Abstract
Clostridium perfringens is a Gram-positive anaerobic spore-forming bacterium capable of producing four major toxins that are responsible for disease symptoms and pathogenesis in a variety of animals, humans, and poultry. The organism is the third leading cause of human foodborne bacterial disease, and C. perfringens is the presumptive etiologic agent of necrotic enteritis among chickens, which in the acute form can cause increased mortality among broiler flocks. Countries that have complied with the ban on antimicrobial growth promoters (AGP) in feeds have had increased incidences of C. perfringens-associated necrotic enteritis in poultry. To address this issue, new antimicrobial agents, putative lysins from the genomes of bacteriophages, are identified. Two putative phage lysin genes (ply) from the clostridial phages phiCP39O and phiCP26F were cloned and expressed in Escherichia coli, and the resultant proteins were purified to near homogeneity. Gene and protein sequencing revealed that the predicted and chemically determined amino acid sequences of the two recombinant proteins were homologous to N-acetylmuramoyl-L-alanine amidases. The proteins were identical in the C-terminal putative cell-wall binding domain, but only 55% identical to each other in the presumptive N-terminal catalytic domain. Both recombinant lysins were capable of lysing both parental phage host strains of C. perfringens as well as other strains of the bacterium in spot and turbidity reduction assays. The observed reduction in turbidity was correlated with up to a 3 log cfu/mL reduction in viable C. perfringens on brain–heart infusion agar plates. However, other member species of the clostridia were resistant to the lytic activity by both assays.
Keywords: Antimicrobial, enzybiotic, peptidoglycan hydrolase, amidase, poultry
Introduction
Clostridium perfringens is a Gram-positive anaerobic spore-forming bacterium that causes a broad spectrum of diseases in humans and animals due to the expression of four major toxins (α, β, ε, and ι) produced by the bacterium, and spores can persist in soil, feces, or the environment (1–3). The organism is the third leading cause of foodborne bacterial disease among humans (4) because not only are foods contaminated with enterotoxigenic C. perfringens at the retail level (5) but also the organism may be carried by healthy adults (6). Economic costs of C. perfringens human food poisoning are estimated to exceed $120 million per year (2,4). Although the number of outbreaks and cases attributed to C. perfringens declined during 2006 compared with the mean annual total during 2001–2005, the continued large number of outbreaks indicates a need for improved attention to cooked meat and poultry (7). Most foodborne disease isolates of C. perfringens are of the toxin type A, possessing and expressing a chromosomal or plasmid copy of the gene for the CPE enterotoxin (cpe gene). Human enteritis strains of C. perfringens harbor the cpe gene chromosomally, whereas livestock-associated strains may carry this gene episomally on a plasmid (1). The genomes of several C. perfringens isolates are now available in public databases (8, 9).
In addition to the expense associated with severe C. perfringens human illness, there is also a cost to the poultry industry. In chickens C. perfringens is the presumptive etiologic agent of necrotic enteritis (10–13). Necrotic enteritis is an acute or chronic enterotoxemia typically characterized by necrotic lesions in the small intestines of poultry. Severe acute cases can have mortality rates as high as 50%. Annett et al. (14) reported that up to 37% of broilers grown in North America are affected by necrotic enteritis, and necrotic enteritis is estimated to cost the U.S. poultry industry $0.05 per broiler (15, 16). Necrotic enteritis has commonly been controlled by antibiotics added to poultry feed (17,18). However, there is evidence that the use of antibiotics in animal feed can potentially lead to antibiotic resistance among human bacterial pathogens (19–22). Countries that have banned the use of antibiotics in animal feed also experienced increases in clostridial necrotic enteritis as well as other animal health problems (23). The increased incidence of necrotic enteritis could potentially have negative consequences for not only animal but also human health (11). Therefore, there is a need to develop alternative antimicrobials as substitutes or to complement currently used antibiotics to control C. perfringens as well as other disease-causing pathogens in animals and humans.
Bacteriophages produce lysins that specifically degrade the peptidoglycan of their host cell wall to allow nascent progeny to be released and infect other cells (24–27). Using lysins to inhibit or kill Gram-positive bacteria is an attractive alternative to using phage therapy due to their species specificity among bacterial isolates and reliable reports that it is highly unlikely that these pathogens develop resistance (28, 29). Although C. perfringens historically has been controlled by antibiotics, with the increasing pressure to ban growth-promoting antimicrobials during poultry production (22, 30) there is an immediate need to develop alternative interventions. The genome sequence of C. perfringens bacteriophages phiCP39O (EU588980) and phiCP26F (GQ443085) revealed the presence of two potential lysins. The purpose of this study was to clone, express, and purify the two lysins (Ply) and determine their activity against C. perfringens isolates as well as other clostridial bacteria.
Materials and Methods
Bacterial Strains, Plasmids, and PCR Cloning of Bacteriophage Lysin Genes
C. perfringens isolates were propagated from cooked meat medium by streaking onto tryptic soy agar plates and incubated overnight in anaerobic jars containing AnaeroGen sachets (Oxoid, Hampshire, U.K.) at 37 °C as described previously (31). All C. perfringens isolates assayed were toxin-typed as reported (31) utilizing methods developed by Meer and Songer (32).
