collection of strains j that share alleles with strain i.
Individuals who have been exposed to a strain that shares alleles with strain i but not strain i itself (that is, ) will become infectious with probability when infected by strain i. The parameter, , is essentially a measure of immune selection. With being the rate of loss of infectiousness (thus, is the infectious period), the dynamics of the proportion of the population infectious with strain i, , are given by
| [3] |
The proportion of the population infectious with strain i′ is entirely analogous.
The dynamics of the pathogen population within host species B follows identical rules.
The per-capita risk of infection for strain i, , takes the form , where and are combinations of transmission parameters that respectively define the rate of transmission from infected individuals in species A and B to a susceptible individual in species A. The quantity defines the basic reproductive number (i) (a measure of fundamental transmission potential of influenza of strain i in species A). In the case where all transmission parameters are equivalent between species and the total number of contacts is constrained, with a proportion p occurring between the species, the expression for can be written as . However, could also assume independent values: for example, when infection is environmentally acquired.
We have simulated these ordinary differential equations (ODE) using a fixed-step, fourth-order Runge Kutta numerical solver. In any ODE model of infection, the fraction of the population that is infectious with any given strain can become arbitrarily small. Consequently, and also because we have not explicitly modeled a mutational process, we have not let the value of any yi drop below 10−50. This is equivalent to each antigenic variant being constantly generated at low levels by mutation.
General Qualitative Behavior.
To investigate the general qualitative behavior of the model, unless otherwise stated, we analyze a period of 500 y to calculate the following three measures.
Single-strain dominance (ε).
Single-strain dominance can be quantified by the measure ε by comparing the relative prevalence of the two most common antigenic variants within single epidemics and then averaging across extended periods of time (10). More formally, averaging across each of P epidemics (where an epidemic is defined as a local maximum in total prevalence),
This therefore takes on values between 0 and 1, with higher values indicating stronger single-strain dominance.
Time between periods of strain dominance.
For each strain, we first identify each period during which it was the most prevalent in the population and record the time during that period when that strain’s prevalence was maximized. We then report the mean length of interval between these times, averaged across all strains.
Definition of strain exclusion.
We describe a variant as being permanently suppressed if that variant never achieves a prevalence greater than 10−4 during the period of analysis.
Analysis of Host Mortality vs. AIV Prevalence.
AIV prevalence might be expected to vary between species, geographical location, and season and also perhaps due to sampling methods. To investigate a potential link between host lifespan and prevalence, we therefore sought data from a single, large study in which many of these factors would have been mitigated. We consequently used data reported from a sizeable study of AIV prevalence in European birds that was conducted over a number of seasons and in which most of the data came from a single country (The Netherlands). We were thus able to restrict our dataset to those species from which more than 200 samples had come from a single location (Table S1). We estimated the relationship between host mortality rate and prevalence with a logistic regression model in R, weighting the data by number of samples. The estimate for the effect of host mortality rate on prevalence was highly significant (P < 0.001), with the model offering a significantly better fit than the null model: χ2(df = 1, n = 15) = 101.26, P < 0.001. We also used the Wilson method to calculate the 95% confidence interval on the binomial probability and plotted both this and the graph of predicted prevalence.
Supplementary Material
Acknowledgments
We thank Tommy Lam, Chris Perrins, Adrian Smith, Don Klinkenberg, Sarah Gilbert, and Nigel Temperton for useful comments. O.G.P. received funding from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement 614725. S.G. is a Royal Society Wolfson Research Fellow and an ERC Advanced Investigator (DIVERSITY).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401849111/-/DCSupplemental.
References
- 1.Olsen B, et al. Global patterns of influenza a virus in wild birds. Science. 2006;312(5772):384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 2.Dugan VG, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4(5):e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ducatez MF, et al. Extent of antigenic cross-reactivity among highly pathogenic H5N1 influenza viruses. J Clin Microbiol. 2011;49(10):3531–3536. doi: 10.1128/JCM.01279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suarez DL. Avian influenza: Our current understanding. Anim Health Res Rev. 2010;11(1):19–33. doi: 10.1017/S1466252310000095. [DOI] [PubMed] [Google Scholar]
- 5.Kuiken T. 2013. Is low pathogenic avian influenza virus virulent for wild waterbirds? Proc Biol Sci 280(1763): 20130990.
