Significance
Beyond its canonical functions in processes such as cell-cycle arrest, apoptosis, and senescence, the tumor suppressor p53 has been increasingly implicated in metabolism. Here, in vitro and in vivo studies establish a role for p53 in gluconeogenesis through a previously unidentified mechanism involving (i) direct activation of the gene encoding the NAD-dependent deacetylase sirtuin 6 (SIRT6), (ii) SIRT6-dependent deacetylation and nuclear exclusion of forkhead box protein O1 (FoxO1), and (iii) down-regulation of FoxO1-activated genes (G6PC and PCK1) that are rate-limiting for gluconeogenesis. These results have implications for proposed tumor-suppressor functions of p53 through regulation of metabolic pathways.
Abstract
In mammalian cells, tumor suppressor p53 plays critical roles in the regulation of glucose metabolism, including glycolysis and oxidative phosphorylation, but whether and how p53 also regulates gluconeogenesis is less clear. Here, we report that p53 efficiently down-regulates the expression of phosphoenolpyruvate carboxykinase (PCK1) and glucose-6-phosphatase (G6PC), which encode rate-limiting enzymes in gluconeogenesis. Cell-based assays demonstrate the p53-dependent nuclear exclusion of forkhead box protein O1 (FoxO1), a key transcription factor that mediates activation of PCK1 and G6PC, with consequent alleviation of FoxO1-dependent gluconeogenesis. Further mechanistic studies show that p53 directly activates expression of the NAD+-dependent histone deacetylase sirtuin 6 (SIRT6), whose interaction with FoxO1 leads to FoxO1 deacetylation and export to the cytoplasm. In support of these observations, p53-mediated FoxO1 nuclear exclusion, down-regulation of PCK1 and G6PC expression, and regulation of glucose levels were confirmed in C57BL/J6 mice and in liver-specific Sirt6 conditional knockout mice. Our results provide insights into mechanisms of metabolism-related p53 functions that may be relevant to tumor suppression.
As the “guardian of the genome,” tumor suppressor p53 has been reported to coordinate diverse cellular responses to a broad range of environment stresses (1) and to play antineoplastic roles by activating downstream target genes involved in DNA damage repair, apoptosis, and cell-cycle arrest (2). Recent studies have indicated broader roles for p53 in mediating metabolic changes in cells under various physiological and pathological conditions (3–7). For example, p53 was reported to influence the balance between glycolysis and oxidative phosphorylation by inducing the p53-induced glycolysis and apoptosis regulator (TIGAR) and by regulating the synthesis of cytochrome c oxidase 2 (SCO2) (3), respectively, thus promoting the switch from glycolysis to oxidative phosphorylation. p53 also may impede metabolism by reducing glucose import (4) or by inhibiting the pentose phosphate pathway (PPP) (5). More recently, context-dependent inhibitory (6) or stimulatory (7, 8) effects of p53 on gluconeogenesis have been reported. It thus is clear that p53 plays important roles in glucose regulation in mammalian cells.
Glucose homeostasis is maintained by a delicate balance between intestinal absorption of sugar, gluconeogenesis, and the utilization of glucose by the peripheral tissues, irrespective of feeding or fasting (9). The gluconeogenesis pathway is catalyzed by several key enzymes that include the first and last rate-limiting enzymes of the process, phosphoenolpyruvate carboxykinase (PCK1) and glucose-6-phosphatase (G6PC), respectively. The expression of both PCK1 and G6PC is controlled mainly at the transcription level. For example, the transcription factor forkhead box protein O1 (FoxO1) activates gluconeogenesis through direct binding to the promoters of G6PC and PCK1 (10). Related, a dominant negative FoxO1 lacking its transactivation domain significantly decreases gluconeogenesis (11) whereas FoxO1 ablation impairs fasting- and cAMP-induced PCK1 and G6PC expression (12). Therefore, factors influencing expression of FoxO1 or its binding activity to the PCK1 and G6PC promoters are potential targets for gluconeogenesis regulation.
The transcription activity of FoxO family members is regulated by a sophisticated signaling network. Various environmental stimuli cause different posttranslational modifications of FoxO proteins, including phosphorylation, acetylation, ubiquitination, and methylation (13–15). The phosphorylation of FoxO proteins is known to be essential for their shuttling between the nucleus and cytoplasm. For example, kinase Akt/PKB phosphorylates FoxO1 at threonine 24 and at serines 256 and 319, which in turn leads to 14-3-3 binding and subsequent cytoplasmic sequestration. The acetylation of FoxO proteins also affects their trafficking and DNA-binding activities (15–17). Sirtuin (SIRT)1, a homolog of the yeast silent information regulator-2 (Sir2), has been identified as a deacetylase for FoxO proteins (15, 17, 18). Of the seven mammalian sirtuins, SIRT1, SIRT6, and SIRT7 are localized to the nucleus (19), and SIRT6 was recently reported to act as a central player in regulating the DNA damage response, glucose metabolism, and aging (20–26). Using a knockout mouse model, it was found that SIRT6 functions as a corepressor of the transcription factor Hif1α to suppress glucose uptake and glycolysis (23). In addition, SIRT6-deficient mice suffer from a variety of degenerative syndromes at ∼4 wk of age (25). These studies reveal essential roles for SIRT6 in the regulation of glucose and lipid homeostasis.
Here, we confirm an important role for p53 in the control of glucose levels through its suppression of gluconeogenesis and establish the underlying mechanism. Thus, in cell-based assays, p53 transcriptionally activates expression of SIRT6, which specifically interacts with FoxO1 to promote its deacetylation and nuclear exclusion and, consequently, loss of FoxO1-induced gluconeogenesis. This p53-modulated glucose decrease is blocked in conditional liver Sirt6 knockout mice during fasting. To our knowledge, our data are the first demonstration that p53 plays an indispensable role in modulating gluconeogenesis through SIRT6-mediated FoxO1 nuclear exclusion.