Oligonucleotide primers were designed to amplify the phiCP26F lysin and phiCP39O lysin genes from bacteriophage DNA (accession numbers EU588980 for the phiCP39O and GQ443085 for the phiCP26F genome) while introducing restriction enzyme sites for subcloning into sequencing and expression vectors (33,34). For cloning the plyCP26F gene into pET-21d, purified phiCP26F DNA was used as a template and amplified with primers plyCP26FexpF (5′ TAC CATGGTGAT AAT TGGAAG TAG ATA T 3′) and plyCP26F-plyCP39OexpR (5′ GTG GTG CTC GAG TAT CTT TTC GGC AAA GCA AT 3′). The NcoI and XhoI restriction sites are underlined in the forward and reverse primers, respectively. For cloning the plyCP39O gene into pET21a, purified phiCP39O DNA was used as a template and amplified with primers plyCP39OexpF (5′ GCA CTA CAT ATG AAA ATA GCT TTA AGA GGT GGA 3′) and plyCP26F-plyCP39OexpR. The NdeI restriction site is underlined in the forward primer for use with the pET-21a plasmid vector. PCR products were purified with spin-columns (Qiagen) and digested with NcoI and XhoI (plyCP26F) or NdeI and XhoI (plyCP39O). The digested PCR products were spin-column purified and cloned into similarly digested vectors pET-21a and pET-21d (Novagen, Inc.). The resulting recombinant constructs were then transformed into Escherichia coli Top 10 cells (Invitrogen) following standard methods (35). The DNA sequences of each resultant recombinant plasmid were verified by automated nucleotide sequencing (36,37).
Expression and Protein Purification of Bacteriophage Lysins
E. coli Rosetta 2(DE3) cells (Novagen, Inc.) cells harboring plasmid constructs were propagated in 200 mL of Luria–Bertani (LB) broth supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol at 37 °C with shaking. Mid log phase cultures (OD600 nm of 0.4–0.6) were placed on ice for 30 min, followed by addition of 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at a final concentration to induce pET vector expression and then incubated at 20 °C for 18 h with shaking (33,34). Cultures were centrifuged at 5000g for 20 min at 4 °C. Cell pellets were frozen at −80 °C, then thawed on ice, and suspended in 4 mL of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8). The suspension was sonicated (15 × 5 s pulses with a 15 s rest on ice between pulses) and centrifuged for 30 min at 5000g at 4 °C. The resulting supernatant was used as clarified lysate to which 1 mL of Ni-NTA (nickel matrix) slurry was added according to the manufacturer's instructions (Qiagen) with gentle shaking for 1 h at 4 °C. The slurry was then applied to a column and allowed to flow through by gravity. The column was washed twice with 4 mL of wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0), and the His-tagged protein (38) was eluted with 2 mL of sample buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). Glycerol was added to the eluted peptide solution (to 10%) and filter sterilized through a 0.2 μm filter.
Bacteriophage Lysin Protein Biochemical Characterizations
Protein concentrations were determined using the Qubit fluorometer (Invitrogen), which utilizes the Quant-it protein fluorescent stain assay (Molecular Probes Inc., Eugene, OR). Eluted proteins and Kaleidoscope protein standards (Invitrogen) were analyzed with 4–20% precise gradient protein gels (Pierce, Rockford, IL) run in Tris–Hepes–SDS buffer at 120 V for 1 h in the Bio-Rad Mini-PROTEAN three-gel apparatus, according to the manufacturer's instructions (39). Gels were stained with Coomassie Brilliant Blue R-250 (Bio-Rad, Hercules, CA) for 1 h, washed overnight in distilled water, and photographed. The protein samples were applied to PVDF by filtration through a ProSorb device (ABI), and Edman degradation amino acid sequencing was performed using standard pulse liquid cycles on an ABI Procise 492 HT sequencer (40). Additionally, recombinant lysin protein sequences were determined by mass spectrometry analyses with some modifications (41,42). All mass spectrometric data were collected using an ABI 4700 Proteomics Analyzer MALDI TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA), using the 4000 Series Explorer software v. 3.6. Data were extracted from the Oracle database, and a peak list was created by GPS Explorer software v. 3.6 (Applied Biosystems) from the raw data generated from the ABI 4700. Analyses were performed as combination MS + MS/MS. Complete proteomics methods are provided in the Supporting Information. Nucleotide sequence editing, analysis, prediction of amino acid sequences, and alignments (43) were conducted using MacVector 7.2 (Accelrys, San Diego, CA) and DNASTAR (Madison, WI) software. The predicted amino acid sequences were searched against the protein database by using BLAST and PSI-BLAST or BLASTP (44, 45) as well as the conserved domain database (46) algorithms.
Bacteriophage Lysin Spot and Turbidity Assays
Purified recombinant protein or the E. coli lysate expressing the proteins was used to determine their ability to lyse C. perfringens on bacterial plates (34, 47). For plate lysis 10 μL of clarified lysate or purified peptide was spotted onto 10 mL of brain–heart infusion (BHI) broth−0.7% agar plates that contained 500 μL of concentrated clostridial cells. Cells were prepared by growing them to mid log phase OD600 nm =0.4–0.6 in BHI broth. Cells (50 mL) were centrifuged (5000g) for 30 min at 4 °C, and the cell pellet was washed in lysin buffer A (LBA 50 mM ammonium acetate, 10 mM CaCl2, 1 mM DTT, pH 6.2). The washed pellet was then suspended in 1 mL of lysin buffer A at a 50× concentration and added to the 50 °C semisolid agar. Spotted plates were allowed to incubate for 1 h at 37 °C and then checked for lysis zones. If no lysis was present, the plates were allowed to incubate overnight at room temperature and checked again for lysis. As a negative control, lysate from E. coli containing pet21a/d plasmids with no inserts was also spotted onto plates.
The reduction in turbidity as a result of target cell lysis due to the activity of phage-derived lysin was measured by monitoring the change in optical density (OD600 nm) over time (32,47). Target cells were grown to mid log phase (OD600 nm = 0.4–0.6) in BHI broth (Becton Dickenson, Franklin Lakes, NJ) and concentrated in lysis buffer A (pH 6.2) to an OD600 nm ∼ 1.0. Assays were completed in 96-well microtiter plates with 344 μL of target cell suspension in each well and 10 μL of purified peptide (∼2.5 μg) for treated samples and 10 μL of buffer for control samples. Change in OD600 nm was recorded after 1 h of incubation at 37 °C Turbidity reduction in the cell samples alone (control) was subtracted from samples with lysin before the specific activity was calculated (change in OD600 nm (mg of protein)−1 min−1).
