Abstract
Designing an ideal biomaterial supportive of multicellular tissue repair is challenging, especially with a poor understanding of the synergy between constituent proteins and growth factors. A brute-force approach, based on screening all possible combinations of proteins and growth factors, is inadequate due to the prohibitively large experimental space coupled with current low-throughput screening techniques. A high-throughput screening platform based on rational and combinatorial strategies for design and testing of proteins and growth factors can significantly impact the discovery of novel tissue-specific biomaterials. Here, we report the development of a flexible high-throughput screening platform, Rapid Assessment of Migration and Proliferation (RAMP), to rapidly investigate cell viability, proliferation, and migration in response to highly miniaturized three-dimensional biomaterial cultures (4–20 μL) with sparingly low cell densities (63–1000 cells per μL for cell arrays; 1 μL of 1000–10,000 cells per μL for migration arrays). The predictions made by RAMP on the efficacy and potency of the biomaterials are in agreement with the predictions made by conventional assays but at a throughput that is at least 100–1000-fold higher. The RAMP assay is therefore a novel approach for the rapid discovery of tissue-specific biomaterials for tissue engineering and regenerative medicine.
Introduction
In 2000 alone, ∼50 million Americans sustained injuries, resulting in an estimated $80 billion in direct healthcare costs.1 Autologous tissue grafts represent a common treatment option for soft tissue trauma and bone defects, resulting in donor site morbidity. For the severely injured, incomplete regeneration may limit patients' ability to rejoin the workforce as well as impact their long-term quality of life. Toward this end, tissue engineering and regenerative medicine aim to develop a replacement biomaterial to support tissue regeneration, and thereby improve functional outcomes. Development of a replacement biomaterial, however, is challenging as the biomaterial needs to be presented with appropriate biomechanical, biochemical, and topographical cues, usually via a scaffold. Ideally, this biomaterial would support the repair of a multicellular native tissue without eliciting an immune response. The experimental discovery space is prohibitively large given the high number of possible constituent proteins and growth factors. A high-throughput screening platform that allows the testing of a large number of candidate biomaterials in a time- and cost-effective manner can significantly impact the development of an ideal tissue-specific biomaterial. High-throughput technologies have enabled significant advances in other fields, such as drug discovery, genetics, and toxicity screening.2–5 High-throughput cell-culture arrays are of particular interest in tissue engineering due to their capacity for rapid culturing, imaging, and analysis of hundreds of samples while reducing experimental costs and increasing reproducibility.
Conventional cell-culture arrays have typically focused on assaying cell response to libraries of proteins and synthetic polymers4 in two dimensions by quantifying metrics, such as viability, proliferation, and differentiation.3,6 While two-dimensional (2D) arrays have some utility in cytotoxicity applications, three-dimensional (3D) arrays will be more advantageous for accurately assessing the cell in a physiologically relevant manner, thus improving their applicability in vivo. Cell response to material composites is also affected by the presentation scheme. Cell behavior (proliferation, gene expression, and spreading) can be different in 2D compared with 3D cultures.7 Although 3D cell-culture arrays have been previously used to evaluate cell viability or differentiation within biomaterials, they do not accommodate the analysis of higher order cellular characteristics (e.g., morphology, migration, etc.) to varied matrix compositions or soluble factors.8–10 This limits their utility for rapid screening of cellular responses to compositionally varied biomaterials, and emphasizes the need to extend such high-throughput methods to functional cell assessments for applications in tissue engineering.
Tissue engineering has broadly been applied to repair injuries and treat diseases that affect a variety of tissues, such as skin, bone, cartilage, liver, and nerves. For example, in the peripheral nervous system (PNS), guidance channels, tubes that bridge the nerve injury generated from a wide array of biomaterials, are used to support the repair of nerve injuries.11 Effective guidance channels must support many cell types in the peripheral nerve, which can contain sensory, motor, or a mix of both neuron populations as well as other resident non-neuronal cells (e.g., glia, endothelial cells, and fibroblasts). To date, engineered guidance channels fail to support repair of large-gap injuries, and therefore autografts remain the gold standard for peripheral nerve injury. During the development of novel guidance channels, nerve regrowth is assessed as a primary metric of repair, but migration and repopulation of other resident cells is also necessary for repair, and is often overlooked. The singular focus on neural regrowth lends itself to only part of the story. If the biomaterial is not supportive of growth and migration for all resident cells vital to the functional regeneration of the injured tissue, then the implanted biomaterial may not be successful.