Results
p53 Transcriptionally Down-Regulates Expression of the Gluconeogenic Genes G6PC and PCK1.
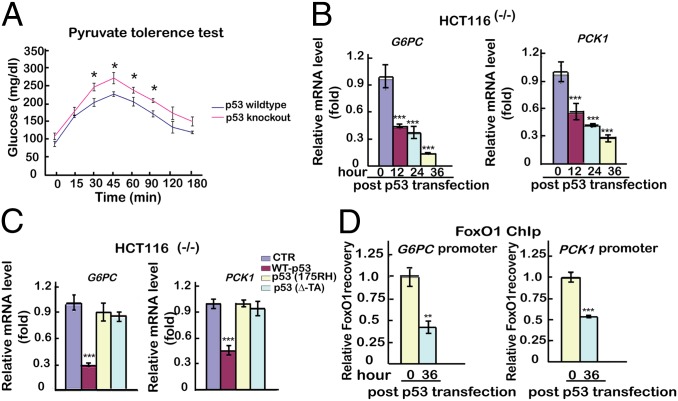
A potential effect of p53 on gluconeogenesis was evaluated by pyruvate tolerance test (PTT) after fasting in p53 wild-type (p53WT) or p53 knockout (p53KO) mice (C57BL/6J). As shown in Fig. 1A, compared with p53WT mice, p53KO mice were more efficient in restoring blood glucose levels. In a further analysis, human hepatic cancer HEPG2 cells (with wild-type p53) and human colon cancer HCT116(−/−) cells (which lack p53) were transfected with either a plasmid expressing wild-type p53 (p53WT) or an empty plasmid. As shown in Fig. 1B and Fig. S1A, time-dependent decreases in G6PC and PCK1 mRNA expression [monitored by quantitative PCR (qPCR)] were observed in response to ectopic p53 expression in both cell lines. The role of p53 in regulating expression of G6PC and PCK1 was confirmed by luciferase reporter assays (Fig. S1B). We next investigated whether the p53-induced inhibition of PCK1 and G6PC expression might be mediated by direct promoter binding using a chromatin immunoprecipitation (ChIP) assay. We did not observe any significant p53 binding or any consensus p53 binding sites in the promoter regions (within 1–2 kb of the transcriptional start site) of these genes, indicating that p53 may act indirectly through other genes/factors to down-regulate expression of G6PC and PCK1.
Fig. 1.
p53 transcriptionally down-regulates expression of the gluconeogenic genes G6PC and PCK1. (A) Pyruvate tolerance tests in normal C57BL/6J mice and p53 knockout mice. Twelve mice were separated into two groups, with six mice per group. Blood glucose concentration of mice was measured at 22 h after fasting. (B) Ectopic p53-induced G6PC and PCK1 expression in HCT116(−/−) cells. Cells were transfected with either an empty plasmid as control (hereafter all transfected empty plasmids serve as controls) or a p53WT-expressing plasmid, and mRNA was analyzed by qPCR at 12 h, 24 h, or 36 h after transfection. mRNA levels of the control sample were set as 1, and relative mRNA levels of the other samples were normalized to this control. (C) Ectopic mutant p53-induced G6PC and PCK1 expression in HCT116(−/−) cells. Cells were transfected with an empty plasmid or with plasmids expressing p53WT, p53(175RH), or p53(ΔTA). Relative mRNA expression levels were measured by qPCR at 36 h. (D) Ectopic p53-induced binding of FoxO1 to G6PC and PCK1 promoters in HCT116(−/−) cells. A qChIP analysis was performed at 36 h after transfection with empty plasmid or plasmid expressing p53WT. The relative recovery of protein binding to the promoters in cells with empty plasmid was set as 1, and the relative recoveries of the other samples were normalized to this control. All experiments were repeated at least three times. The data are shown as the mean ± SD, with *P < 0.05, **P < 0.01, and ***P < 0.001.
To assess whether p53 transcription activity is required for the down-regulation of G6PC and PCK1, the functional assays were repeated with p53 mutants that lack either the transactivation domain [p53(∆TA)] or the ability to bind DNA [p53(175RH)] (27). Unlike p53WT, none of these p53 mutants decreased expression of G6PC or PCK1 (Fig. 1C and Fig. S1C), suggesting that p53 transcription activity is indispensable for the down-regulation of gluconeogenic G6PC and PCK1 in both HCT116 and HEPG2 cells.
Next, we sought to determine whether p53 regulates gluconeogenesis indirectly through effects on the expression or the promoter-binding activity of FoxO1, HNF4, or CREB, which bind to the G6PC and PCK1 promoters. As shown in Fig. S1D, expression of ectopic p53 did not decrease the mRNA expression levels (monitored by qPCR) of FoxO1, HNF4, or CREB. Interestingly, however, a ChIP analysis revealed that overexpression of p53 significantly decreased binding of FoxO1, but not HNF4 or CREB, to the G6PC and PCK1 promoters (Fig. 1D and Fig. S1E). These results indicate that p53 may inhibit gluconeogenesis by influencing the capacity of FoxO1 to bind the G6PC and PCK1 promoters.
Activation of p53 Promotes the Translocation of FoxO1 from the Nucleus to the Cytoplasm.
