Abstract
Various diseases, injuries, and congenital abnormalities may result in degeneration and loss of organs and tissues. Recently, tissue engineering has offered new treatment options for these common, severe, and costly problems in human health care. Its application is often based on the usage of differentiated stem cells. However, despite intensive research and growing knowledge, many questions remain unresolved in the process of cell differentiation. The aim of this study was to find standardized cell models for analyzing molecular mechanisms of cell differentiation. We investigated the multipotency of three standardized murine embryonic fibroblast cell cultures using histological staining, western blotting, and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). Our results demonstrated that NIH-3T3 and mouse embryonic fibroblast (MEF) cells were able to differentiate into adipogenic, chondrogenic, and osteogenic lineages expressing typical differentiation markers. Interestingly, Flp-In-3T3 cells did not differentiate into any of the three mesenchymal lineages, although this cell line is genetically closely related to NIH-3T3. The results were confirmed by histological staining. Flp-In-3T3, NIH-3T3, and MEF cells have usually been used for DNA transfections, recombinant protein expression, and as “feeder cells.” Unlike mesenchymal stem cells (MSCs) and mesenchymal progenitor cells (MPCs), they are easy to obtain and to expand and are less prone to change their structure and morphology, even at higher passages. Our results suggest that Flp-In-3T3, MEF, and NIH-3T3 cells are highly suitable to be used as models to analyze molecular mechanisms of cell differentiation.
Introduction
One of the most common, severe, and costly problems in health care is the loss or failure of organs and tissues (Niklason and Langer, 2001). Approximately 8 million patients with these disorders are treated annually in the United States (Vacanti and Langer, 1999). In this regard, tissue engineering, which is offering a very promising treatment for these clinical obstacles, is one of the fastest growing fields in medicine today. The process of cell differentiation, a major component of tissue engineering, offers opportunities to develop biological substitutes for research and application in reconstructive medicine.
It is well documented that MSCs are capable of differentiating into bone, cartilage, and fat (Boeuf and Richter, 2010; Haynesworth, et al., 1992; Janderova et al., 2003; Li et al., 2005a; Moosmann et al., 2005; Vaananen, 2005; Yoo, et al., 1998). But mesenchymal stem cell (MSC) harvesting procedures are invasive, expensive, and laborious. In addition, the in vitro expansion capacity of isolated MSCs is limited and leads to changes in cellular phenotype (Derubeis and Cancedda, 2004; Vacanti et al., 2005). The populations may differ between single isolation processes and need exact definition to ensure reproducibility. Despite the rapid growth of knowledge in that field, uncertainties remain with respect to the determinants of cell differentiation. In particular, the requirements and conditions for effective induction of chondrogenesis and for the production of a stable cartilaginous tissue are not well understood (Boeuf and Richter, 2010). Also the methods and factors of inducing cell differentiation like differentiation media, differentiation environments, bioreactors, and many other factors need further investigation and must be improved. To clarify the unresolved questions in that area, detailed studies with standardized cell models are required, which must be at least multipotent, easy to handle, have a high expansion capacity, and do not change their phenotype easily. The aim of this study was to find standardized cell models, which were suitable for certain cell differentiation processes. Therefore, we chose three mouse embryonic fibroblast (MEF) cell types—Flp-In-3T3, MEF, and NIH-3T3.
NIH-3T3 cells are MEFs derived from a cell line that was isolated and initiated at the New York University School of Medicine Department of Pathology in 1962. They were obtained from desegregated NIH Swiss mouse embryo fibroblasts by George Todaro and Howard Green. 3T3 stands for “3-day transfer, inoculum 3×105 cells” and is derived from the original cell transfer and inoculation protocol. This cell line has since become a standard fibroblast cell line and is usually used for DNA transfection studies (Bai et al., 2011). It is not known which tissues gave rise to the NIH-3T3 cells because whole embryos were minced and cell cultures were then prepared (Todaro and Green, 1963).
MEFs were derived from 14-day-old mouse embryos (C57/BL6 52 strain). It is also not known what tissues contributed to the generation of MEFs because of their isolation from aliquots of fine minced whole mouse embryos. MEF cells are often used as “feeder cells” in human embryonic stem cell (ESC) research (Invitrogen, 2010).
The Flp-In-3T3 cell line was generated from NIH-3T3 cells by transfection with the vector pFRT/lacZeo. Thus, these cells contain a Flp recombination target (FRT) for the Flp recombinase–mediated targeted integration of a Flp-In expression vector and zeocin resistance. The pFRT/lacZeo vector allows the generation of stable cell lines with high transcriptional activity (Invitrogen, 2010).
To test for the multipotency of these cell lines, we treated them with adipogenic, chondrogenic, and osteogenic media. The results were detected by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR), western blotting, and histological staining.
Materials and Methods
Cell culturing
Flp-In-3T3 (Invitrogen, Carlsbad, CA, USA), MEF, and NIH-3T3 cells [American Type Culture Collection (ATCC), Manassas, VA, USA] were stored frozen at 123.15° K, thawed for use, and kept under standard cell culture conditions. In brief, cells were detached with trypsin [0.25% trypsin (wt/vol)] (Biochrom, Berlin, Germany) and 0.5 mμ EDTA in phosphate-buffered saline (PBS) (Biochrom, Berlin, Germany) and replated in culture medium consisting of Dulbecco's Modified Eagle Medium (DMEM) High-Glucose (Biochrom, Berlin, Germany) with 1% penicillin/streptomycin (Biochrom, Berlin, Germany), 1% sodium pyruvate (Biochrom, Berlin, Germany), and 10% fetal calf serum (FCS) (Biochrom, Berlin, Germany) for NIH-3T3 and MEF cells (Invitrogen, 2010; Todaro and Green, 1963). The medium for Flp-In-3T3 culture contained 10% donor calf serum (DCS) (Biochrom, Berlin, Germany) instead of FCS (Invitrogen, 2010).
Isolation of murine MSCs
Bone marrow–derived murine MSCs were isolated according to established protocols based on the plastic adherence of these cells (Lennon and Caplan, 2006; Meirelles Lda and Nardi, 2003). Bone marrow was obtained from 6-week-old male BALB/c mice, purchased either from Charles River or the central animal facility at Hannover Medical School. Femurs and tibias were dissected free of surrounding tissue and bone marrow was flushed out with Hanks' Balanced Salt Solution (HBSS, PAA, Cölbe, Germany) containing 1000 U/mL heparin (Sigma, Taufkirchen, Germany). The resulting bone marrow was disrupted by passing through a 20-gauge needle, centrifuged at 300×g for 7 min, and resuspended in 3 mL of DMEM (PAA, Cölbe, Germany). The cell suspension was applied to a density gradient (LSM 1077, 1.077 g/mL; PAA, Cölbe, Germany) (Aurich, et al., 2007) and centrifuged again (500×g, 30 min). The interphase containing bone marrow–derived mononuclear cells was transferred to a 15-mL tube (Greiner, Frickenhausen, Germany) and washed three times with 10 mL of DMEM containing 10% FCS (both PAA, Cölbe, Germany). Cells were plated at a density of 1×105 on cell culture dishes (100 mm diameter; Greiner, Frickenhausen, Germany) and incubated at 37°C in a humidified incubator with an atmosphere of 95% air, 5% CO2 until confluent.