- 6.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaidet N, et al. Evidence of infection by H5N2 highly pathogenic avian influenza viruses in healthy wild waterfowl. PLoS Pathog. 2008;4(8):e1000127. doi: 10.1371/journal.ppat.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Songserm T, et al. Domestic ducks and H5N1 influenza epidemic, Thailand. Emerg Infect Dis. 2006;12(4):575–581. doi: 10.3201/eid1204.051614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henning J, et al. Scavenging ducks and transmission of highly pathogenic avian influenza, Java, Indonesia. Emerg Infect Dis. 2010;16(8):1244–1250. doi: 10.3201/eid1608.091540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pantin-Jackwood M, Swayne DE, Smith D, Shepherd E. 2013. Effect of species, breed and route of virus inoculation on the pathogenicity of H5N1 highly pathogenic influenza (HPAI) viruses in domestic ducks. Vet Res 44:62-9716-44-62. [DOI] [PMC free article] [PubMed]
- 11.Yuan R, et al. Pathogenicity and transmission of H5N1 avian influenza viruses in different birds. Vet Microbiol. 2014;168(1):50–59. doi: 10.1016/j.vetmic.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Kim JK, Negovetich NJ, Forrest HL, Webster RG. Ducks: The “Trojan horses” of H5N1 influenza. Influenza Other Respi Viruses. 2009;3(4):121–128. doi: 10.1111/j.1750-2659.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sturm-Ramirez KM, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79(17):11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulse-Post DJ, et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci USA. 2005;102(30):10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178(1):63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shortridge KF. Poultry and the influenza H5N1 outbreak in Hong Kong, 1997: Abridged chronology and virus isolation. Vaccine. 1999;17(Suppl 1):S26–S29. doi: 10.1016/s0264-410x(99)00102-4. [DOI] [PubMed] [Google Scholar]
- 17.Guan Y, et al. H5N1 influenza viruses isolated from geese in Southeastern China: Evidence for genetic reassortment and interspecies transmission to ducks. Virology. 2002;292(1):16–23. doi: 10.1006/viro.2001.1207. [DOI] [PubMed] [Google Scholar]
- 18.Lam TT, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502(7470):241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25(30):5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 20.Chen R, Holmes EC. Avian influenza virus exhibits rapid evolutionary dynamics. Mol Biol Evol. 2006;23(12):2336–2341. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- 21.Perdue ML. In: Avian Influenza. Swayne DE, editor. Ames, IA: Blackwell; 2008. pp. 23–41. [Google Scholar]
- 22.Koel BF, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342(6161):976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 23.Gupta S, Ferguson N, Anderson R. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science. 1998;280(5365):912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- 24.Recker M, Pybus OG, Nee S, Gupta S. The generation of influenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc Natl Acad Sci USA. 2007;104(18):7711–7716. doi: 10.1073/pnas.0702154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikramaratna PS, Sandeman M, Recker M, Gupta S. The antigenic evolution of influenza: Drift or thrift? Philos Trans R Soc Lond B Biol Sci. 2013;368(1614):20120200. doi: 10.1098/rstb.2012.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latorre-Margalef N, et al. Heterosubtypic immunity to influenza A virus infections in mallards may explain existence of multiple virus subtypes. PLoS Pathog. 2013;9(6):e1003443. doi: 10.1371/journal.ppat.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210(8):1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause JC, et al. Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J Virol. 2010;84(6):3127–3130. doi: 10.1128/JVI.02184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter DM, et al. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J Virol. 2013;87(3):1400–1410. doi: 10.1128/JVI.02257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422(6930):428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 31.Bedford T, Rambaut A, Pascual M. Canalization of the evolutionary trajectory of the human influenza virus. BMC Biol. 2012;10:38. doi: 10.1186/1741-7007-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 2006;314(5807):1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 33.Levin SA, Dushoff J, Plotkin JB. Evolution and persistence of influenza A and other diseases. Math Biosci. 2004;188(1-2):17–28. doi: 10.1016/j.mbs.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Gog JR, Grenfell BT. Dynamics and selection of many-strain pathogens. Proc Natl Acad Sci USA. 2002;99(26):17209–17214. doi: 10.1073/pnas.252512799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YJ, et al. Continuing evolution of H9 influenza viruses in Korean poultry. Virology. 2007;359(2):313–323. doi: 10.1016/j.virol.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 36.Moon HJ, et al. Active reassortment of H9 influenza viruses between wild birds and live-poultry markets in Korea. Arch Virol. 2010;155(2):229–241. doi: 10.1007/s00705-009-0577-4. [DOI] [PubMed] [Google Scholar]