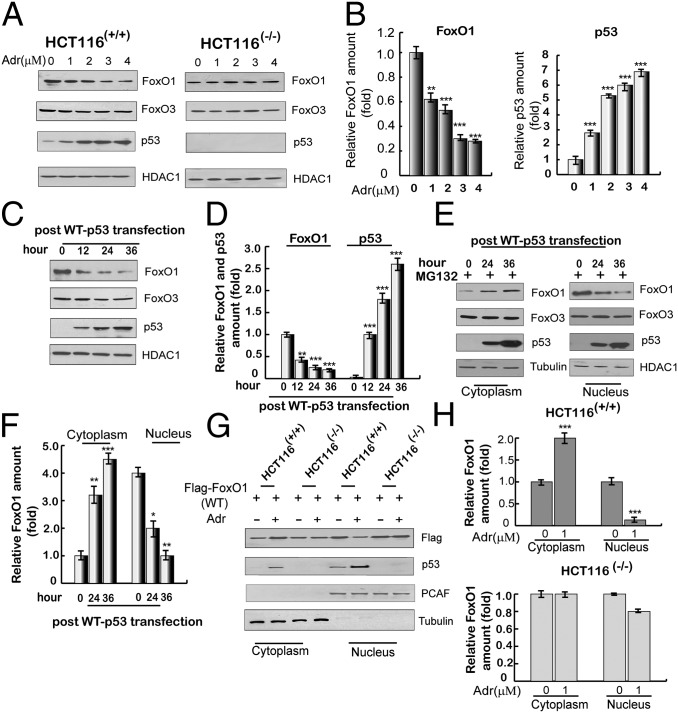
To investigate a potential connection between FoxO1 levels and p53, HCT116(+/+) (with wild-type p53) and HCT116(−/−) cell lines were exposed to adriamycin to activate p53. As shown in Fig. 2 A and B, the endogenous levels of nuclear FoxO1 were selectively decreased in HCT116(+/+) cells relative to HCT116(−/−) cells in response to adriamycin, which was negatively correlated with the p53 levels. To further confirm the role of p53 in the reduction of nuclear FoxO1 levels, HCT116(−/−) cells were transfected with a plasmid expressing p53. As shown in Fig. 2C, nuclear FoxO1 levels were significantly decreased in the p53-transfected HCT116(−/−) cells and negatively correlated with the p53 expression levels (Fig. 2D). In addition, as shown in Fig. 2 E and F, this decrease in the nuclear FoxO1 level was associated with a concomitant increase in the cytoplasmic FoxO1 level. Moreover, adriamycin-mediated activation of p53 in HCT116(+/+) cells also led to a decrease in nuclear FoxO1 and a concomitant increase in cytoplasmic FoxO1 (Fig. S2A). To further validate this result, HCT116(−/−) and HCT116(+/+) cells were separately transfected with a flag-tagged FoxO1-expressing plasmid and then treated with adriamycin. As shown in Fig. 2 G and H, ectopic flag-FoxO1 was expressed at a higher level in the nucleus than in the cytoplasm in both cell lines in the absence of adriamycin. Notably, however, treatment with adriamycin led to increased cytoplasmic and decreased nuclear flag-FoxO1 in HCT116(+/+) cells, which correlated with increased p53 expression, but no changes in the levels of cytoplasmic and nuclear flag-FoxO1 in HCT116(−/−) cells.
Fig. 2.
Activation of p53 promotes translocation of FoxO1 from the nucleus to the cytoplasm. (A) Adriamycin-induced expression of FoxO1 and FoxO3 proteins in HCT116(+/+) and HCT116(−/−) cells. Nuclear extracts were subjected to immunoblotting with indicated antibodies at 12 h after treatment with indicated concentrations of adriamycin. HDAC1 was used as a loading control. (B) Quantitation of FoxO1 and p53 expression levels. Immunoblots in A were scanned and normalized to HDAC1. (C) Ectopic p53-induced expression of nuclear FoxO1 and FoxO3 in HCT116(−/−) cells. Cells were transfected with an empty plasmid or a plasmid expressing p53, and nuclear extracts were analyzed by immunoblotting with indicated antibodies. (D) Quantitation of FoxO1 and p53 expression levels. Immunoblots in C were scanned and normalized to HDAC1. (E) Nuclear versus cytoplasmic localization of FoxO1 and FoxO3 following ectopic p53 expression in HCT116(−/−) cells. Subcellular fractions were analyzed by immunoblotting at 24 h or 36 h after transfection with an empty plasmid or a plasmid expressing p53 in the presence of MG132 (2 μM, for 12 h). (F) Quantitation of relative levels of cytoplasmic and nuclear FoxO1. Immunoblots in E were scanned and normalized to HDAC1. (G) p53 induction-dependent redistribution of ectopic FoxO1 in HCT116(+/+) and HCT116(−/−) cells. Cells were transfected with flag-tagged FoxO1 in the presence or absence of adriamycin (1 μM, for 12 h before harvest, and the same for below). At 36 h posttransfection, cytoplasmic and nuclear lysates were subjected to immunoblotting with antibodies indicated on the right. PCAF and tubulin were used as loading controls for nuclear and cytosolic proteins, respectively. (H) Quantitation of relative amounts of cytoplasmic and nuclear flag-FoxO1. Immunoblots in G were scanned and normalized to either PCAF or tubulin. All data above are shown as the mean ± SD, n = 3. *P < 0.05, **P < 0.01, and ***P < 0.001.