Media were changed every third day unless otherwise specified. Culture medium for primary cultures was MesenCult (StemCell Technologies) (Schumann et al., 2009). To prepare secondary subcultures from these cells, the cell layers were gently rinsed with HBSS and then incubated with trypsin/EDTA (PAA, Cölbe, Germany). Cells released from the culture surface were washed with two changes of secondary culture (DMEM with 10% FCS, 1000 IU/mL penicillin and 0.1 mg/mL streptomycin; all PAA, Cölbe, Germany) and aliquoted on new 100 mm diameter cell culture dishes at the desired plating density using secondary culture medium. The cells were used at passage 9.
Flow cytometry
The cell-surface phenotype was analyzed by incubating the cells with antibodies against CD105, CD73 and CD90, SCa-1, CD11b, and CD34 (BD Pharmingen, Franklin Lakes, NJ, USA) for 30 min at 4°C. The cells were washed twice, resuspended in Isoton® II (Beckman-Coulter, Krefeld, Germany) to a final concentration of 5×106 cells/mL. The analyses were performed using an FC-500 flow cytometer equipped with CXP software (Beckman- Coulter).
Differentiation of the three cell models into adipogenic, chondrogenic, and osteogenic lineages
The induction of MEFs into adipogenic, osteogenic, and chondrogenic lineages was performed according to previously reported methods with some modifications as stated below (Zuk et al., 2001). The differentiation protocols were performed at three independent time points and analyzed separately. Representative results were depicted in the corresponding figures as stated below.
Adipogenic differentiation
After the cells had reached 100% density, the medium was replaced by adipogenic medium: DMEM F12 (Biochrom, Berlin, Germany), 10% FCS, 393 ng/mL dexamethasone (Sigma, St. Louis, MO, USA), 50 μg/mL ascorbic acid-2-phosphate (Sigma, St. Louis, MO, USA), 10 μL/mL penicillin/streptomycin, 111.1 μg/mL 3-isobutyl-1-methylxanthine (IBMX) (Sigma, St. Louis, MO, USA), 1 μg/mL insulin (Sigma, St. Louis, MO, USA), and 35.8 μg/mL indomethacin (Sigma, St. Louis, MO, USA).
Chondrogenic differentiation
Cells were seeded in culture plates in standard growth medium until confluence. For chondrogenic differentiation, the Flp-In-3T3, MEF, and NIH-3T3 cells were centrifuged to form a pelleted mass and then placed in the chondrogenic medium, consisting of DMEM F12 containing 39.3 ng/mL dexamethasone, 50 μg/mL ascorbic acid-2-phosphate, 10 μL/mL penicillin/streptomycin, 10 μL/mL insulin-transferrin-selenium (ITS+) (Sigma, St. Louis, MO, USA), 22.5 μL/mL sodium pyruvate, 40 μL/mL proline (Biochrom, Berlin, Germany), and 1 μL/mL transforming growth factor-β1 (TGF-β1; Promocell, Heidelberg, Germany).
Osteogenic differentiation
Cells were seeded in culture plates in standard growth medium until confluence. For osteogenic differentiation, the medium was replaced by osteogenic medium, consisting of DMEM F12 containing 10% FCS, 39.3 ng/mL dexamethasone, 50 μg/mL ascorbic acid-2-phosphate, 10 μL/mL penicillin/streptomycin, and 756 μg/mL β-glycerol (Sigma, St. Louis, MO, USA).
Histological staining
Oil Red O staining
After 2 weeks of adipogenic induction, cells were rinsed twice with PBS, fixed with 4% (wt/vol) paraformaldehyde (Sigma, Taufkirchen, Germany) in PBS, washed with distilled water, rinsed in 60% (vol/vol) isopropanol, and covered with a Oil Red O solution (Serva, Heidelberg, Germany) (0.3% in 60% isopropanol). After 10 min, cultures were briefly rinsed in 60% isopropanol, thoroughly washed with distilled water, and air dried at room temperature. Cells were counted and analyzed with Excel (Microsoft).
Alcian Blue staining
After 3 weeks, pellets of chondrogenically induced cells were rinsed twice with PBS, fixed with 4% formalin (Roth, Karlsruhe, Germany), and embedded in paraffin (Roth). The samples were cut in 5-μm-thick sections, which were stained for the presence of glucosaminoglycans with Alcian Blue (Fluka, Taufkirchen, Germany) (1% (wt/vol) in 3% (vol/vol) acetic acid). Morphological examination was performed microscopically.
Alizarin Red S staining
After 4 weeks, cells were rinsed twice with PBS, fixed with 4% (wt/vol) paraformaldehyde (Sigma, Taufkirchen, Germany) in PBS, and rinsed with distilled water. To stain for calcium deposits, cells were covered with a 0.5% aqueous solution of Alizarin Red S (Sigma; pH 4) for 60 min. Specimens were rinsed thoroughly with distilled water and air dried at room temperature.
The stained cells were observed under an Olympus microscope (CK40) and analyzed using cell^D software.
Western blot analysis
All materials were purchased from Sigma (Taufkirchen, Germany), if not indicated otherwise. The cells were lysed for blotting in radioimmunoprecipitation assay (RIPA) buffer [10 mM Tris (pH 8), 150 mM NaCl, 1% (vol/vol) Nonidet P-40, 0.5% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) sodium dodecyl sulfate (SDS), 1 mM phenylmethylsulfonyl fluoride (PMSF), 4 μg/mL aprotinin, 1 mM sodium orthovanadate] supplemented with the protease inhibitor PMSF. Bone, cartilage, and fat tissue samples obtained from mice for positive controls had been minced previously and lysed in RIPA supplemented with PMSF. A 25-μg amount of protein was loaded on a SDS polyacrylamide gel (PAGE) and transferred to Amersham Hybond ECL Nitrocellulose Membrane (GE-Healthcare, Buckinghamshire UK). The membranes were blocked in 5% (wt/vol) skim milk and 0.03% Tween-20 in PBS, and incubated with the following primary antibodies: Goat polyclonal to β-actin, rabbit polyclonal to resistin, mouse monoclonal to adiponectin, rabbit monoclonal to bone alkaline phosphatase, mouse monoclonal to Runt-related transcription factor 2 (RUNX-2), rabbit polyclonal to elastin, and goat polyclonal to aggrecan. The antibodies were purchased from Abcam (Cambridge, UK), with the exception of aggrecan, which was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Secondary antibodies used were: Alexa Fluor 680 goat-anti-rabbit (excitation/emission maxima: 679/702 nm) (Life Technologies Europe B.V, Zug 34, Switzerland), LI-COR 680 donkey-anti-mouse (excitation/emission maxima: 679/696 nm), and LI-COR 800 donkey-anti-goat (excitation/emission maxima: 778/795 nm) (Lincoln, NE, USA). The blots were analyzed via direct infrared fluorescence detection using the Odyssey Infrared Scanner from LI-COR. The results were confirmed at two independent time points.
qRT-PCR
Total cellular RNA was isolated from cultured cells using a NucleoSpin RNA II Kit (Macherey-Nagel, Düren, Germany) following the provided protocol. Bone, cartilage, and fat samples, obtained from mice as positive controls, were minced to extract total RNA using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The RNA concentrations were determined by spectrophotometry (NanoDrop, Wilmington, DE, USA). RNA quality was verified on a 2% Tris-borate-EDTA (TBE) gel supplemented with 0.5 μL of ethidium bromide (Sigma, Taufkirchen, Germany). Reverse transcription was carried out with 1 μg of total mRNA using iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA). A 5-μL amount of diluted reverse-transcribed cDNA [reverse transcription (RT) reaction, 1:100 in high-performance liquid chromatography (HPLC) water] was amplified in a 15-μL PCR assay volume, using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA), target primers (Table 1), and HPLC water.