Effect of pH on Bacteriophage Lysin Activity and Determination of Minimum Bactericidal Concentration (MBC)
The activity of phage-derived lysins in response to pH was measured as described in the turbidity assay method but with the pH of LBA adjusted using either glacial acetic acid or ammonium hydroxide (48). The pH with the highest activity was arbitrarily set at 100%, and the activities at other pH values were expressed relative to this optimal activity.
MBCs were determined using the method employed by Makobongo et al. (49) with slight modification. Clostridial cells were grown in BHI broth to mid log phase (OD600 nm = 0.4–0.6). Cells were then diluted into fresh BHI broth (pH 7.4) to an OD600 nm of 0.05 and aliquoted into wells of a 96-well microtiter plate containing the phage lysin at concentrations ranging from 0 to 66.67 μg/mL. The microtiter plate was incubated under anaerobic conditions at 37 °C for 20 h. The MBCs were determined by enumeration of starting and ending colony-forming units (cfu) on BHI plates. The MBC was defined as the lowest concentration of the phage lysin that killed 99.9% of the starting cfu (0.1 % cfu survival). All assays were performed in triplicate using independent preparations of each bacteriophage lysin. Data were analyzed using ezANOVA (http://sph.sc.edu/comd/rorden/ezanova/index.html), and means were compared using the TukeyHSDmethod with a significance level of 0.05.
Results
Bacteriophage Lysin Expression
Genes encoding the bacteriophage endolysins PlyCP39O and PlyCP26F were PCR amplified using the purified bacteriophage genomic DNAs as templates with oligonucleotide primers that included an introduced NdeI or NcoI restriction site for PlyCP39O and PlyCP26F, respectively, and an altered stop site to include an XhoI restriction site for cloning. Subsequently, the amplification products were cloned into sequencing and expression plasmids to transform E. coli. Because the bacteriophage host C. perfringens is a Gram-positive organism, Rosetta strains (Novagen, Inc.) were utilized for gene expression as these bacteria also contain plasmids that provide appropriate tRNA genes to alleviate codon bias when expressing heterologous proteins in E. coli (50). For the plyCP39O (EU588980) the Met start site was intact, whereas an ATG was incorporated into the plyCP26F start site primer because the N-terminal amino acid in the original plyCP26F gene encoded a predicted Val for the bacteriophage protein (GQ443085).
Nucleotide sequencing of the cloned bacteriophage DNA confirmed the presence of both endolysin genes in their respective expression vectors encoding proteins with predicted amino acid sequences for each of the bacteriophage endolysins. The genes were then expressed as C-terminal, His-tagged proteins and purified by Ni-column chromatography. The purified proteins were analyzed by gel electrophoresis that resulted in the detection of two proteins, each with a relative mobility corresponding to a molecular size of approximately 25000 (Figure 1A). This agrees with the predicted sizes of approximately 25100 for both bacteriophage lysin proteins. Both the E. coli lysates containing the expressed lysin and the purified proteins were capable of producing a clear “plaque” on a plate of confluent C. perfringens following a spot assay (Figure 1B).
Figure 1.
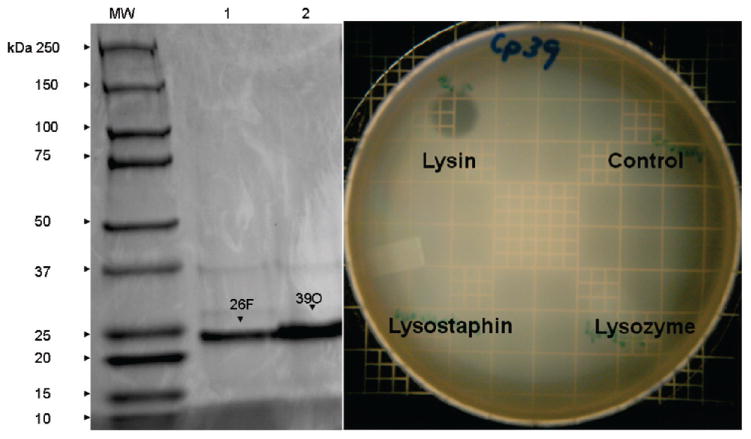
Gel electrophoresis of purified recombinant PlyCP26F and PlyCP39O accompanied by spot assays of C. perfringens on confluent plates. (A) SDS-PAGE is shown in a gel of 2.35 μg of nickel column-purified phage-derived lysin proteins. Lanes: MW, molecular weight markers; 1, Ply 26F;2, Ply 39O. Arrows indicate phage-derived lysins. (B) Spot assays were conducted by applying either bacterial lysates or the purified protein on lawns of C. perfringens as well as 10 μL of 10 mg/mL lysozyme and lysostaphin.
Characterization of Recombinant Bacteriophage Lysins
The expressed bacteriophage lysin proteins were extracted as single bands following SDS-PAGE and subjected to N-terminal amino acid sequencing and analysis by mass spectrometry. This was done to confirm the peptides were not only expressed but also in frame and not truncated or altered from the predicted amino acid sequence. Both techniques confirmed that the primary amino acid sequences of the proteins were as predicted from the nucleotide sequence of each cloned gene (Supporting Information). BLAST and domain database analyses of the bacteriophage lysin protein sequences resulted in identity to closely related proteins encoded in a variety of other Gram-positive bacteria (data not shown). Each of the two proteins reported herein were predicted to be homologous with N-acetylmuramoyl-L-alanine amidases or MurNAc-LAA (also known as peptidoglycan aminohydrolase, NAMLA amidase, NAMLAA, amidase 3, and peptidoglycan amidase; EC 3.5.1.28). These peptidoglycan hydrolases cleave the amide bond between N-acetylmuramoyl and L-amino acids in bacterial cell wall glycopeptides (51). Amidases have also been reported among bacteriophages wherein MurNAc-LAAs are endolysins, which break down bacterial peptidoglycan during the terminal stage of the phage reproduction cycle (26, 27).