- 37.Park KJ, et al. Rapid evolution of low pathogenic H9N2 avian influenza viruses following poultry vaccination programs. J Gen Virol. 2011;92(Pt 1):36–50. doi: 10.1099/vir.0.024992-0. [DOI] [PubMed] [Google Scholar]
- 38.Kida H, Kawaoka Y, Naeve CW, Webster RG. Antigenic and genetic conservation of H3 influenza virus in wild ducks. Virology. 1987;159(1):109–119. doi: 10.1016/0042-6822(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 39.Shortridge KF, et al. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252(2):331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- 40.Swayne DE, Perdue ML, Beck JR, Garcia M, Suarez DL. Vaccines protect chickens against H5 highly pathogenic avian influenza in the face of genetic changes in field viruses over multiple years. Vet Microbiol. 2000;74(1–2):165–172. doi: 10.1016/s0378-1135(00)00176-0. [DOI] [PubMed] [Google Scholar]
- 41.Magor KE. Immunoglobulin genetics and antibody responses to influenza in ducks. Dev Comp Immunol. 2011;35(9):1008–1016. doi: 10.1016/j.dci.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Fereidouni SR, et al. Dynamics of specific antibody responses induced in mallards after infection by or immunization with low pathogenicity avian influenza viruses. Avian Dis. 2010;54(1):79–85. doi: 10.1637/9005-073109-Reg.1. [DOI] [PubMed] [Google Scholar]
- 43.Barber MR, et al. Identification of avian RIG-I responsive genes during influenza infection. Mol Immunol. 2013;54(1):89–97. doi: 10.1016/j.molimm.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoye BJ, et al. Reconstructing an annual cycle of interaction: Natural infection and antibody dynamics to avian influenza along a migratory flyway. Oikos. 2011;120:748–755. [Google Scholar]
- 45.Ip HS, et al. Prevalence of influenza A viruses in wild migratory birds in Alaska: Patterns of variation in detection at a crossroads of intercontinental flyways. Virol J. 2008;5:71. doi: 10.1186/1743-422X-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pybus OG, et al. The ecology and age structure of a highly pathogenic avian influenza virus outbreak in wild mute swans. Parasitology. 2012;139(14):1914–1923. doi: 10.1017/S0031182012000261. [DOI] [PubMed] [Google Scholar]
- 47.Munster VJ, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3(5):e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Germundsson A, Madslien KI, Hjortaas MJ, Handeland K, Jonassen CM. 2010. Prevalence and subtypes of influenza A viruses in wild waterfowl in Norway 2006-2007. Acta Vet Scand 52:28-0147-52-28. [DOI] [PMC free article] [PubMed]
- 49.Tonnessen R, et al. 2013. Molecular and epidemiological characterization of avian influenza viruses from gulls and dabbling ducks in Norway. Virol J 10:112-422X-10-112.
- 50.Garamszegi LZ, Møller AP. Prevalence of avian influenza and host ecology. Proc Biol Sci. 2007;274(1621):2003–2012. doi: 10.1098/rspb.2007.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veits J, et al. Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. Proc Natl Acad Sci USA. 2012;109(7):2579–2584. doi: 10.1073/pnas.1109397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munster VJ, Fouchier RAM. Avian influenza virus: Of virus and bird ecology. Vaccine. 2009;27(45):6340–6344. doi: 10.1016/j.vaccine.2009.02.082. [DOI] [PubMed] [Google Scholar]
- 53.Swayne DE. Impact of vaccines and vaccination on global control of avian influenza. Avian Dis. 2012;56(4) Suppl:818–828. doi: 10.1637/10183-041012-Review.1. [DOI] [PubMed] [Google Scholar]
- 54.Hoye BJ, Munster VJ, Nishiura H, Klaassen M, Fouchier RA. Surveillance of wild birds for avian influenza virus. Emerg Infect Dis. 2010;16(12):1827–1834. doi: 10.3201/eid1612.100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boon ACM, Webby RJ. In: Vaccines for Pandemic Influenza. Current Topics in Microbiology and Immunology. Compans RW, Orenstein WA, editors. Vol 333. Germany: Springer, Heidelberg; 2009. pp. 25–40. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.