Table 1.
Primers for Detecting Specific Genetic Markers of Adipogenic, Chondrogenic, and Osteogenic Differentiation Listed with Expected Product Length and Optimal Annealing Temperature
| Gene | Primers | Product length (bp) | Melting temperature (°C) | GenBank no. |
|---|---|---|---|---|
| PPARγ | F, 5′-atgtgtcgccttcttgctct-3′ | 180 | 57.3 | NM_008904 |
| R, 5′-atctactgcctggggacctt-3′ | 59.4 | |||
| Resistin | F, 5′-gctgagggtctggaaatgaa-3′ | 223 | 57.3 | NM_022984 |
| R, 5′-ggccagcctggactatatga-3′ | 59.4 | |||
| Adiponectin | F, 5′-gttgcaagctctcctgttcc-3′ | 192 | 59.4 | NM_009605 |
| R, 5′-tctccaggagtgccatctct-3′ | 59.4 | |||
| SOX9 | F, 5′-agctcaccagaccctgagaa-3′ | 200 | 59.4 | NM_011448 |
| R, 5′-tcccagcaatcgttaccttc-3′ | 57.3 | |||
| Aggrecan | F, 5′-aggactgaaatcagcggaga-3′ | 188 | 57.3 | NM_007424 |
| R, 5′-agggacatggttgtttctgc-3′ | 57.3 | |||
| Collagen(II) | F, 5′-agagacctgaactgggcaga-3′ | 200 | 59.4 | NM_031163 |
| R, 5′-gcaccattgtgtaggacacg-3′ | 59.4 | |||
| RUNX2 | F, 5′-cccagccacctttacctaca-3′ | 150 | 59.4 | NM_009820 |
| R, 5′-tatggagtgctgctggtctg-3′ | 59.4 | |||
| BALP | F, 5′-gacacaagcattcccactatgtctg-3′ | 103 | 64.6 | NM_007431 |
| R, 5′-tgcatgtccccgggctcaaag-3′ | 63.7 | |||
| Sclerostin | F, 5′-cgcctcaccggcttccaca-3′ | 82 | 61.5 | NM_024449 |
| R, 5′-gcggcttgcgacccttctgc-3′ | 63.7 | |||
| ACTB | F, 5′-cgtcgacaacggctccg-3′ | 110 | 67.7 | NM_007393 |
| R, 5′-ccaccatcacaccctggt-3′ | 69.5 | |||
| TBP | F, 5′-gtgcgtcaggcgttcggtgg-3′ | 113 | 65.5 | NM_013684 |
| R, 5′-aatagtgatgctgggcactgcgga-3′ | 64.4 | |||
| B2M | F, 5′-gcctgtatgctatccagaaaacccc-3′ | 114 | 64.6 | NM_009735 |
| R, 5′-tgtgaggcgggtggaactgtg-3′ | 63.7 | |||
| RPL32 | F5′aacccagaggcattgacaac3′ | 152 | 64.6 | NM_172086 |
| F5′cacctccagctccttgacat3′ | 63.7 | |||
| P53 | F5′ctggctgccatattccagat3′ | 197 | 57.3 | NM_022032 |
| R, 5′-cccaaaaggtcatcctcgta-3′ | 57.3 |
Resistin and adiponectin were used as markers for adipogenic differentiation, aggrecan and collagen (II) for chondrogenic differentiation, and Runt-related transcription factor 2 (RUNX-2), bone alkaline phosphatase (BALP), and sclerostin for osteogenic differentiation.
Reference primer pairs: ACTB, actin beta; TBP, TATA box–binding protein; B2M, β2-microglobulin; RPL32, ribosomal protein L32. p53 was detected as an additional investigation point.
PPARγ, peroxisome proliferator-activated receptor γ; SOX9, sex determining region Y-box 9.
Gene expression was measured by qRT-PCR performed with a Bio-Rad iCycler PCR machine. The data were analyzed by the qRT-PCR data analysis software qbasePlus (Biogazelle, Zwijnaarde, Belgium). The most stably expressed genes were identified among a set of eight normalization genes using the GeNorm algorithm implemented in the software. qbasePlus allows the usage of multiple normalization genes that are required for robust normalization. Subsequently, expression levels of investigated genes were normalized to the expression levels of β-actin (ACTB), TATA-box binding protein (TBP), β2-microglobulin (B2M), and ribosomal protein L32 (RPL32), and reported as arbitrary units of gene expression. Representative results were visualized on a 2% TBE gel supplemented with 0.5 μL of ethidium bromide. Bands were detected using a gel documentation system (Vilber Lourmat, Eberhardzell, Germany). All experiments were performed in triplicate and repeated at two independent time points.
Results
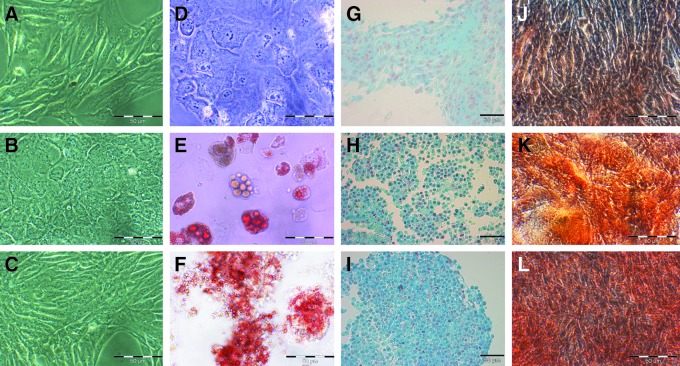
Three standardized cell models of murine fibroblasts (NIH-3T3, Flp-In-3T3, and MEF) were used due to their excellent properties, such as high expansion capacity, easy handling, and good availability. The cells were expanded in vitro and showed a typical morphological appearance (Fig. 1A–C).
FIG. 1.
Photomicrographs of differentiated and undifferentiated Flp-In-3T3, MEF, and NIH-3T3 cell cultures revealing their morphological changes during the differentiation processes (D–L) and showing their multilineage potential. Cell cultures were stained with Oil Red O after 2 weeks of adipogenic induction. Accumulated lipid droplets were observable in adipogenicallly differentiated MEF and NIH-3T3 cultures (E, F). Chondrogenically induced Flp-In-3T3 (G), MEF (H), and NIH-3T3 (I) cells were cultured for 3 weeks in pellets and stained with Alcian Blue. Arrows mark rounded (chondrocyte-type) cells in differentiated and accordingly positively stained MEF and NIH-3T3 cell cultures. Flp-In-3T3, MEF, and NIH-3T3 cells were treated with osteogenic medium for 4 weeks and stained with Alizarin Red S. MEF and NIH-3T3 cells revealed calcium deposits and stained positively with Alizarin Red S. Treated Flp-In-3T3 cells revealed no chondrogenic, adipogenic, or osteogenic resemblance in their morphology and did not stain positively (D, G, J). Scale bars, 50 μm.
To determine their multilineage differentiation potential, cells from each line were split into three groups, which were incubated in established induction media optimized for adipogenic, chondrogenic, and osteogenic differentiation.