To verify that the p53-induced translocation of FoxO1 from the nucleus to the cytoplasm was not caused by the degradation of nuclear FoxO1, leptomycin B (LMB), a potent and specific nuclear export inhibitor, was used after ectopic expression of p53 in HCT116(−/−) cells or after treatment of HCT116(+/+) cells with adriamycin. In each case the p53-dependent reduction of FoxO1 in the nucleus was almost reversed when LMB was added (Fig. S2B); and, correspondingly, adriamycin did not induce changes in the cytoplasmic FoxO1 level in the presence of LMB (Fig. S2C). In further analyses, and before adriamycin treatment, HCT116(+/+) cells were transfected either with a plasmid expressing flag-tagged FoxO1 or with a plasmid expressing a flag-tagged mutant [flag-FoxO1(3A)] that is unable to translocate to the cytoplasm (28). As expected, flag-FoxO1 exhibited a visible shuttling from the nucleus to the cytoplasm in the presence of adriamycin whereas flag-FoxO1(3A) did not (Fig. S2D).
In confirmation of the p53-induced nuclear export of FoxO1 as a general phenomenon, similar results were observed when HEPG2 cells and murine embryonic fibroblasts (MEFs) were analyzed under the same conditions (Fig. S2 E and F). Finally, we investigated whether nuclear FoxO1 exclusion occurred in a p53 transcription-dependent manner by transfection of HCT116(−/−) cells with plasmids that express either p53 or p53 mutants. Unlike p53, the transcription-defective p53(175RH) and p53(∆TA) mutants did not promote the nuclear exclusion of FoxO1 (Fig. S2G). Altogether, these results suggest that p53 induces the translocation of FoxO1 from the nucleus to the cytoplasm in a transcription-dependent manner.
The Deacetylation of FoxO1 Is a Prerequisite for Its Translocation.
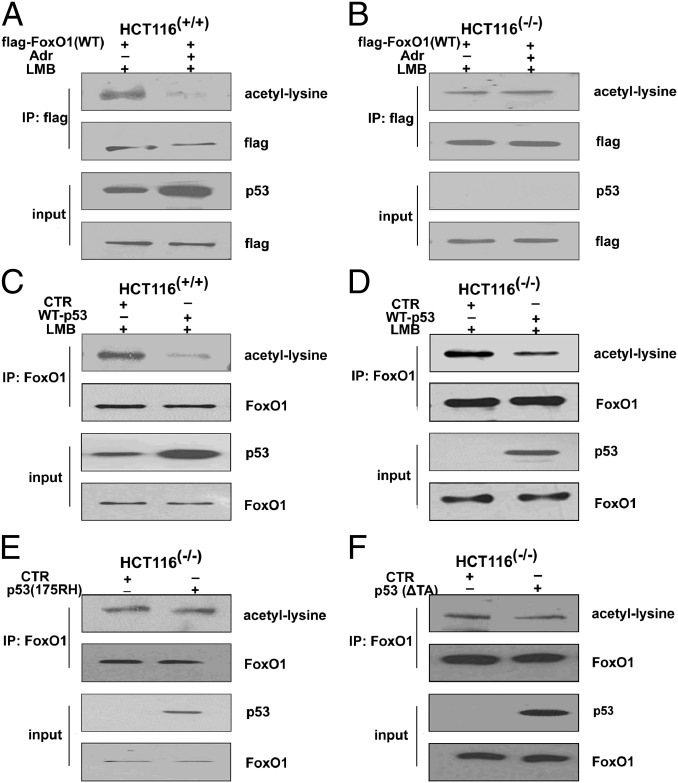
To determine the mechanism of p53-induced FoxO1 nuclear/cytoplasmic shuttling, we performed an immunoprecipitation (IP) assay to score possible changes in the phosphorylation, acetylation, and ubiquitination status of nuclear FoxO1 in HCT116(+/+) cells in response to adriamycin treatment. Although we did not observe obvious changes in pan-phosphorylation or ubiquitination of FoxO1 in these cell lines, the exogenous nuclear FoxO1 exhibited a significant decrease in acetylation when HCT116(+/+) cells, but not HCT116(−/−) cells, were exposed to adriamycin (Fig. 3 A and B). In confirmation of this result, the levels of endogenous acetylated FoxO1 were significantly decreased when p53 was ectopically expressed either in HCT116(−/−) or HCT116(+/+) cells (Fig. 3 C and D) or in HEPG2 cells (Fig. S3A).
Fig. 3.
The deacetylation of FoxO1 is a prerequisite for its translocation. (A) HCT116(+/+) and (B) HCT116(−/−) cells were transfected with a flag-tagged FoxO1 plasmid with or without adriamycin treatment (1 μM, for 12 h) in the presence of LMB (0.5 ng/mL, for 6 h). At 36 h, nuclear proteins were immunoprecipitated with anti-flag antibody and probed with indicated antibodies. (C) HCT116(+/+) and (D) HCT116(−/−) cells were transfected with an empty plasmid or a plasmid expressing p53 in the presence of LMB (0.5 ng/mL, for 6 h). At 36 h, nuclear proteins were immunoprecipitated with anti-FoxO1 antibody and then probed with indicated antibodies. (E and F) HCT116(−/−) cells were transfected with an empty plasmid, a plasmid expressing p53(175RH), or a plasmid expressing p53(∆TA), and, at 36 h, nuclear proteins were immunoprecipitated with anti-FoxO1 antibody and probed with indicated antibodies.
In addition, neither p53(175RH) nor p53(∆TA) had such an effect on the deacetylation of nuclear FoxO1 (Fig. 3 E and F), indicating that p53 transcriptional activity is required for FoxO1 deacetylation. To determine whether histone deacetylases are involved in the p53-modulated deacetylation of FoxO1, effects of the class I and II HDAC inhibitor trichostatin A (TSA) and the sirtuin inhibitor nicotinamide (NA) were tested. Neither inhibitor induced changes in the FoxO1 mRNA or protein levels (Fig. S3B). However, both inhibitors were able to reverse the nuclear exclusion of FoxO1 in response to p53 activation or overexpression (Fig. S3 C and D). Similar results were obtained in HEPG2 cells (Fig. S3E). Together, these data suggest that the deacetylation of FoxO1 is a critical posttranslational modification that is necessary for its nuclear exclusion following p53 activation.