The lysin genes were cloned from two bacteriophages that had host ranges restricted to individual isolates of C. perfringens. Overall, the genomes of these two bacteriophages shared >95% nucleotide sequence similarity (data not shown). Despite the high overall sequence similarity between the two bacteriophage genomes, the lysins' predicted protein amino acid sequences were identical only at the C-terminus cell-wall binding domain and shared only 56% identity to each other at the N-terminal catalytic domain (Figure 2). Specifically, residues 164–213 of PlyCP39O were 100% identical to residues 163–212 of PlyCP26F. The variable (56%) N-terminal regions of the proteins to position 162 were both predicted as homologous to N-acetylmuramoyl-L-alanine amidases. Furthermore, PlyCP39O and PlyCP26F were more closely related to the published amidase-type lysins reported from Clostridium difficile bacteriophages (52–55) rather than from the only other lysin reported for a C. perfringens bacteriophage (47). Specifically, the lysin proteins reported herein shared 24% overall identity to CD119, 21% identity to the C2 and CD27, and only 16% overall identity to CP3626 and were much shorter in length compared to the other bacteriophage lytic proteins (Figure 2).
Figure 2.
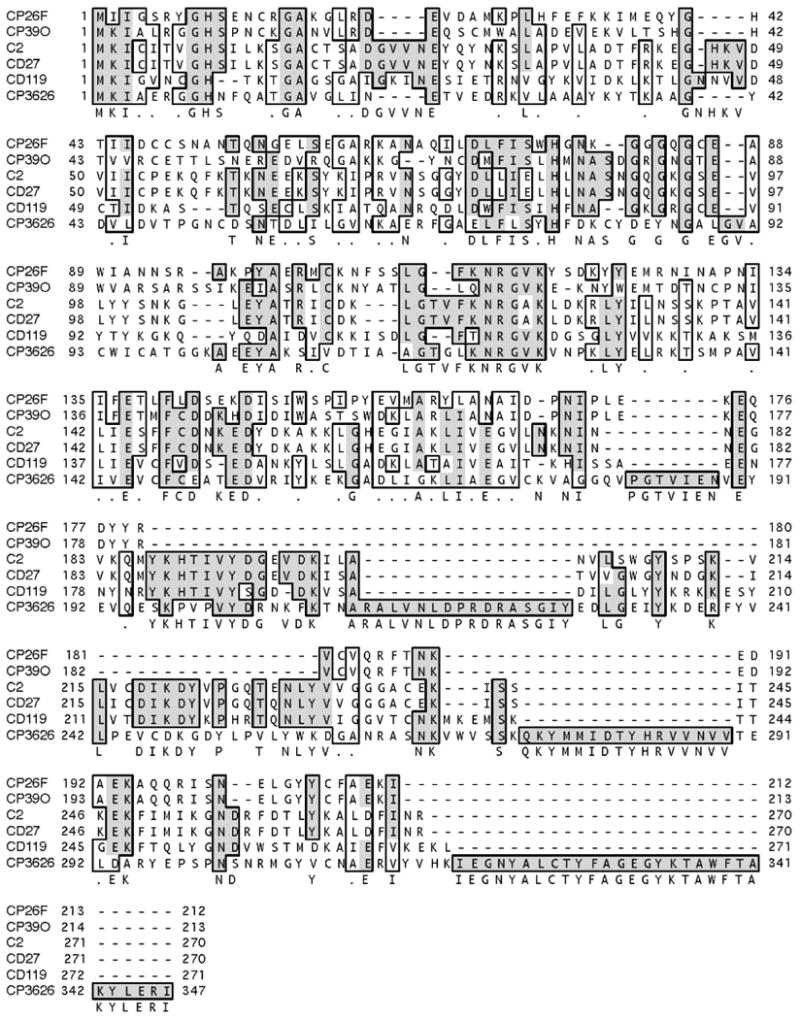
Primary amino acid sequence alignment of the recombinant PlyCP26F and PlyCP39O compared with other clostridial bacteriophage lysins. Bacteriophage lysin proteins were from Clostridium perfringens (CP) or C. difficile (C or CD) as reported in the text. Alignment of amino acid sequences was completed using Clustal W in MacVector, and boxing represents conserved residues shared among proteins.
Turbidity Assays, pH Range, and Minimum Bacteriocidal Concentration of Recombinant Lysins
Both recombinant lysins were used to measure lysis of C. perfringens by turbidity reduction assays (Figure 3). The recombinant bacteriophage lysins were capable of lysing both parental bacteriophage host strains of C. perfringens as well as 13 other poultry isolates of the bacterium in turbidity reduction assays. That is, PlyCP39O was capable of lysing host Cp39 and host Cp26, whereas PlyCP26F could reduce turbidity if utilizing isolates Cp26 or Cp39 for the assay. This was despite the fact that the predicted N-terminal enzymatic regions differed by 55% in their amino acid sequences. The reduction in turbidity yielded up to a 3 log cfu/mL reduction in viable C. perfringens on BH agar plates following the 1 h incubation. Although all C. perfringens isolates assayed were lysed by the proteins, neither was capable of lysing other members of the genus Clostridium examined during this investigation (Table 1). The optimal pH range varied from 5.5 to 9.9 for PlyCP26F (Figure 4A), whereas the pH maximum was 8.2 for PlyCP39O (Figure 4B).