Histological staining illustrated morphological changes in differentiated cell cultures
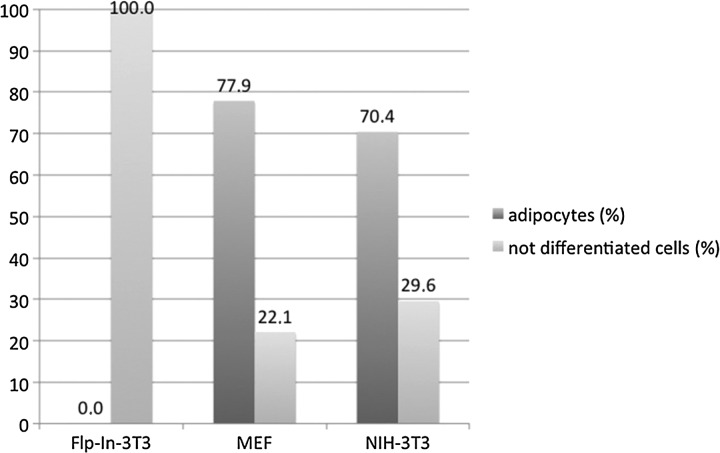
To visualize morphological changes after incubation of cell cultures with differentiation media, we used histological stains for each differentiation lineage (Fig 1). After 2 weeks of incubation in adipogenic medium, NIH-3T3 and MEF cultures had adopted a more rounded shape and displayed intracellular droplets, whereas Flp-In-3T3 cells did not show any morphological changes. The droplets accumulated over time, filling out large proportions of the observed cells. Adipogenic differentiation was assessed by Oil Red O staining for neutral lipids and triglycerides (Fig. 1E, F). In contrast to treated NIH-3T3 and MEF, Flp-In-3T3 cells (cultured for the same time period in adipogenic culture medium) were not stained for fat vacuoles (Fig. 1D). Undifferentiated control cells did not stain positively for Oil Red O. The percentages of Oil-Red-O–positive MEF cells were counted to 77.9% and NIH-3T3 cells to 70.4% in the adipogenically induced cultures (Fig. 2).
FIG. 2.
Percentages of Oil Red O–positive cells. Cells were adipogenically induced for 2 weeks and stained for lipid vesicles. The dark columns represent cells with Oil Red O vesicles, whereas the light columns represent Oil Red O–negative cells counted in the corresponding samples. No Oil Red O–positive cells could be found in the uninduced control cells (data not shown) such as in induced Flp-In-3T3 cells. (Dark grey bars) adipocytes (%); (light grey bars) cells not differentiated (%).
After 3 weeks of induction in chondrogenic media, NIH-3T3 and MEF cells cultured in pellets displayed radical changes in their macroscopic appearance. Formation of nodules and ridges with a high density of cells and extracellular matrix were observed. Flp-In-3T3 cells remained unchanged throughout the experiments. Cells from induced NIH-3T3 and MEF cultures were stained with Alcian Blue, indicating the presence of sulfated proteoglycans typical for chondrocytes. The blue color was most prominent in the areas of high cell density. In both cultures, this resulted in large amounts of intensively stained extracellular matrix (Fig. 1G–I). Percentages could not be determined because the cells were cultured in three-dimensional cultures and Alcian Blue stains extracellular proteoglycans. Undifferentiated controls and induced Flp-In-3T3 cells revealed no Alcian Blue staining.
After 4 weeks, NIH-3T3 and MEF cultures, which had been induced with osteogenic medium, displayed clusters with high cell density and large amounts of extracellular matrix, which were positive in Alizarin Red S staining (Fig. 1J–L). Untreated control cells and induced Flp-In-3T3 cells maintained their phenotypes and did not stain positively in all evaluations. The high density of the osteogenic cell cultures and the large amounts of extracellular matrix hampered the cell count, and therefore we could not determine any percentages.
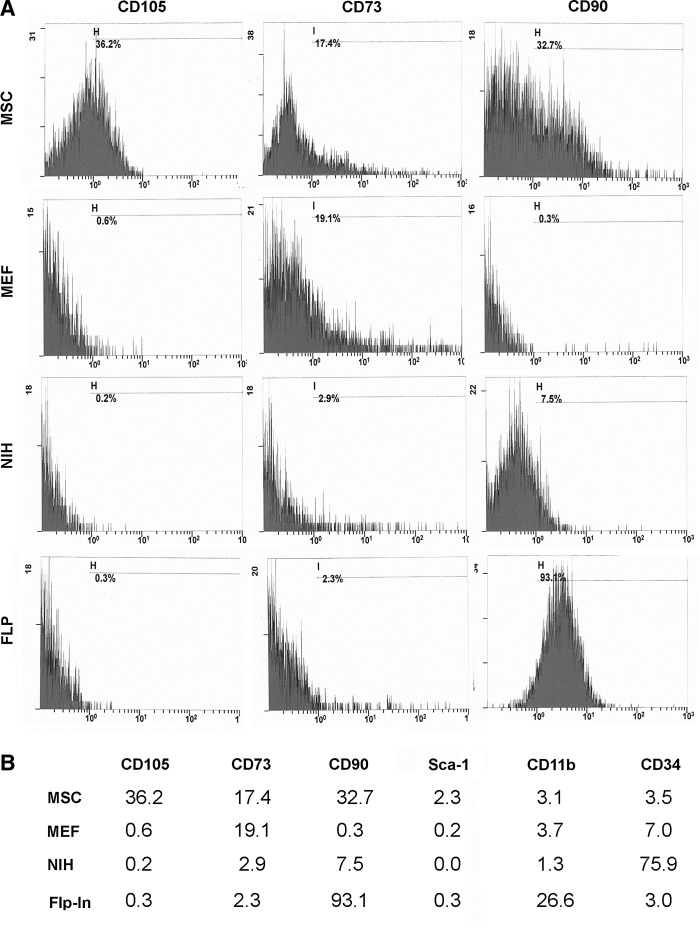
Surface markers were evaluated to reveal the similarity of the three cell lines to MSCs
We used antibodies directed against CD90, CD73, CD105, Sca-1, CD11b, and CD34 to characterize the expression of surface proteins. We found that most of the cell-surface markers typical for MSCs were not expressed on the MEFs, although Flp-In-3T3 cells expressed CD90. Only 7.5% of the closely related NIH-3T3 cells expressed CD90, although 75.9% of the cells expressed CD34 (Barth and Westhoff, 2007). MEF cells do not express CD90, but 19.1% of the cells were positive for CD73 (Fig. 3). Surprisingly, we found that MSCs derived from murine bone marrow only in part expressed CD90, CD73, CD 105, and Sca-1, but that may be due to the high passage number of the cells (Fig. 3B).
FIG. 3.
Flow cytometric analysis of selected cell-surface markers. The percentages of positive cells are given as indicated. NIH-3T3, Flp-In-3T3, MEF, and MSC cells were used in their undifferentiated state.
Detection of typical marker proteins in differentiated cultures
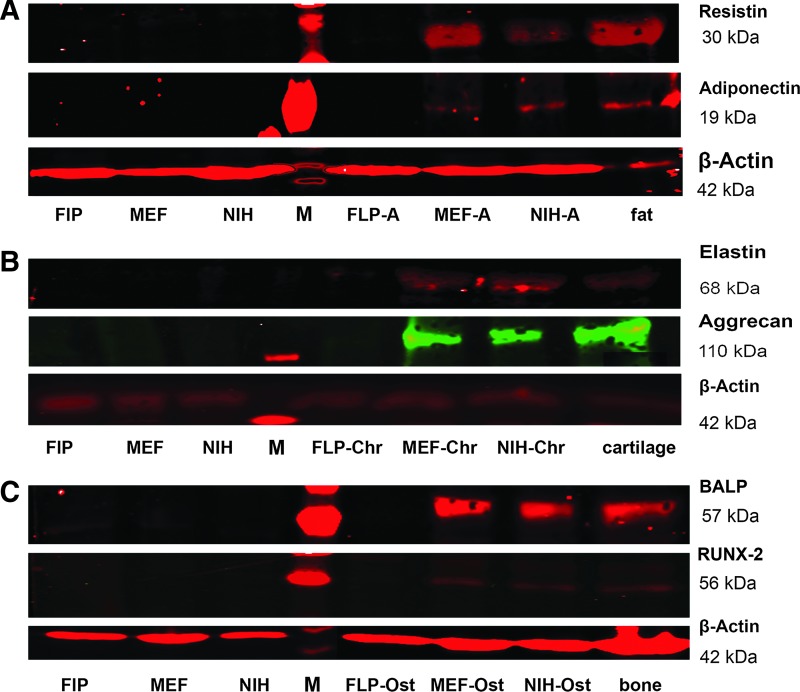
The next step was to investigate the expression of marker proteins typical for each lineage using western blot analysis. Western blot results correlated with our histological findings and indicated production of specific protein markers for adipogenic, chondrogenic, and osteogenic differentiation (Fig. 4).