The Interplay Between FoxO1 and Class III HDAC SIRT6 Promotes FoxO1 Nuclear Exclusion.
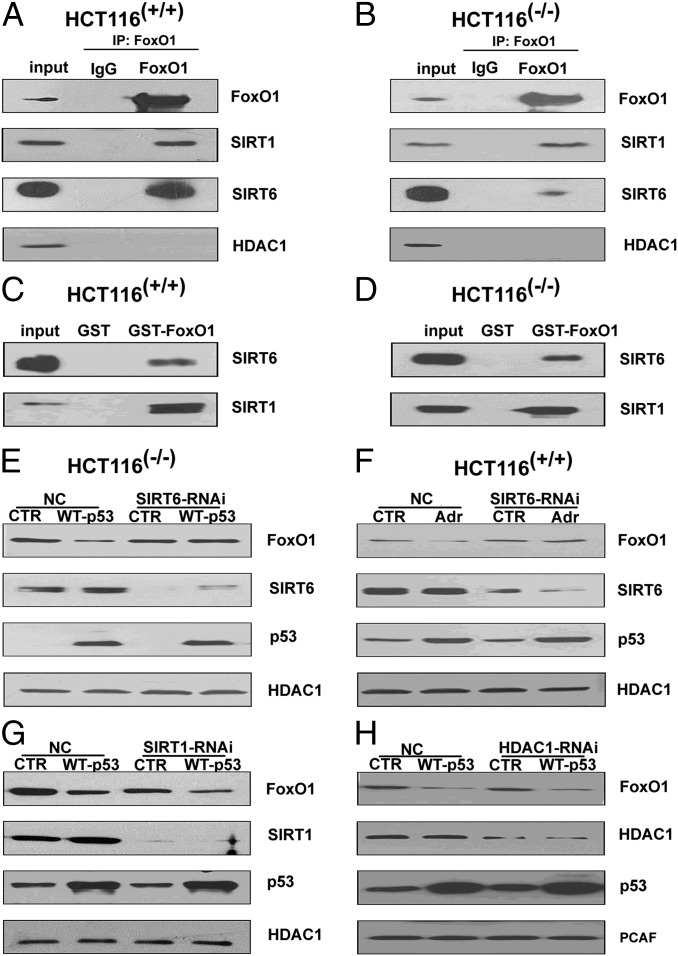
To determine which HDAC(s) interacts with FoxO1, nuclear extracts were subjected to Co-IP assays that assessed possible interactions of FoxO1 with HDAC1, SIRT1, or SIRT6 (the main nuclear HDACs). The results in Fig. 4 A and B demonstrate an interaction of FoxO1 with SIRT1 and SIRT6, but not with HDAC1, in HCT116(−/−) and HCT116(+/+) cells. In view of these results, a likely explanation for the observed inhibitory effect of TSA on p53-induced FoxO1 nuclear exclusion (above) is that we also observed an inhibitory effect of TSA on Class III HDAC expression (Fig. S4A). To provide additional evidence consistent with the interaction between FoxO1 and either SIRT1 or SIRT6, GST-pull down assays were performed. As shown in Fig. 4 C and D, SIRT1 and SIRT6 from both HCT116 cell lines were both bound to GST-FoxO1 but not to GST alone. SIRT1 and SIRT6 from HEPG2 cells also bound to GST-FoxO1 and could be immunoprecipitated with FoxO1 (Fig. S4B).
Fig. 4.
The interaction between FoxO1 and SIRT6 promotes FoxO1 nuclear exclusion. (A and B) Analysis of endogenous FoxO1 interactions with SIRT1, SIRT6, or HDAC1 in HCT116(+/+) cells and HCT116(−/−)cells. Anti-FoxO1 immunoprecipitates of nuclear extracts were probed with the indicated antibodies. (C and D) Analysis of recombinant FoxO1 interactions with SIRT1 and SIRT6 in the nuclear extracts of HCT116(+/+) and HCT116(−/−) cells. GST pull-down assays with bound proteins probed with the indicated antibodies. (E and F) Effects of SIRT6 knockdown on nuclear exclusion of FoxO1 following ectopic p53 expression in HCT116(−/−) cells or adriamycin-induced p53 expression in HCT116(+/+) cells. Nuclear extracts from CTR or SIRT6 RNAi-treated cells were subjected to immunoblotting with the indicated antibodies. (G and H) Effects of SIRT1 or HDAC1 knockdown on nuclear exclusion of FoxO1 following ectopic p53 expression in HCT116(+/+) cells. Nuclear extracts from CTR and SIRT1 RNAi- and HDAC1 RNAi-treated cells were subjected to immunoblotting with the indicated antibodies.
In subsequent studies, SIRT6 knockdown by RNAi almost completely blocked the nuclear exclusion of FoxO1 by p53 following ectopic expression in HCT116(−/−) cells or induction by adriamycin in HCT116(+/+) cells (Fig. 4 E and F and Fig. S4 C and D). An identical phenomenon was observed in HEPG2 cells (Fig. S4E). In contrast, no alterations in p53-triggered FoxO1 shuttling was observed following RNAi-mediated knockdown of either HDAC1 or SIRT1 in HCT116(−/−) cells expressing ectopic p53 (Fig. 4 G and H). In a further analysis, the ectopic expression of flag-SIRT6 in HCT116(−/−) and HCT116(+/+) cells resulted in a dramatic decrease of the FoxO1 level in the nucleus and a corresponding increase in the FoxO1 level in the cytoplasm (Fig. S4F). However, the enzymatically inactive SIRT6(133HY) failed to facilitate the nuclear export of FoxO1 (Fig. S4G). Therefore, SIRT6 is a critical factor influencing the capacity of FoxO1 to shuttle from the nucleus to the cytoplasm.