Figure 3.
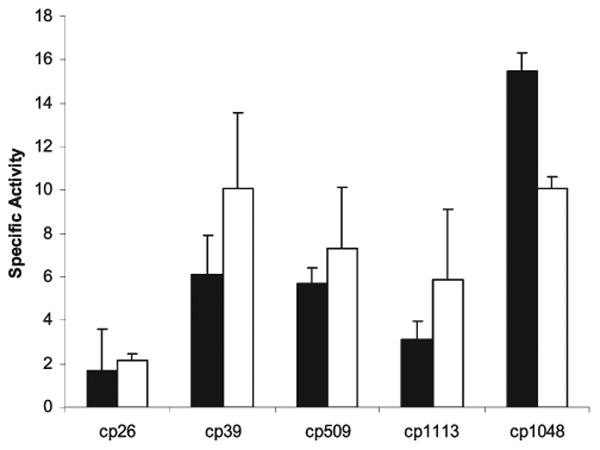
Turbidity reduction assay results for recombinant bacteriophage lysins active against Clostridium perfringens at pH 6.2. Specific activity (change in OD 600 nm mg −1 min −1) utilized 2.35 μg of phage-derived lysin protein per sample; the lysins are represented by PlyCP26F (black bars) and PlyCP39O (white bars).
Table 1. Non-perfringens Clostridial Isolates That Were Utilized as Substrates in PlyCP26F and PlyCP39O Lytic Activity Assays.
| C. absonum ATCC 27555 | C. noveyi ATCC 19402 |
| C. bifermentans ATCC 638 | C. paraputrificum ATCC 25780 |
| C. difficile ATCC 43255 | C. septicum ATCC 12464 |
| C. histolyticum ATCC 19401 | C. sordellii ATCC 9714 |
| C. innocuum ATCC 14501 | C. sporogenes ATCC 3584 |
| C. limosum ATCC 25620 | C. tetani ATCC 19406 |
Figure 4.
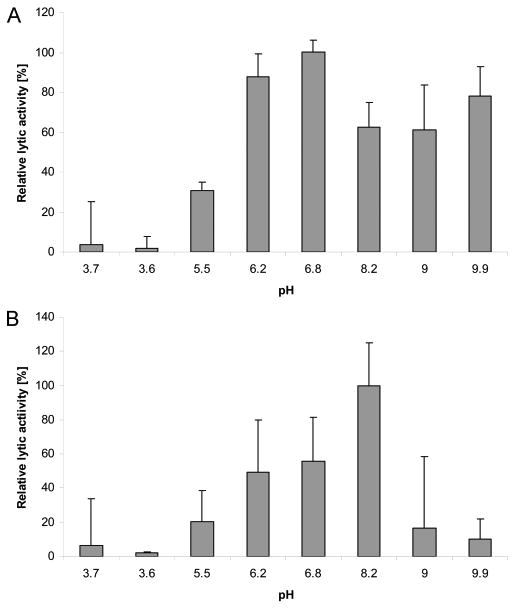
Biological activity pH profiles for bacteriophage lysins active against Clostridium perfringens. Lytic activity was measured over pH values from <4 to 9.9. The optimal activities were 6.8 and 8.2 for PlyCP26F (A) and PlyCP39O (B), respectively. The average activity at the optimal pH was arbitrarily set as 100%.
Both phage lysins had bacteriacidal activity against all C. perfingens isolates tested. The results of the MBC assays are consistent with other antimicrobial studies on Gram-positive anaerobes in which the activity was observed over a range of 1–2-fold dilutions (56, 57). The MBC ranged from 16.67 to 33.33 μg/mL against both isolates Cp26 and Cp39, whereas the MBCs for the other isolates ranged from 33.33 to 66.67 μg/mL (Table 2). It may appear as though the slightly lower MBC range for isolates Cp26 and Cp39 is due to the fact that the phage lysins were derived from phages that specifically targeted these two isolates. However, there was no significant difference (α = 0.05) between the mean MBC for each of the isolates. Furthermore, there was no difference in MBC for a given isolate between Ply26F and Ply39O. This suggests that despite the differences in the N-terminal domain of the two peptides there is enough similarity to ensure activity.
Table 2. Antimicrobial Activity of Ply26F and Ply39O against Clostridium perfringens Isolates Expressed as the Minimum Bacteriacidal Concentration (MBC).
| C. perfringensb | genotype | MBCa | |
|---|---|---|---|
|
| |||
| Ply26F | Ply39O | ||
| Cp26 | α, β2 | 16.67–33.33 (22.22 ± 5.56) | 16.67–33.33 (22.22 ± 5.56) |
| Cp39 | α, β2 | 16.67–33.33 (22.22 ± 5.56) | 16.67–33.33 (22.22 ± 5.56) |
| Cp509 | α, β2, netB | 33.33–66.67 (50.00 ± 16.67) | 33.33–66.67 (50.00 ± 16.67) |
| Cp1048 | α, β2 | 33.33–66.67 (55.56 ± 11.11) | 33.33–66.67 (55.56 ± 11.11) |
| Cp1113 | α, β2, netB | 33.33–66.67 (55.56 ± 11.11) | 33.33–66.67 (55.56 ± 11.11) |
| CpA12916 | α, enterotoxin | 190 (190)c | 190 (190)c |
| Cp3626 | enterotoxin | 190 (190)c | 95 (95)c |
The MBC in μg/mL is defined as lowest concentration to result in 99.9% killing. All assays were performed in triplicate using independent preparations of the antimicrobial, and results are expressed as a range with the mean (the standard error in parentheses.
Bacterial isolates are as reported under Materials and Methods.
This value is omitted for samples in which the results were the same for all three reps.