FIG. 4.
Western blot analyses showing differentiation markers of adipogenic, chondrogenic, and osteogenic lineages. The expression of specific proteins for each lineage were analyzed in differentiated cells as well as in cells not differentiated and in protein samples derived from fat, cartilage, and bone as positive controls. (A) Adipogenic lineage, adiponectin and resistin. (B) Chondrogenic lineage, elastin and aggrecan. (C) Osteogenic lineage, bone alkaline phosphatase and RUNX2. M represents the marker lane.
Resistin and adiponectin were detected in the protein fractions of adipogenically treated MEFs and NIH-3T3 cells (Fig. 4A). In the chondrogenic lineage of NIH-3T3 and MEF cells, elastin and aggrecan were found to be expressed (Fig. 4B). The presence of the specific osteogenic proteins RUNX-2 and BALP in induced NIH-3T3 and MEF cultures was indicative of an osteogenic differentiation (Fig. 4C). All specific protein markers used for adipogenic, chodrogenic, and osteogenic differentiation could not be detected in uninduced cells and in induced Flp-In-3T3 cells.
qRT-PCR data confirmed previous results
To quantify expression of specific genetic markers of adipogenic, chondrogenic, and osteogenic differentiation, nine gene-specific and four normalization gene primer pairs were designed (Table 1), and quantitative PCR analysis was performed. The expression levels of investigated genes were normalized to expression levels of control samples and reported as arbitrary units of gene expression (Fig. 5).
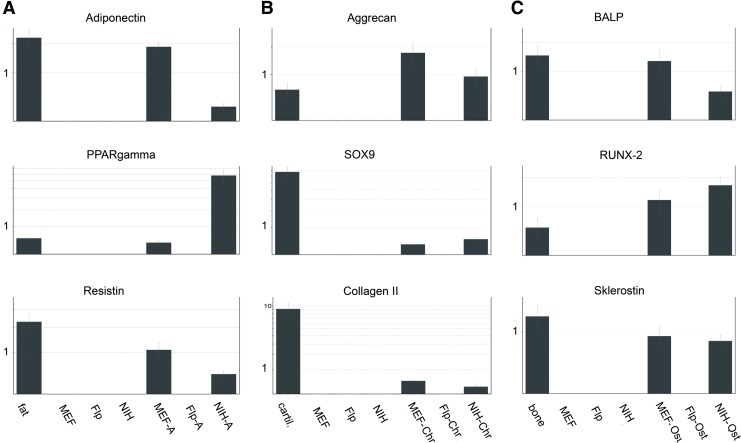
FIG. 5.
qRT-PCR data normalized with respect to ACTB, RPL32, TBP, and B2M expression. Note the quantitative differences observable between the differentiated cell lines. (A) Results of qRT-PCR assays on Flp-In-3T3, MEF, and NIH-3T3 cells induced to adipogenic lineage. The three cell lines were induced into adipogenesis for 14 days. Upregulated genes were involved in lipid metabolism during adipogenesis. Note the higher fold changes in resistin and adiponectin in MEF cells than in NIH-3T3 cells after induction with adipogenic medium. (B) Results of quantitative real-time PCR assays on Flp-In-3T3, MEF, and NIH-3T3 cells induced in chondrogenic lineage. The three cell lines were induced into chondrogenesis under pellet culture for 21 days. Upregulated genes were involved in cartilage metabolism during chondrogenesis. (C) Results of quantitative real-time PCR assays on Flp-In-3T3, MEF, and NIH-3T3 cells induced in osteogenic direction. The three cell lines were induced into osteogenesis for 28 days. Upregulated genes involved in bone metabolism during osteogenesis. Flp, Flp-In-3T3 cells; NIH, NIH-3T3 cells; A, adipogenically differentiated; Chr, chondrogenically differentiated; Ost, osteogenically differentiated.
Primer pairs specific for peroxisome proliferator-activated receptor-γ (PPARγ), adiponectin, and resistin were used to determine whether Flp-In-3T3, MEF, and NIH-3T3 cells underwent adipogenesis (Bajpai et al., 2012; Deng and Scherer, 2010).
Adipogenic differentiation of NIH-3T3 cells was demonstrated by an increase in PPARγ expression. MEF cultures and the positive control also expressed PPARγ, but to lesser extents. In contrast to PPARγ, the target genes adiponectin and resistin levels were higher in MEF cultures than in differentiated NIH-3T3 cultures and positive controls (Fig. 5). Specifically, the genes encoding for collagen type II, aggrecan, and sex determining region Y-box 9 (SOX9) are known to be potential markers for chondrogenic differentiation (Bajpai et al., 2012; Mizuno et al., 2002; Todaro and Green, 1963). As shown in Figure 5B, the level of SOX9 expression in MEF cells was significantly lower (0.2-fold) than in NIH-3T3 cells. Expression pattern of aggrecan and collagen II were different so that we could detect higher levels in treated MEF than in NIH-3T3 cells.
The expression of osteoblast-specific genes, bone alkaline phosphatase (BALP), RUNX2, and sclerostin in Flp-In-3T3, MEF, and NIH-3T3 cellswas used to verify osteogenic differentiation (Bajpai et al., 2012; Winkler et al., 2003). The level of RUNX2-specific gene expression in osteogenically induced NIH-3T3 cells was 0.5-fold higher than in MEF cells. In contrast to that, BALP and sclerostin expression was lower in NIH-3T3 cells than in MEF cells and positive controls (Fig. 5C).
Interestingly, all target genes for validation of adipogenic, chondrogenic, and osteogenic differentiation were not detected in untreated cells and induced Flp-In-3T3 cells (Fig. 5).