To identify the nuclear export fragment of FoxO1, plasmids encoding flag-tagged FoxO1 and derived fragments were analyzed for nuclear exclusion following p53 induction by adriamycin in HCT116(+/+) cells (Fig. S4H). The persistence of p53-induced FoxO1 export with FoxO1(1-537), but not with FoxO1(1-417), raised the possibility that the region containing amino acids 417–537 might harbor an acetylation site(s) necessary for SIRT6 interaction. To test this possibility, the resident lysines at positions 423, 446, 463, and 516 were individually mutated to arginines in FoxO1, and corresponding mutants were analyzed as above. Of these mutants, only FoxO1(423KR) appeared to show a deficiency in nuclear exclusion in response to adriamycin treatment (Fig. S4I), thus indicating that lysine 423 of FoxO1 may be the p53-dependent SIRT6 deacetylation site.
p53 Activates SIRT6 Expression.
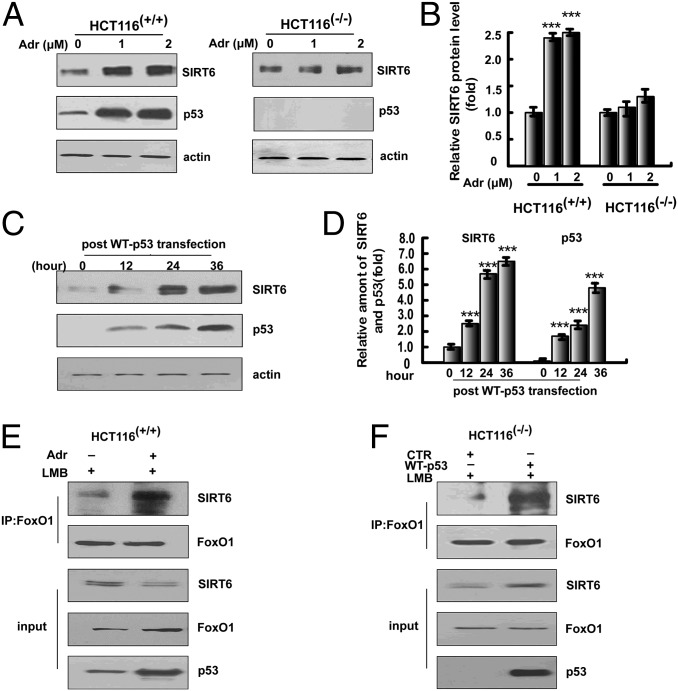
The above data led us to ask whether there is a direct interaction between SIRT6 and p53. As a Co-IP assay failed to reveal any interaction between p53 and SIRT6, we investigated a potential role for p53 in activating SIRT6. An initial ChIP assay confirmed p53 binding to the SIRT6 promoter in both human and MEF cells (Fig. S5A). A subsequent qPCR analysis (Fig. S5B) revealed an adriamycin-induced increase of SIRT6 mRNA that was greater in HCT116(+/+) cells than in HCT116(−/−) cells. Consistent with these results, SIRT6 protein levels were also remarkably increased in HCT116(+/+) cells but not in HCT116(−/−) cells (Fig. 5 A and B). SIRT6 protein levels were also significantly increased in HCT116(−/−) cells in response to ectopic p53 expression (Fig. 5 C and D), further suggesting that p53 activation is required for the increased SIRT6 expression. The activation of SIRT6 expression by intact p53, but not by p53(175RH) or p53(∆TA), also indicates a requirement for p53 transcriptional activity (Fig. S5C). The induction of SIRT6 by adriamycin treatment of HCT116(+/+) cells and by ectopic p53 expression in HCT116(−/−) cells was also accompanied by a significantly enhanced intracellular interaction between FoxO1 and SIRT6 (Fig. 5 E and F and Fig. S5D). These data suggest that p53 activates the expression of SIRT6, which in turn leads to an elevated association of SIRT6 with FoxO1.
Fig. 5.
p53 activates SIRT6 expression. (A) Activation p53 is associated with enhanced SIRT6 expression. Cell lysates of HCT116(+/+) or HCT116(−/−) cells treated with adriamycin (0 μM, 1 μM, or 2 μM for 12 h) were subjected to immunoblotting with anti-SIRT6 antibody or anti-p53 antibody. Protein was extracted from the same numbers of cells, which were digested in a specific buffer (SI Materials and Methods). (B) Quantitation of the relative SIRT6 protein levels in A. (C) Overexpression of p53 activates SIRT6 expression. Immunoblotting was performed to determine the expression of SIRT6 and p53 in HCT116(−/−) cells at 12 h, 24 h, or 36 h after p53WT transfection. (D) Quantitation of the relative levels of SIRT6 and p53 in C. All data above are shown as the mean ± SD, with ***P < 0.001. (E and F) p53 activation increases the interaction between SIRT6 and FoxO1. HCT116 (+/+) cells were treated with or without adriamycin (1 μM for 12 h) in the presence of LMB (0.5 ng/mL for 6 h before harvest). HCT116(−/−) cells were transfected with an empty plasmid or a plasmid expressing p53 in the presence of LMB (0.5 ng/mL for 6 h before harvest) for 36 h. Nuclear proteins were extracted for Co-IP with anti-FoxO1 antibody and probed with anti-SIRT6 antibody.
p53 Acts in Concert with SIRT6 to Regulate Blood Glucose Levels of Mice.