Collagen type I, the most abundant protein in peripheral nerves, is supportive of neuronal growth; however, Schwann cells, the glial support cells in the PNS, are not supported within this biomaterial.12,13 Since Schwann cell repopulation is necessary to support axonal growth and ultimately to remyelinate the injured nerve, a biomaterial required for peripheral nerve repair would need to support both neurons and Schwann cells.11,14 It is unlikely that a single factor can adequately support all resident PNS cells; however, it is likely that a synergistic combination of matrix proteins and soluble growth factors has the potential to facilitate tissue regeneration. The rate-limiting step is the ability to rapidly screen relevant cell responses in three dimensions to a variety of candidate matrix proteins and soluble growth factors, thereby identifying an optimum tissue-specific biomaterial. This approach, ideally, would evaluate both, basic functions (viability, proliferation, etc.) as well as more complex cell behavior (spreading, migration, etc.), in a rapid manner.
To achieve this goal, we have designed and developed Rapid Assessment of Migration and Proliferation (RAMP), a high-throughput cell-array-based platform to test naturally occurring matrix proteins within a 3D hydrogel. The RAMP assay employs a spatially addressable, miniaturized, 3D cell-culture array to determine functional outputs, such as number, spreading, and migration, rapidly screened with a high-resolution flatbed scanner. These cell arrays can be evaluated at a single time point to evaluate differences in spreading or over time to examine changes in cell number (e.g., proliferation and toxicity screening) and migration. We envision that this assay will identify biomaterials that are growth supportive (GS, reported as “positive hits”), but could be expanded to discovery of biomaterials that are not supportive of cellular processes or are growth inhibitive (GI, reported as “negative hits”). More subtle differences can subsequently be identified using higher resolution imaging modalities.
Here, we demonstrate the application of RAMP assay for identifying GS and GI biomaterials, while biomaterials that provide intermediate cell support are not pursued further as they are less likely to find much utility. While the need to investigate GS biomaterials is obvious, it is equally advantageous to identify GI biomaterials. During development, both GS and GI cues work in concert to direct cells to form organized tissues. A highly tuned biomaterial that incorporates a combination of GS and GI biomaterials to control migration and proliferation of the many beneficial and potentially detrimental cell populations would provide a “smart” scaffold to support tissue-specific repair.
The challenge of increasingly complex biomaterial optimization needs to be met with an effective screening assay. The studies described in this article exemplify the use of the RAMP assay for the rapid design and discovery of optimal biomaterials. We provide a proof of concept using Schwann cells and two collagen-based hydrogels (collagen type I, GI; a composite of collagen type I and Matrigel™ [4:1], GS). We have evaluated the sensitivity of RAMP assay to rapidly assess Schwann cell number, spreading, and migration within these two similar yet functionally diverse biomaterials, and subsequently confirmed the results of RAMP with conventional macroscale assays. While other assays exist to evaluate/screen for cell proliferation and viability on or within a 3D biomaterial, to our knowledge, this is the first assay to evaluate cell migration within a 3D biomaterial in a rapid format.
Materials and Methods
Isolation of primary Schwann cells
Primary Schwann cells were isolated from the sciatic nerve of postnatal day-2 Sprague Dawley rat pups (Taconic Farms, Inc.) as previously described.15–17 Briefly, the sciatic nerve was removed and plated in 1-mm lengths in six-well culture dishes containing base medium (Dulbecco's modified Eagle medium [DMEM; Mediatech, Inc.] supplemented with 10% fetal bovine serum [FBS; Hyclone], 2 mM L-glutamine [Hyclone], and 50 U/mL penicillin/streptomycin [Mediatech, Inc.]) under standard culture conditions (37°C, 5% CO2). Isolated cell cultures were treated with antimitotic agents (10−5 M cytosine arabinoside; Sigma Aldrich) for 72 h to remove the highly mitotic fibroblasts. Contaminant fibroblasts were targeted with complement-mediated cell lysis.18 Purified Schwann cells were expanded in growth medium (base medium supplemented with 6.6 mM forskolin [Sigma Chemical] and 10 μg·mL−1 bovine pituitary extract [BD Biosciences]). Purity was assessed by S100 immunostaining, a Schwann-cell-specific marker, with cells used from passages 3–10 at >97% purity.