Discussion
Herein we report the cloning and expression of two bacteriophage lytic proteins that were 100% identical in their C-terminal putative cell-wall binding domains but were only 55% identical in their N-terminal amino acid sequences for the predicted amidase catalytic domains. This is despite the fact that the genes were identified in the genomes from two closely related C. perfringens viruses that shared >95% overall nucleotide sequence identity between the two genomes (accession numbers EU588980 for the phiCP39O and GQ443085 for the phiCP26F). The structure of Gram-positive bacteriophage lysins is highly conserved, with N-terminal enzymatic domains usually separated by an approximate six amino acid linker region followed by the C-terminal cell-wall binding domain (26, 27). The putative cell wall binding domains were identical in both proteins, which may explain why both proteins were capable of lysing a variety of C. perfringens isolates. The lysins were predicted to be N-acetylmuramoyl-L-alanine amidases by BLAST or domain database analyses, and these lytic proteins have been reported in a variety of bacteriophages (26,27). Also of interest was that our two C. perfringens bacteriophage proteins were more closely related to lysins reported for C. difficile bacteriophages (52–55) rather than to the other lysin PlyCP3626 previously reported for a C. perfringens bacteriophage (47).
Spot and turbidity assays were utilized to demonstrate lysis of C. perfringens isolates relative to other clostridia. Despite the sequence differences between the two proteins in the putative active domains, the proteins were capable of lysing a variety of poultry isolates in our C. perfringens collection along with human strains obtained from sources such as ATCC. However, the lysins were not active against any other clostridial species examined to date. This species specificity of the bacteriophage lysins reported herein is consistent with other results (47). The turbidity reduction assays did not decrease optical density to the extent that others have reported (47);however, spot assays on plates always resulted in complete lysis zones of C. perfringens. This species specificity of the bacteriophage lysins argues more favorably for use as an antimicrobial to the relative broad-range effects on the gastrointestinal bacterial flora of currently utilized antibiotics in feeds (58, 59) that can potentially lead to antibiotic resistance issues during poultry production (60, 61).
The bacterium C. perfringens causes a wide variety of diseases in humans and animals as well as being the third leading cause of bacterial foodborne disease in humans (2, 4). The bacterium is the presumptive etiologic agent of necrotic enteritis in chickens, which is one of the leading causes of losses during poultry production (11,13) and which has increased in prevalence among countries that have withdrawn antibiotic growth promoters in animal feeds, resulting in estimated losses of U.S. $2 billion internationally (16,23). The use of bacteriophages to control gastrointestinal bacterial diseases has been proposed for use during food-animal production (62). Also, a bacteriophage cocktail has recently been approved by the U.S. FDA to control Listeria spp. on ready-to-eat meats and poultry (63, 64), which was proven effective to potentially reduce this bacterium by up to 5 logs on solid foods (65) and produce (66). However, the bacteriophage mixtures containing different viral isolates recently approved by the FDA were also reported to be ineffective because the Listeria spp. bacteriophage treatment did not infect all of the examined isolates (67) and bacteriophage resistance among Listeria spp. could be a problem in poultry-processing plants (68).
There is an increase in bacterial mutation rates due to bacteriophages (69), and various phage resistance mechanisms occur among bacteria (70, 71). Bacteriophages can act as agents for horizontal gene transfer (72,73) among foodborne bacteria (74,75), and recently it was demonstrated that the intergeneric transfer of toxin genes may also be mediated by phages (76). These phenomena all argue for use of phage lytic mechanisms (77) rather than intact, live-phage therapy. The bacteriophage lysins reported herein were identified among a group of phages that exhibited extremely high sequence similarity but had host restriction among C. perfringens isolates (data not shown). Because antibiotics can themselves induce horizontal transfer of resistance genes (78, 79), the use of a more precise mechanism such as bacteriophage lysins as proposed by several investigators (26–29) may be a superior method to employ for the control of virulent or antibiotic-resistant bacteria in the future.
Supplementary Material
Acknowledgments
We thank Johnna Garrish for excellent technical support. We acknowledge primary amino acid sequencing and mass spectrometry analyses of the recombinant proteins by Rebekah Woolsey and Dr. Kathleen Schegg at the University of Nevada, Reno (UNR), Proteomics Center supported by NIH Grant P20 RR-016464 from the INBRE Program of the National Center for Research Resources.
Funding was provided by the Agricultural Research Service (ARS), USDA CRIS Project 6612-3200-046-00D.
Footnotes
Supporting Information Available: Primary amino acid sequences of PlyCP39O and PlyCP26F and results of BLAST analysis and PFAM identification of PlyCP39O and PlyCP26F. This material is available free of charge via the Internet at http://pubs.acs.org.