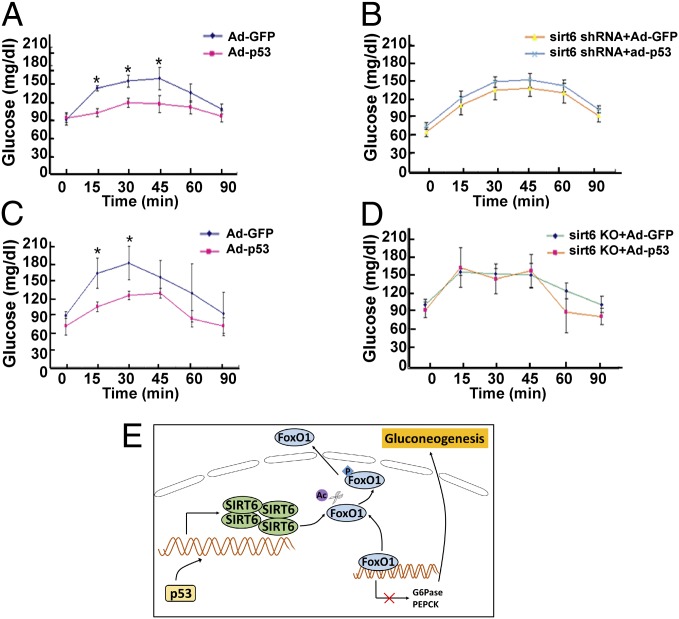
We next sought to determine the role of p53 and SIRT6 in suppressing glucose levels in mice. To this end, C57BL/6J mice were injected with a recombinant p53-expressing adenovirus (Ad-p53), leading to overexpression in liver (Fig. S6A), and blood glucose levels were monitored by PTT assay after fasting. Notably, mice expressing ectopic p53 were significantly less efficient than control mice in restoring blood glucose levels (Fig. 6A); and the p53-induced inhibitory effects on blood glucose levels were effectively prevented when the mice were coinjected with a Sirt6-shRNA (Fig. 6B). In a related series of experiments, ectopic p53 consistently prevented the recovery of blood glucose levels induced by pyruvate (Fig. 6C); and this p53-induced inhibition of glucose levels was alleviated in liver-specific Sirt6-KO mice (Fig. 6D). Consistent with these results, ectopic expression of p53 led to reduced G6PC and PCK1 expression in liver (Fig. S6B) whereas concomitant knockdown of SIRT6 (Fig. S6C) reversed these effects. Taken together, these data suggest that p53 effectively down-regulates gluconeogenesis in mice through coordination with SIRT6.
Fig. 6.
p53 acts cooperatively with SIRT6 to regulate murine blood glucose levels. Glucose levels based on pyruvate tolerance tests on C57BL/6J mice (A) or Sirt6 shRNA-pretreated C57BL/6J mice (B) that were injected with an empty plasmid or a recombinant adenovirus plasmid expressing p53 (Ad-WT-p53) after 22 h of fasting. Twenty-four C57BL/6J mice were separated into four groups, with six mice per group. Data are reported as the mean ± SD with *P < 0.05. Glucose levels based on pyruvate tolerance tests on control C57BL/6J mice (C) or conditional liver Sirt6-knockout mice (D) in response to the injection of Ad-GFP or Ad-WT-p53. Twenty-four mice were separated into four groups, with six mice per group. Data are shown as the mean ± SD with *P < 0.05. (E) A hypothetic model showing how p53 coordinates with SIRT6 to regulate gluconeogenesis.
Discussion
Beyond recent reports of variable context-dependent effects of p53 on gluconeogenesis (6–8), our results confirm a clear role for p53 in the down-regulation of gluconeogenesis both in vitro and in vivo and, importantly, establish a previously unidentified underlying mechanism of action. We first established a p53-dependent down-regulation of the key gluconeogenic enzymes G6PC and PCK1 not only in a colon cancer cell line (HCT116) but also in liver (a major gluconeogenic tissue) and in a liver cancer cell line (HEPG2). In relation to the key role of the transcription factor FoxO1 in activation of these genes, their down-regulation was related to a p53-dependent nuclear exclusion of FoxO1. This phenomenon in turn was linked to p53-mediated activation of the SIRT6 gene, which encodes an NAD+-dependent deacetylase that interacts with FoxO1 and, presumably through direct modification, leads to FoxO1 deacetylation with consequent nuclear exclusion and down-regulation of G6PC and PCK1 transcription. These results are consistent with the previously reported down-regulation of gluconeogenesis through telomere dysfunction-induced activation of p53 (6), although the underlying mechanism was reported to be an inhibitory effect of p53 on the expression of transcriptional coactivators (PGC-1α and PGC-1β) implicated in FoxO1 function. Consistent with our combined results, emphasizing ultimate (common) p53 effects through FoxO1, it has been established that FoxO1 ablation impairs fasting- and cAMP-induced glycolysis and gluconeogenesis and that PGC-1α is unable to induce gluconeogenesis in FoxO1-deficient cells (12). Relatedly, SIRT6 has been reported to down-regulate gluconeogenesis through an enhancement of GCN5-mediated acetylation and consequent inhibition of PGC-1α, raising the possibility of a complementary pathway for p53 effects on gluconeogenesis (26).