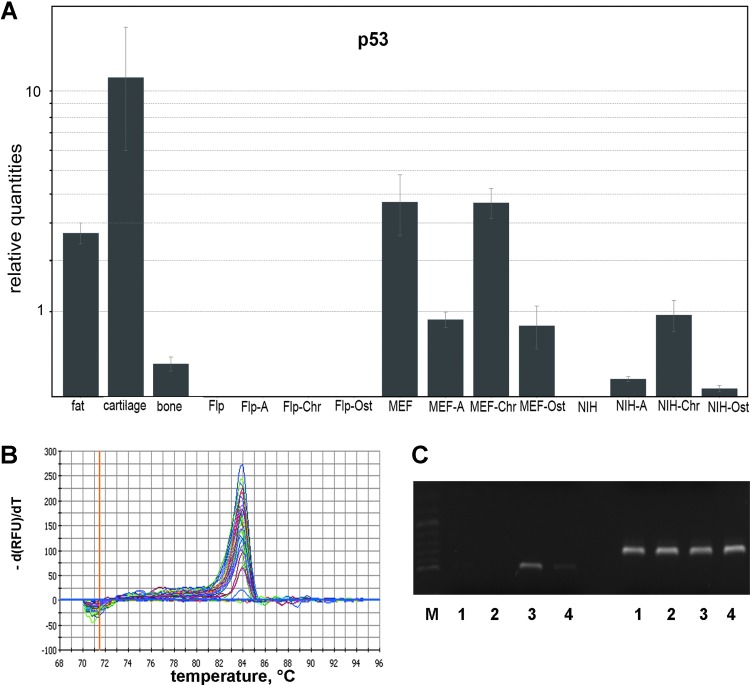
According to previous studies showing that p53 acts as an important general regulator that controls various cellular gene networks, we investigated the expression of p53 in induced and uninduced Flp-In-3T3, MEF, and NIH-3T3 cells (Molchadsky et al., 2008). As the most widely mutated gene in human tumorigenesis, p53 is involved in many cellular programs, such as cell cycle control, apoptosis, and genomic stability (Rivlin et al., 2011). Despite the clear establishment of its tumor suppressor activity, less attention was given to its function in the process of cell differentiation. As shown in Figure 6A MEF cells expressed p53 in the adipogenically, chondrogenically, and osteogenically differentiated states as well as in the undifferentiated state. Uninduced Flp-In-3T3 and NIH-3T3 cells expressed either no p53 or at levels beyond detection. Cycle threshold (Ct) values from a representative experiment are given in Table 2, demonstrating reproducibility of the results over three independent repetitions. In contrast to treated Flp-In-3T3 cells, the expression of p53 in NIH-3T3-derived adipocytes, chondrocytes, and osteocytes increased. The specificity of the PCR reaction was surveyed by melting curve analysis (Fig. 6B). Additionally, the PCR reactions were analyzed by electrophoretical separation of the final products (Fig. 6C). Although there is a faint band visible for Flp-In-3T3 cells, the synthesis of the PCR product was below the level of detection in qRT-PCR.
FIG. 6.
Expression of the p53 gene compared among the three cell lines in their differentiated and undifferentiated state. (A) Normalized data obtained by quantitative real-time PCR. ACTB, RPL32, TBP, and B2M were used for normalization. A, adipogenically differentiated; Chr, chondrogenically differentiated; Ost, osteogenically differentiated. (B) Representative melting curve obtained using primers specific for p53 as indicated in Table 1. (C) Representative PCR products obtained by quantitative PCR analyses. Left lanes, p53; right lanes, RPL32. Lane 1, NIH-3T3; lane 2, Flp-In-3T3; lane 3, MEF; lane 4, bone; lane M, marker.
Table 2.
Ct Values Obtained for Undifferentiated Flp-In-3T3, NIH-3T3, and MEF Cells Using Primers Specific for p53 As Stated in Table 1
| Sample | Ct value | Sample | Ct value | Sample | Ct value | Sample | Ct value |
|---|---|---|---|---|---|---|---|
| Flp | N/A | MEF | 27.7 | NIH | N/A | ||
| Flp | N/A | MEF | 28.6 | NIH | N/A | ||
| Flp | 35.8 | MEF | 27.4 | NIH | 34.7 | ||
| Fat | 28.9 | Flp-A | N/A | MEF-A | 30.4 | NIH-A | 32.1 |
| Fat | 29.2 | Flp-A | N/A | MEF-A | 30.7 | NIH-A | 32.3 |
| Fat | 28.9 | Flp-A | N/A | MEF-A | 30.9 | NIH-A | 32.6 |
| Cartilage | 29.6 | Flp-Chr | 38 | MEF-Chr | 27.9 | NIH-Chr | 30.3 |
| Cartilage | 31.6 | Flp-Chr | 35.3 | MEF-Chr | 28.1 | NIH-Chr | 30.5 |
| Cartilage | 31.3 | Flp-Chr | 35.4 | MEF-Chr | 28.4 | NIH-Chr | 31.2 |
| Bone | 32.4 | Flp-Ost | N/A | MEF-Ost | 29.4 | NIH-Ost | 33.6 |
| Bone | 31.7 | Flp-Ost | N/A | MEF-Ost | 31.1 | NIH-Ost | 33.9 |
| Bone | 32.4 | Flp-Ost | N/A | MEF-Ost | 30.4 | NIH-Ost | 32.2 |
The values are from a representative example from three independent runs.
Flp, Flp-In-3T3; MEF, mouse embryonic fibroblasts; NIH, NIH-3T3; A, adipogenically differentiated; Chr, chondrogenically differentiated; Ost, osteogenically differentiated.
Discussion
In the present study, we investigated the multilineage differentiation potential of standardized murine embryonic fibroblasts cell types Flp-In-3T3, MEF, and NIH-3T3 in vitro employing histological staining (Fig. 1), gene (Fig. 5), and protein expression analysis (Fig. 4). The purpose of this work was to: (1) Find new cell populations that will provide new opportunities to investigate the process of cell differentiation, and (2) reveal the specificity of NIH-3T3, MEF, and Flp-In-3T3 cells according to their differentiation potentials. Our data suggest differences between the differentiation properties of the three cell lines. Although Flp-In-3T3 cells did not show any signs of differentiation after induction using common differentiation media, NIH-3T3 and MEF cells were able to differentiate into all of the induced mesenchymal lineages. We found a common set of genes that were upregulated during differentiation toward all three mesenchymal lineages in both NIH-3T3 and MEF cells. Although the multilineage potential of MEF and NIH-3T3 cells was similar according to cell morphology and histology, some minor differences in marker gene expression in qRT-PCR occurred after induction of diverse differentiation pathways. The transcription factors PPARγ, SOX9, and RUNX2 are transcription factors expressed during early adipogenic, chondrogenic, and osteogenic differentiation, their expression levels are known to decrease after completion of differentiation (Franceschi, et al., 2009; MacArthur et al., 2008). These three factors were detected in higher concentrations in treated NIH-3T3 cells than in MEF cells (Fig. 5). Furthermore, the data from qRT-PCR demonstrated the expression of more mature marker genes in MEF cells than in NIH-3T3 cells, e.g., adiponectin and resistin as markers of adipogenesis, aggrecan and collagen II as markers of chondrogenesis and BALP, and sclerostin as markers of osteogenesis (Bajpai et al., 2012; Deng and Scherer, 2010; Li et al., 2005b). These results may indicate a temporal difference in the process of differentiation between NIH-3T3 and MEF cells on the basis of qRT-PCR. Subsequent time-resolved studies might confirm this impression. Time differences with respect to the upregulation of marker genes were also observed in processed lipoaspirate cells and MSCs during their differentiation into mesenchymal lineages by Zuk et al. (2002). Muraglia et al. (2007) and Sudo et al. (2007) described a hierarchy of lineage commitment of MSCs and MPC-like cells, which demonstrated differences between MSC and MPC-like populations derived from several tissues. This altered expression of genes and the hierarchical nature of different types of stem cells indicate once again that the differentiation behavior of our three cell lines resembles that of stem cells.
We found that the tested profile of cell-surface markers does not correspond to MSCs (Fig. 3). However, the cell-surface markers do not necessarily predict the potential of the cell populations to differentiate into specific lineages (Sudo et al., 2007). Gene expression analysis seems to be more useful for comparing the pattern of gene expression with regard to the differentiation properties of each cell type (Chang et al., 2006; Panepucci et al., 2004).