Literature Cited
- 1.Rood JI. Virulence genes of Clostridium perfringens. Annu Rev Microbiol. 1998;52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 2.McClane BA. Clostridium perfringens. In: Doyle MP, Beuchat LR, Montville TJ, editors. Food Microbiology: Fundamentals and Frontiers. 2nd. ASM Press; Washington, DC: 2001. pp. 351–372. [Google Scholar]
- 3.Smedley JG, 3rd, Fisher DJ, Sayeed S, Chakrabarti G, McClane BA. The enteric toxins of Clostridium perfringens. Rev Physiol Biochem Pharmacol. 2004;152:183–204. doi: 10.1007/s10254-004-0036-2. [DOI] [PubMed] [Google Scholar]
- 4.Olsen SJ, MacKinon LC, Goulding JS, Bean NH, Slutsker L. Surveillance for foodborne-disease outbreaks- United States, 1993–1997. Morb Mortal Wkly Rep. 2000;49:1–51. [PubMed] [Google Scholar]
- 5.Wen Q, McClane BA. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl Environ Microbiol. 2004;70:2685–2691. doi: 10.1128/AEM.70.5.2685-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman RJ, Sayeed S, Li J, Genheimer CW, Hiltonsmith MF, Wilkins TD, McClane BA. Clostridium perfringens toxin genotypes in the feces of healthy North Americans. Anaerobe. 2008;14:102–108. doi: 10.1016/j.anaerobe.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC) Surveillance for foodborne disease outbreaks – United States, 2006. MMWR Morb Mortal Wkly Rep. 2009;58:609–615. [PubMed] [Google Scholar]
- 8.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16:1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4(2) doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Immerseel F, De Buck J, Pasmans F, Huyghebaert G, Haesebrouck F, Ducatelle R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 2004;33:537–549. doi: 10.1080/03079450400013162. [DOI] [PubMed] [Google Scholar]
- 12.Van Immerseel F, Rood JI, Moore RJ, Titball RW. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 2009;17:32–36. doi: 10.1016/j.tim.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Cooper KK, Songer JG. Necrotic enteritis in chickens: a paradigm of enteric infection by Clostridium perfringens type A. Anaerobe. 2009;15:55–60. doi: 10.1016/j.anaerobe.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Annett CB, Viste JR, Chirino-Trejo M, Classen HL, Middleton DM, Simko E. Necrotic enteritis: effects of barley, wheat and corn diets on proliferation of Clostridium perfringens type A. Avian Pathol. 2002;31:599–602. doi: 10.1080/0307945021000024544. [DOI] [PubMed] [Google Scholar]
- 15.Van der Sluis W. Clostridial enteritis is an often underestimated problem. World Poul. 2000;16:42–43. [Google Scholar]
- 16.McDevitt RM, Brooker JD, Acamovic T, Sparks NHC. Necrotic enteritis; a continuing challenge for the poultry industry. World Poult Sci J. 2006;62:221–247. [Google Scholar]
- 17.Devriese LA, Daube G, Hommez J, Haesebrouck F. In vitro susceptibility of Clostridium perfringens isolated from farm animals to growth-enhancing antibiotics. J Appl Bacteriol. 1993;75:55–57. doi: 10.1111/j.1365-2672.1993.tb03407.x. [DOI] [PubMed] [Google Scholar]
- 18.Watkins KL, Shryock TR, Dearth RN, Saif YM. In vitro antimicrobial susceptibility of Clostridium perfringens from commercial turkey and broiler chicken origin. Vet Microbiol. 1997;54:195–200. doi: 10.1016/s0378-1135(96)01276-x. [DOI] [PubMed] [Google Scholar]
- 19.Witte W. Medical consequences of antibiotic use in agriculture. Science. 1998;279:996–997. doi: 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- 20.Wegener HC, Aarestrup FM, Jensen LB, Hammerum AM, Bager F. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg Infect Dis. 1999;5:329–335. doi: 10.3201/eid0503.990303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapin A, Rule A, Gibson K, Buckley T, Schwab K. Airborne multi-drug resistant bacteria isolated from a concentrated swine feeding operation. Environ Health Perspect. 2005;113:137–142. doi: 10.1289/ehp.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyles CL. Antimicrobial resistance in selected bacteria from poultry. Anim Health Res Rev. 2008;9:149–158. doi: 10.1017/S1466252308001552. [DOI] [PubMed] [Google Scholar]
- 23.Casewell M, Friis C, Marco E, McMullin P, Phillips I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J Antimicrob Chemother. 2003;52:159–161. doi: 10.1093/jac/dkg313. [DOI] [PubMed] [Google Scholar]
- 24.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernhardt TG, Wang IN, Struck DK, Young R. Breaking free: “protein antibiotics” and phage lysis. Res Microbiol. 2002;153:493–501. doi: 10.1016/s0923-2508(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 26.Hermoso JA, García JL, García P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Fischetti VA. Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol. 2008;11:393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 29.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 30.Teuber M. Veterinary use and antibiotic resistance. Curr Opin Microbiol. 2001;4:493–499. doi: 10.1016/s1369-5274(00)00241-1. [DOI] [PubMed] [Google Scholar]
- 31.Siragusa GR, Danyluk MD, Hiett KL, Wise MG, Craven SE. Molecular subtyping of poultry-associated type A Clostridium perfringens isolates by repetitive-element PCR. J Clin Microbiol. 2006;44:1065–1073. doi: 10.1128/JCM.44.3.1065-1073.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meer RR, Songer JG. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res. 1997;58:702–705. [PubMed] [Google Scholar]
- 33.Pritchard DG, Dong S, Baker JR, Engler JA. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology. 2004;150:2079–2087. doi: 10.1099/mic.0.27063-0. [DOI] [PubMed] [Google Scholar]
- 34.Donovan DM, Dong S, Garrett W, Rousseau GM, Moineau S, Pritchard DG. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl Environ Microbiol. 2006;72:2988–2996. doi: 10.1128/AEM.72.4.2988-2996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell DW. Molecular Cloning, A Laboratory Manual. 3rd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 36.Smith LM, Sanders JZ, Kaiser RJ, Hughs P, Dodd C, Connell CR, Heines C, Kent SBH, Hood LE. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:673–681. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 37.Kristensen T, Voss H, Schwager C, Stegemann J, Sproat B, Ansorge W. T7 DNA polymerase in automated dideoxy sequencing. Nucleic Acids Res. 1988;16:3487–3496. doi: 10.1093/nar/16.8.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crowe J, Döbeli H, Gentz R, Hochuli E, Stüber D, Henco K. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol Biol. 1994;31:371–387. doi: 10.1385/0-89603-258-2:371. [DOI] [PubMed] [Google Scholar]