Array assembly and sterilization
Sylgard 184 poly(dimethylsiloxane) (PDMS; Dow Corning) sheets were generated by pouring the base (15 g) and curing agent at 10:1 wt/wt ratio into a 128×86 mm2 rectangular dish (Nunc) and allowing the PDMS to cure at room temperature for 48 h prior to use. About 75×25 mm2 rectangles were excised from the PDMS sheets. A dermal punch was used to create wells from the excised PDMS to create gaskets with inner diameters (IDs) of 2, 3, or 4 mm. The PDMS gaskets were scrubbed clean with detergent (Alconox), rinsed with diH2O for 5 min in a sonicator bath, dried with filtered air, and UV sterilized. Cleaned, acid-etched coverglass (50×24 mm2) was coated with 0.5% w/v poly-2-hydroxyethyl methacrylate (pHEMA; Sigma Aldrich) to prevent cell attachment to the glass.19 The gasket was placed on the pHEMA-coated glass and a multiwell assembly was formed that was UV sterilized prior to use.
Cell population arrays
To determine assay sensitivity to cell concentration, spot size, and biomaterial composition, serial dilutions of purified Schwann cells ranging from 63 to 1000 cells per μL were suspended in bovine collagen type I (MP Biomedical) biomaterials, with or without growth-factor-reduced Matrigel (BD Biosciences), as adapted from Dewitt et al.13 These materials were chosen because Schwann cell migration and spreading are not supported within 3D collagen hydrogels, while the inclusion of Matrigel that is comprised of collagen IV, laminin, and other soluble and insoluble factors serves to support these cell functions.13,20
Phenol red free (PRF) 1× DMEM, PRF growth medium, FBS, 0.1 N NaOH, and stock 4 mg·mL−1 bovine collagen were prepared in a 3:6:4:7:20 ratio, respectively, resulting in a final concentration of 2 mg·mL−1 collagen for each concentration of Schwann cells. Cell-laden collagen and collagen–Matrigel hydrogels were pipetted into the arrays, in replicates of eight for each well size. To avoid artifacts from imaging, the biomaterial height was kept constant (∼1 mm) irrespective of the well ID, yielding hydrogel volumes of 4, 9, and 16 μL for wells of 2, 3, or 4 mm, respectively. Schwann-cell-laden constructs were incubated for 10 min at 37°C and then checked for even cell dispersion throughout the biomaterials and covered in PRF Schwann cell growth medium and maintained in culture for 1–7 days. Acellular collagen and collagen–Matrigel biomaterials served as controls.
Migration arrays
Step-wise injection of cells during polymerization helped retain Schwann cells locally within the center of the hydrogel. First, acellular collagen was pipetted into an array with 4-mm-ID wells and allowed to partially polymerize for 2 min at room temperature. A second layer of collagen was pipetted on top of the partially polymerized acellular collagen layer and incubated for an additional 2 min. Schwann cells (0–10,000 cells per μL) were suspended in collagen and 1 μL of the suspension was injected on top of partially polymerized, double-layered hydrogel, incubated at 37°C for 10 min to complete polymerization, and then covered in PRF growth medium and cultured for 1–7 days in vitro. Step-wise injection of the cells during polymerization helped retain the Schwann cells locally within the center of the hydrogel. More traditional macromigration studies were performed concurrently in a 48-well-plate assay for comparison.
Fluorescent staining and imaging
Cell proliferation and migration array samples were fixed with 4% w/v paraformaldehyde and 4% w/v sucrose (Sigma-Aldrich) in a buffer solution containing 60 mM piperazine-1,4-bis(2-ethanesulfonic acid), 25 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, 10 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetra acetic acid, 2 mM MgSO4, and sufficient KOH to achieve pH 7.0 in diH2O for 1 h. Samples were rinsed with 1× phosphate-buffered saline (PBS; Cambrex) for three times and permeabilized with 0.01% v/v Triton-X (Sigma-Aldrich) in PBS. Due to the high purity of the isolated Schwann cell cultures (>97%), cell spreading was visualized using phalloidin, rather than a Schwann-cell-specific stain. The samples were incubated at room temperature in phalloidin-conjugated tetramethylrhodamine B isothiocyanate (TRITC-phalloidin; Sigma-Aldrich) diluted 1:500 and 4′,6-diamidino-2-phenylindol (DAPI; Invitrogen) diluted 1:1000 in 5% w/v bovine serum albumin (Sigma-Aldrich) for 1 h. Samples were rinsed in PBS, and stored overnight at 15°C prior to imaging.