The precise biological mechanism involved in the process of cell differentiation is generally not understood in full detail. As a consequence, there have been intense debates about the process and mechanism of cell differentiation and a variety of different opinions and hypotheses on the plasticity of various other cell-populations than the MSCs, e.g., fibroblasts derived from various tissues (Junker et al., 2010; Lennon and Caplan, 2006; Sabatini et al., 2005; Zuk et al., 2002). As previously mentioned, the Flp-In-3T3 cell line was generated from the NIH-3T3 cell line by transfection with the vector pFRT/lacZeo to create a cell line that can be stably transformed using the Flp-In™ system (Invitrogen). Despite their genetic similarity to NIH-3T3 cells, these cells did not differentiate into any of the three mesenchymal lineages. The reason for this difference is not clear, but it is possible that the clonal expansion process, which was essentially performed to generate Flp-In-3T3 cells, had a negative influence on the differentiation capacity of these cells. Higher passage numbers and overgrown cultures might be the reason for this, although other causes directly depending on the genetic modification by a vector system are also possible. It has been described in the literature that clonal expansion of MSCs results in different differentiation capacities of the cell clones (Jeon et al., 2011; Russell et al., 2011). Their discrete differentiation potential and the similarity between Flp-In-3T3 and NIH-3T3 cells offer a new way to investigate the process of cell differentiation.
The properties and functions of p53, a tumor suppressor and cellular transcription factor in cellular immortalization, have been well documented (Blattner et al., 1999; Harvey and Levine, 1991; Levine, 1997; Metz et al., 1995). The involvement of p53 in the process of cell differentiation is still discussed controversially. Some studies suggest that p53 accelerates cell differentiation, whereas others propose that it suppresses differentiation (Armesilla-Diaz et al., 2009; Molchadsky et al., 2008; Qin et al., 2007). The results of our study confirm both statements. Unlike MEF cells, which downregulated the expression of p53 during the adipogenic, and osteogenic differentiation, we could detect higher levels of p53 in NIH-3T3 cells after adipogenic and osteogenic induction. Flp-In-3T3 and NIH-3T3 cells (Fig. 6) expressed p53 at very low levels, so that we could not detect p53 in both cell types using qRT-PCR. After the cells were induced with differentiation medium, we could not determine any changes in the expression of p53 in Flp-In-3T3 cells, which as previously mentioned did also not show any signs of differentiation. Chondrogenic differentiation of MEF cells seemed to have little impact on p53 expression, whereas an increase of p53 expression was observed in chondrogenic NIH-3T3 cells.
Molchadsky et al. noticed that many authors, who described differing results with respect to the regulative properties of p53 during cell differentiation, used different cell types in their studies (Metz et al., 1995; Molchadsky et al., 2008). Armesilla-Diaz et al. used MSCs and proposed negative regulation of p53 in the process of cell differentiation, and Qin et al. claimed that p53 plays a positive role in the regulation of cell differentiation when using hESCs (Armesilla-Diaz, et al. 2009; Qin et al. 2007). Our data suggest that p53 may be involved, at least in a specific concentration, in the process of differentiation. Its function according to cell differentiation may depend on the cell type. This could be one of the reasons for the slow differentiation rate of NIH-3T3 cells compared to MEF cells. Further efforts to characterize the precise cell cycle dynamics in relation to addition of differentiation media, the nature of the factors involved, and acquisition of the chondrogenic, adipogenic, and osteoblastic phenotypes will help to clarify the role of p53 in these mesenchymal lineages systems.
Conclusions
In conclusion, it could be shown that under appropriate culture conditions MEFs and NIH-3T3 cells are able to differentiate effectively into adipogenic, chondrogenic, and osteogenic lineages even in higher passages. Expression of typical marker genes as well as characteristic morphological changes could be observed. Interestingly, Flp-In-3T3 cells are not able to differentiate into any of these lineages using the same differentiation protocol. Although further characterization of Flp-In-3T3, MEF, and NIH-3T3 populations within their multilineage properties is necessary, our results suggest that these three cell lines may be another source of multipotent cells with multilineage potential. Flp-In-3T3 cells can be used as a negative control. Their cultivation and selective differentiation should provide further understanding of the process of cell differentiation into multiple tissues.
Acknowledgments
The work was supported in part by Biofabrication for NIFE (Niedersachsen Vorab). We thank Christina Allmeling for fruitful discussions and Jörn Kuhbier for good suggestions concerning the differentiation protocols.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Armesilla-Diaz A., Elvira G., and Silva A. (2009). p53 regulates the proliferation, differentiation and spontaneous transformation of mesenchymal stem cells. Exp. Cell Res. 315, 3598–3610 [DOI] [PubMed] [Google Scholar]
- Aurich I., Mueller L.P., Aurich H., Luetzkendorf J., Tisljar K., Dollinger M.M., Schormann W., Walldorf J., Hengstler J.G., Fleig W.E., and Christ B. (2007). Functional integration of hepatocytes derived from human mesenchymal stem cells into mouse livers. Gut 56, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X.P., Zheng H.X., Fang R., Wang T.R., Hou X.L., Li Y., Chen X.B., and Tian W.M. (2011). Fabrication of engineered heart tissue grafts from alginate/collagen barium composite microbeads. Biomed. Mater. 6, 045002. [DOI] [PubMed] [Google Scholar]
- Bajpai V.K., Mistriotis P., and Andreadis S.T. (2012). Clonal multipotency and effect of long-term in vitro expansion on differentiation potential of human hair follicle derived mesenchymal stem cells. Stem Cell Res. 8, 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P.J., and Westhoff C.C. (2007). CD34+ fibrocytes: Morphology, histogenesis and function. Curr. Stem Cell Res. Ther. 2, 221–227 [DOI] [PubMed] [Google Scholar]
- Blattner C., Tobiasch E., Litfen M., Rahmsdorf H.J., and Herrlich P. (1999). DNA damage induced p53 Stabilization: No indication for an involvement of p53 phosphorylation. Oncogene 18, 1723–1732 [DOI] [PubMed] [Google Scholar]
- Boeuf S., and Richter W. (2010). Chondrogenesis of mesenchymal stem cells: Role of tissue source and inducing factors. Stem Cell Res.Ther. 1, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.J., Tobiasch E., Litfen M., Rahmsdorf H.J., and Herrlich P. (2006). Characterization of two populations of mesenchymal progenitor cells in umbilical cord blood. Cell Biol. Int. 30, 495–499 [DOI] [PubMed] [Google Scholar]
- Deng Y., and Scherer P.E. (2010). Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann. N.Y. Acad. Sci. 1212, E1–E19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derubeis A.R., and Cancedda R. (2004). Bone marrow stromal cells (BMSCs) in bone engineering: Limitations and recent advances. Ann. Biomed. Eng. 32, 160–165 [DOI] [PubMed] [Google Scholar]