- 39.Hames BD. One-dimensional polyacrylamide gel electrophoresis. In: Hames BD, Rickwood D, editors. Gel Electrophoresis of Proteins: A Practical Approach. 2nd. Oxford University Press; New York: 1990. pp. 1–147. [Google Scholar]
- 40.Niall HD. Automated Edman degradation: the protein sequenator. Methods Enzymol. 1973;27:942–1010. doi: 10.1016/s0076-6879(73)27039-8. [DOI] [PubMed] [Google Scholar]
- 41.Hunt DF, Yates JR, 3rd, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 43.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmer M, Vukov N, Scherer S, Loessner MJ. The murein hydrolase of the bacteriophage phi3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl Environ Microbiol. 2002;68:5311–5317. doi: 10.1128/AEM.68.11.5311-5317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoong P, Schuch R, Nelson D, Fischetti VA. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J Bacteriol. 2004;186:4808–4812. doi: 10.1128/JB.186.14.4808-4812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makobongo MO, Kovachi T, Gancz H, Mor A, Merrell DS. In vitro anti-bacterial activity of acyl-lysyl oligomers against. Helicobacter pylori Antimicrob Agents Chemother. 2009;53:4231–4239. doi: 10.1128/AAC.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in. Escherichia coli Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 51.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 52.Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology. 2007;153:676–685. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- 53.Goh S, Riley TV, Chang BJ. Isolation and characterization of temperate bacteriophages of. Clostridium difficile Appl Environ Microbiol. 2005;71:1079–1083. doi: 10.1128/AEM.71.2.1079-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Govind R, Fralick JA, Rolfe RD. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J Bacteriol. 2006;188:2568–2577. doi: 10.1128/JB.188.7.2568-2577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008;190:6734–6740. doi: 10.1128/JB.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Critchley IA, Green LS, Young CL, Bullard JM, Evans RJ, Price M, Jarvis TC, Guiles JW, Janjic N, Ochsner UA. Spectrum of activity and mode of action of REP3123, a new antibiotic to treat Clostridium difficile infections. J Antimicrob Chemother. 2009;63:954–963. doi: 10.1093/jac/dkp041. [DOI] [PubMed] [Google Scholar]
- 57.Roberts SA, Shore KP, Paviour SD, Holland D, Morris AJ. Antimicrobial susceptibility of anaerobic bacteria in New Zealand: 1999–2003. J Antimicrob Chemother. 2006;57:992–998. doi: 10.1093/jac/dkl052. [DOI] [PubMed] [Google Scholar]
- 58.Wise MG, Siragusa GR. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J Appl Microbiol. 2007;102:1138–1149. doi: 10.1111/j.1365-2672.2006.03153.x. [DOI] [PubMed] [Google Scholar]
- 59.Thompson K, Burkholder K, Patterson J, Applegate TJ. Microbial ecology shifts in the ileum of broilers during feed withdrawal and dietary manipulations. Poult Sci. 2008;87:1624–1632. doi: 10.3382/ps.2007-00324. [DOI] [PubMed] [Google Scholar]
- 60.Guan J, Wasty A, Grenier C, Chan M. Influence of temperature on survival and conjugative transfer of multiple antibiotic-resistant plasmids in chicken manure and compost microcosms. Poult Sci. 2007;86:610–613. doi: 10.1093/ps/86.4.610. [DOI] [PubMed] [Google Scholar]
- 61.Miranda JM, Vazquez BI, Fente CA, Barros-Velazquez J, Cepeda A, Franco CM. Evolution of resistance in poultry intestinal Escherichia coli during three commonly used antimicrobial therapeutic treatments in poultry. Poult Sci. 2008;87:1643–1648. doi: 10.3382/ps.2007-00485. [DOI] [PubMed] [Google Scholar]
- 62.Johnson RP, Gyles CL, Huff WE, Ojha S, Huff GR, Rath NC, Donoghue AM. Bacteriophages for prophylaxis and therapy in cattle, poultry and pigs. Anim Health Res Rev. 2008;9:201–215. doi: 10.1017/S1466252308001576. [DOI] [PubMed] [Google Scholar]
- 63.Lang LH. FDA approves use of bacteriophages to be added to meat and poultry products. Gastroenterology. 2006;131:1370. doi: 10.1053/j.gastro.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Bren L. Bacteria-eating virus approved as food additive. FDA Consum. 2007;41:20–22. [PubMed] [Google Scholar]
- 65.Guenther S, Huwyler D, Richard S, Loessner MJ. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl Environ Microbiol. 2009;75:93–100. doi: 10.1128/AEM.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leverentz B, Conway WS, Camp MJ, Janisiewicz WJ, Abuladze T, Yang M, Saftner R, Sulakvelidze A. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl Environ Microbiol. 2003;69:4519–4526. doi: 10.1128/AEM.69.8.4519-4526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Y, Liu Y, Zhang Y, Cripe J, Conway W, Meng J, Hall G, Bhagwat AA. Isolation and characterization of Listeria monocytogenes isolates from ready-to-eat foods in Florida. Appl Environ Microbiol. 2006;72:5073–5076. doi: 10.1128/AEM.00435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim JW, Siletzky RM, Kathariou S. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl Environ Microbiol. 2008;74:6623–3660. doi: 10.1128/AEM.01282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pal C, Macia MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- 70.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 71.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–424. doi: 10.1016/s1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 73.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brabban AD, Hite E, Callaway TR. Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne Pathog Dis. 2005;2:287–303. doi: 10.1089/fpd.2005.2.287. [DOI] [PubMed] [Google Scholar]
- 75.Kelly BG, Vespermann A, Bolton DJ. The role of horizontal gene transfer in the evolution of selected foodborne bacterial pathogens. Food Chem Toxicol. 2009;47:951–968. doi: 10.1016/j.fct.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 76.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 77.Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol. 2004;22:185–191. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- 78.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 79.Hastings PJ, Rosenberg SM, Slack A. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 2004;12:401–404. doi: 10.1016/j.tim.2004.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