The precise control of FoxO1 subcellular localization and function can be modulated by acetylation and deacetylation by various acetyltransferases and HDACs, respectively, in response to different environmental stresses (29). For example, fasting-induced glucagon secretion triggers HDAC3 recruitment to the nucleus, with consequent FoxO1 deacetylation at lysine residues 259/262/271 and activation of gluconeogenic gene expression (30). Among the nuclear-localized sirtuins, SIRT1 was previously identified as a regulator of FoxO proteins and shown to deacetylate both FoxO1 and FoxO3 in vitro (17, 18). FoxO1 also interacts with SIRT1 in the nucleus of several cancer cell lines, leading to an inability of FoxO1 to induce apoptosis (31) or to up-regulate gluconeogenesis (32). Our current results, showing that deacetylation of FoxO1 is a prerequisite for FoxO1 nuclear exclusion and down-regulation of gluconeogenic genes upon p53 activation, are consistent with these observations. However, whereas SIRT1 and SIRT6 both interact with FoxO1, p53-modulated FoxO1 nuclear exclusion was found to be dependent on SIRT6, but not SIRT1 (or HDAC1), in this study.
Among a variety of other functions, SIRT6 was previously connected to glucose metabolism. For example, SIRT6 acts as a corepressor of the transcription factor Hif1-α to suppress glycolysis (23). Conversely, the deletion of Sirt6 in mice results in severe hypoglycemia (33) whereas the liver-specific deletion of Sirt6 leads to increased glycolysis and triglyceride synthesis (23, 34). Our study adds further evidence that SIRT6 plays an important role in glucose metabolism by connecting p53 transcription activity and gluconeogenesis. Our data also reemphasize a previously established role for SIRT6 in regulating the acetylation state and nuclear localization of FoxO proteins, albeit in a divergent manner. Thus, the Caenorhabditis elegans SIRT6/7 homolog SIR-2.4 was implicated in DAF-16 deacetylation and consequent nuclear localization and function in stress responses (35); and the effect was reported to be indirect and to involve a stress-induced inhibition by SIR-2.4 of CBP-mediated acetylation of DAF-16 that is independent of its deacetylase activity. These results, emphasizing context-dependent SIRT6 mechanisms, contrast with the SIRT6 deacetylase activity requirement for FoxO1 nuclear exclusion in the present study and a likely direct effect of SIRT6 on FoxO1 deacetylation based on their direct interaction, the SIRT6 deacetylase activity requirement, and precedent (15, 17, 18) from direct SIRT1-mediated deacetylation of FoxO proteins.
Despite a high genetic diversity, cancer cells exhibit a common set of functional characteristics, one being the “Warburg effect”: i.e., continuous high glucose uptake and a higher rate of glycolysis than that in normal cells (36). To favor the rapid proliferation requirement for high ATP/ADP and ATP/AMP ratios, cancer cells use large amounts of glucose. p53, as one of the most important tumor suppressors, exerts its antineoplastic function through diverse pathways that include the regulation of glucose metabolism. Thus, p53 regulates glucose metabolism by activation of TIGAR (3), which lowers the intracellular concentrations of fructose-2,6-bisphosphate and decreases glycolysis. On the other hand, p53 activation causes down-regulation of several glycolysis-related factors such as phosphoglycerate mutase (PGM) (37) and the glucose transporters (4). Expression of p53 also can limit the activity of IκBα and IκBβ, thereby restricting the activation of NFκB and dampening the expression of glycolysis-promoting genes such as GLUT3 (38). As a reverse glycolysis pathway, gluconeogenesis generates glucose from noncarbohydrate precursors and is conceivably essential for tumor cell growth. However, the current study further supports the notion (6) that p53 is also involved in a gluconeogenesis inhibition pathway, which in this case is executed by enhanced SIRT6 expression and subsequent FoxO1 nuclear exclusion. These results raise the interesting possibility that an inhibition of gluconeogenesis may contribute to the tumor-suppressive function of p53.
Materials and Methods
Procedures for cellular extraction have been detailed previously (13). Procedures for immunoblot analysis and immunoprecipitation assays, GST-pulldown, RNAi, mutagenesis, ChIP, and pyruvate tolerance test are detailed in SI Materials and Methods. Animals used in this paper are approved by the National Institute of Health, and Peking University Health Science Center.
Supplementary Material
Acknowledgments
This work was supported by National Key Basic Research Program of China Grants 2011CB504200, 2012CB517501, and 2013CB911001; National Natural Science Foundation of China Grants 31070691, 81321003, and 91319302; Minister of Education of China “111 Project” (to W.-G.Z.); and the National Institutes of Health Grant CA129325 (to R.G.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411026111/-/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the LIGHT: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 4.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64(7):2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 5.Jiang P, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13(3):310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein I, et al. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer Metab. 2013;1(1):9. doi: 10.1186/2049-3002-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang SJ, et al. p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle. 2013;12(5):753–761. doi: 10.4161/cc.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin HV, Accili D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 2011;14(1):9–19. doi: 10.1016/j.cmet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilling MM, Oeser JK, Boustead JN, Flemming BP, O’Brien RM. Gluconeogenesis: Re-evaluating the FOXO1-PGC-1alpha connection. Nature. 2006;443(7111):E10–E11. doi: 10.1038/nature05288. [DOI] [PubMed] [Google Scholar]
- 11.Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108(9):1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6(3):208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12(7):665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 14.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32(2):221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura YI, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2(3):153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101(27):10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 19.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCord RA, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany, NY Online) 2009;1(1):109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 25.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 26.Dominy JE, Jr, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell. 2012;48(6):900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kern SE, et al. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252(5013):1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 28.Kops GJ, et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398(6728):630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J Mol Cell Biol. 2011;3(5):276–282. doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
- 30.Mihaylova MM, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145(4):607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24(5):1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao C, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. 2010;285(47):36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H-S, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang WC, et al. C. elegans SIRT6/7 homolog SIR-2.4 promotes DAF-16 relocalization and function during stress. PLoS Genet. 2012;8(9):e1002948. doi: 10.1371/journal.pgen.1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 37.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65(1):177–185. [PubMed] [Google Scholar]
- 38.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10(5):611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.