- Franceschi R.T., Ge C., Xiao G., Roca H., and Jiang D. (2009). Transcriptional regulation of osteoblasts. Cells Tissues Organs 189, 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey D.M., and Levine A.J. (1991). p53 Alteration is a common event in the spontaneous immortalization of primary BALB/C murine embryo fibroblasts. Genes Dev. 5, 2375–2385 [DOI] [PubMed] [Google Scholar]
- Haynesworth S.E., Goshima J., Goldberg V.M., and Caplan A.I. (1992). Characterization of cells with osteogenic potential from human marrow. Bone 13, 81–88 [DOI] [PubMed] [Google Scholar]
- Invitrogen. Growth and Maintenance of Flp-in Cell Lines (2010). Available at www.lifetechnologies.com/us/en/home/references/protocols/proteins-expression-isolation-and-analysis/protein-expression-protocol/flp-in-system-for-generating-constitutive-expression-cell-lines.html#prot5
- Janderova L., McNeil M., Murrell A.N., Mynatt R.L., and Smith S.R. (2003). Human mesenchymal stem cells as an in vitro model for human adipogenesis. Obes. Res. 11, 65–74 [DOI] [PubMed] [Google Scholar]
- Jeon M.S., Yi T.G., Lim H.J, Moon S.H., Lee M.H., Kang J.S., Kim C.S., Lee D.H., and Song S.U. (2011). Characterization of mouse clonal mesenchymal stem cell lines established by subfractionation culturing method. World J. Stem Cells 3, 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker J.P., Sommar P., Skog M., Johnson H., and Kratz G. (2010). Adipogenic, chondrogenic and osteogenic differentiation of clonally derived human dermal fibroblasts. Cells Tissues Organs 191, 105–118 [DOI] [PubMed] [Google Scholar]
- Lennon D.P., and Caplan A.I. (2006). Isolation of rat marrow-derived mesenchymal stem cells. Exp. Hematol. 34, 1606–1607 [DOI] [PubMed] [Google Scholar]
- Levine A.J. (1997). P53, the cellular gatekeeper for growth and division. Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- Li W.J., Tuli R., Huang X., Laquerriere P., and Tuan R.S. (2005a). Multilineage differentiation of human mesenchymal stem cells in a three-dimensional nanofibrous scaffold. Biomaterials 26, 5158–5166 [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Y., Kang H., Liu W., Liu P., Zhang J., Harris S.E., and Wu D. (2005b). Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 280, 19883–19887 [DOI] [PubMed] [Google Scholar]
- MacArthur B.D., Please C.P., and Oreffo R.O. (2008). Stochasticity and the molecular mechanisms of induced pluripotency. PLoS One 3, e3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles Lda S., and Nardi N.B. (2003). Murine marrow-derived mesenchymal stem cell: Isolation, in vitro expansion, and characterization. Br. J. Haematol. 123, 702–711 [DOI] [PubMed] [Google Scholar]
- Metz T., Harris A.W., and Adams J.M. (1995). Absence of p53 allows direct immortalization of hematopoietic cells by the Myc and Raf oncogenes. Cell 82, 29–36 [DOI] [PubMed] [Google Scholar]
- Mizuno H., Zuk P.A., Zhu M., Lorenz H.P., Benhaim P., and Hedrick M.H. (2002). Myogenic differentiation by human processed lipoaspirate cells. Plast. Reconstr. Surg. 109, 199–209; discussion 210–211. [DOI] [PubMed] [Google Scholar]
- Molchadsky A., Shats I., Goldfinger N., Pevsner-Fischer M., Olson M., Rinon A., Tzahor E., Lozano G., Zipori D., Sarig R., and Rotter V. (2008). p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One 3, e3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann S., Hutter J., Moser C., Krombach F., and Huss R. (2005). Milieu-adopted in vitro and in vivo differentiation of mesenchymal tissues derived from different adult human CD34-negative progenitor cell clones. Cells Tissues Organs 179, 91–101 [DOI] [PubMed] [Google Scholar]
- Muraglia A., Cancedda R., and Quarto R. (2000). Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J. Cell Sci. 113, 1161–1166 [DOI] [PubMed] [Google Scholar]
- Niklason L.E., and Langer R. (2001). Prospects for organ and tissue replacement. JAMA 285, 573–576 [DOI] [PubMed] [Google Scholar]
- Panepucci R.A., Siufi J.L., Silva W.A., Jr., Proto-Siquiera R., Neder L., Orellana M., Rocha V., Covas D.T., and Zago M.A. (2004). Comparison of gene expression of umbilical cord vein and bone marrow-derived mesenchymal stem cells. Stem Cells 22, 1263–1278 [DOI] [PubMed] [Google Scholar]
- Qin H., Yu T., Qing T., Liu Y., Zhao Y., Cai J., Li J., Song Z., Qu X., Zhou P., Wu J., Ding M., and Deng H. (2007). Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J. Biol. Chem. 282, 5842–5852 [DOI] [PubMed] [Google Scholar]
- Rivlin N., Brosh R., Oren M., and Rotter V. (2011). Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2, 466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell K.C., Lacey M.R., Gilliam J.K., Tucker H.A., Phinney D.G., and O'Connor K.C. (2011). Clonal analysis of the proliferation potential of human bone marrow mesenchymal stem cells as a function of potency. Biotechnol. Bioeng. 108, 2716–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini F., Petecchia L., Tavian M., Jodon de Villeroché V., Rossi G.A., and Brouty-Boyé D. (2005). Human bronchial fibroblasts exhibit a mesenchymal stem cell phenotype and multilineage differentiating potentialities. Lab. Invest. 85, 962–971 [DOI] [PubMed] [Google Scholar]
- Schumann P., Tavassol F., Lindhorst D., Stuehmer C., Bormann K.H., Kampmann A., Mülhaupt R., Laschke M.W., Menger M.D., Gellrich N.C., and Rücker M. (2009). Consequences of seeded cell type on vascularization of tissue engineering constructs in vivo. Microvasc. Res. 78, 180–190 [DOI] [PubMed] [Google Scholar]
- Sudo K., Kanno M., Miharada K., Ogawa S., Hiroyama T., Saijo K., and Nakamura Y. (2007). Mesenchymal progenitors able to differentiate into osteogenic, chondrogenic, and/or adipogenic cells in vitro are present in most primary fibroblast-like cell populations. Stem Cells 25, 1610–1617 [DOI] [PubMed] [Google Scholar]
- Todaro G.J., and Green H. (1963). I. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaananen H.K. (2005). Mesenchymal stem cells. Ann. Med. 37, 469–479 [DOI] [PubMed] [Google Scholar]
- Vacanti J.P., and Langer R. (1999). Tissue engineering: The design and fabrication of living replacement devices for surgical reconstruction and transplantation. Lancet 354(Suppl 1), SI32–SI34 [DOI] [PubMed] [Google Scholar]
- Vacanti V., Kong E., Suzuki G., Sato K., Canty J.M., and Lee T. (2005). Phenotypic changes of adult porcine mesenchymal stem cells induced by prolonged passaging in culture. J. Cell. Physiol. 205, 194–201 [DOI] [PubMed] [Google Scholar]
- Winkler D.G., Sutherland M.K., Geoghegan J.C., Yu C., Hayes T., Skonier J.E., Shpektor D., Jonas M., Kovacevich B.R., Staehling-Hampton K., Appleby M., Brunkow M.E., and Latham J.A. (2003). Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 22, 6267–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.U., Barthel T.S., Nishimura K., Solchaga L., Caplan A.I., Goldberg V.M., and Johnstone B. (1998). The chondrogenic potential of human bone-marrow-derived mesenchymal progenitor cells. J. Bone Joint Surg. Am. 80, 1745–1757 [DOI] [PubMed] [Google Scholar]
- Zuk P.A., Zhu M., Mizuno H., Huang J., Futrell J.W., Katz A.J., Benhaim P., Lorenz H.P., and Hedrick M.H. (2001). Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 7, 211–228 [DOI] [PubMed] [Google Scholar]
- Zuk P.A., Zhu M., Ashjian P., De Ugarte D.A., Huang J.I., Mizuno H., Alfonso Z.C., Fraser J.K., Benhaim P., and Hedrick M.H. (2002). Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell 13, 4279–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]