The arrays (cell population and migration) were imaged on a Typhoon Trio flatbed scanner (GE Healthcare Biosciences; 10 μm resolution) in the fluorescence mode (532 nm laser, 580 BP 546 emission filter) to rapidly assess the total cell signal along the z-axis on a xy-plane image. In parallel, the arrays were also imaged using an Olympus IX81 inverted microscope (Olympus) with a 4× dry objective and representative images were acquired from the center of each culture.
Data analysis and statistics
Cell array scans were assessed as 8-bit images using the NIH ImageJ toolkit (National Institute of Health) to quantify average intensity per culture condition as a measure of cell number or the signal area as a measure of migration. The average signal intensity per test condition (m=8 replicates per array) in the proliferation assay was measured and presented as a raw value for each data set (n=3). Average signal intensity was also normalized to the maximum average signal intensity value in any single array to account for variability across replicates. Migration was quantified using the ImageJ freehand selection tool by tracing the perimeter of the cell-laden area, and then quantifying the average intensity and area of each culture condition (n=3 separate trials, m=12 replicates). Migration data at any time point was normalized to similar values prior to the onset of significant migration (8 h for the 4-mm-ID arrays and 24 h for the 48-well-plate arrays). In parallel, individual wells within the cell arrays were imaged using a fluorescence microscope to quantify cell number, spreading, and migration. Images were analyzed using ImageJ to determine the total cell number (DAPI signal) and cell spreading (phalloidin signal). Cell spreading was normalized to cell number to determine the average cell spreading area for each sample.
Data obtained from flatbed scanning and fluorescent microscopy were rank-ordered (1 corresponding to inferior performance). The average intensity measured by the scanner was compared to results obtained with conventional macroscale assays, microscopy and image analysis. Two-way multiple-replicate ANOVA was performed using Excel (Microsoft) to determine statistical significance of spot size, cell density, composition, culture time, and migration (p-values <0.05). Student's t-test was performed to determine statistical significance between individual conditions (p-value <0.05). Error bars are represented as standard deviation.
Results
Assay sensitivity to cell density in collagen-based biomaterials
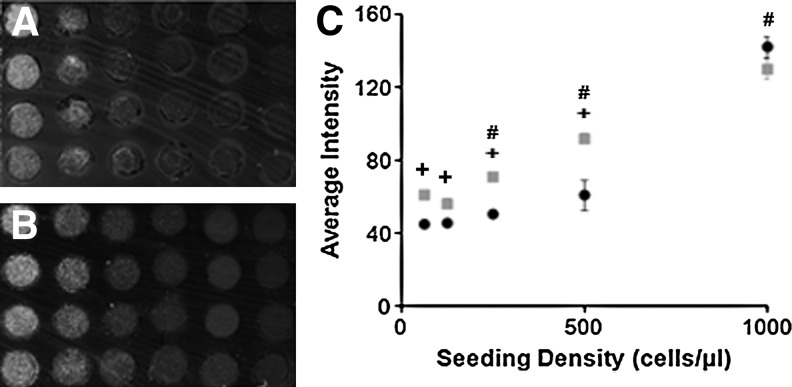
RAMP sensitivity to cell number was evaluated using 3D hydrogels over a range of cell densities (63–1000 cells per μL). For both collagen-based biomaterials, collagen alone and collagen–Matrigel composite, the scanned array image exhibits an increase in signal with increasing seeding density that was well dispersed throughout the biomaterial (Fig. 1A, B). Quantitative measurements of signal intensity from the scanned array image confirm a significant increase in signal with increasing seeding density within the range of 250–1000 cells per μL (Fig. 1C). Thus, RAMP can rapidly detect gross differences in cell number within a 3D hydrogel.
FIG. 1.
Rapid Assessment of Migration and Proliferation (RAMP) arrays imaged by the flatbed fluorescent scanner are sensitive to differences in seeding density and matrix composition. Representative scans for phalloidin-stained Schwann cells cultured for 24 h in collagen (A) and collagen–Matrigel™ (B) were analyzed. (C) The average intensity increases significantly with increasing seeding density within both biomaterials for 250–1000 cells per μL densities (#p<0.05, n=3, m=8). Intensity differences between the two model biomaterials can be detected at individual seeding densities from 63 to 500 cells per μL with collagen (black circles) exhibiting a significantly lower average intensity relative to the collagen–Matrigel biomaterial (gray squares) (+p<0.05, n=3, m=8). Error bars represent standard deviation.
Rapid assessment of morphological differences in collagen-based 3D biomaterials
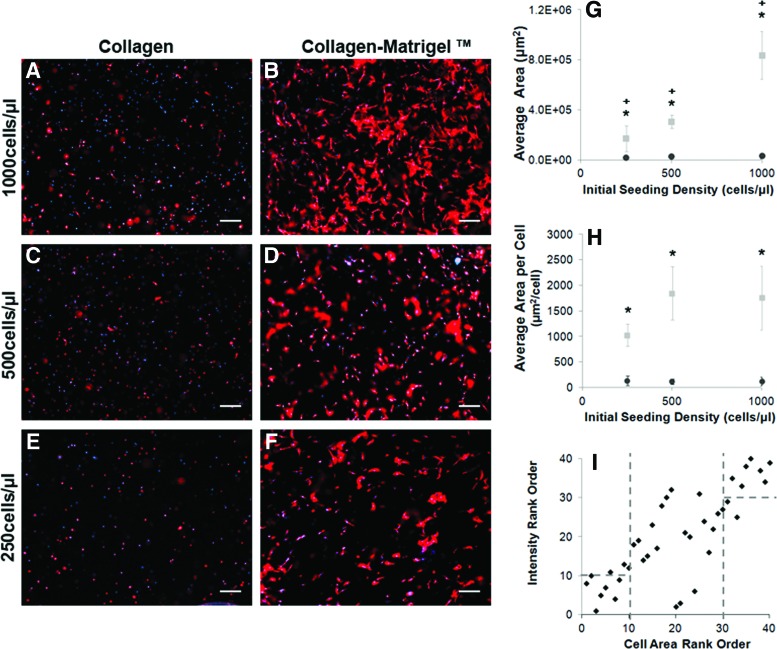
The established sensitivity of the RAMP assay to cell density (Fig. 1C) can be expanded to detect differences in cell morphology due to biomaterial composition. Significant differences between the model collagen biomaterials, collagen and collagen–Matrigel composite, were detected for individual densities within the range of 63–500 cells per μL. The minimum cell density required for cell detection in 3D using the scanner was evaluated by first scanning the array and identifying spots that did not have a significantly greater signal than acellular controls, and then confirmed using microscopy to visualize and quantify the actual cell densities based on DAPI signal. The minimum cell density required for detection by the scanner (Fig. 1) was 451 cells per μL in collagen and 267 cells per μL in collagen–Matrigel, as based on actin signal, suggesting differences in cell spreading within the biomaterials. Thus, the RAMP assay is sensitive enough to detect differences in cell response between different collagen-based biomaterials, and the threshold values are cell-type dependent. Conventional fluorescence microscopy was used to verify results (sensitivity to number and composition) from RAMP and demonstrated even cell dispersion across randomly selected focal planes (Fig. 2A–G). As expected, a higher overall actin signal based on cell area was measured in response to Matrigel and increased seeding density (Fig. 2A–G). Spreading was limited in the collagen-only biomaterials and no significant increase in Schwann cell area with increasing seeding density was observed even after 3 or 7 days in culture. In both biomaterials, Schwann cells were well dispersed and did not form clusters within the wells.
FIG. 2.
Traditional microscopy confirms RAMP analysis of Schwann cell response to model biomaterials. Arrays were imaged using microscopy and changes in cell morphology were analyzed. Schwann cells were stained with phalloidin to visualize the actin cytoskeleton (red) and the nuclei were labeled with DAPI (blue). Cells were seeded within the collagen (A, C, E) or collagen–Matrigel (B, D, F) biomaterials at densities ranging from 250 to 1000 cells per μL and were cultured for 3 days. Schwann cells remained largely rounded in the collagen-only construct, while the collagen–Matrigel biomaterial supported a spread morphology at all cell densities examined. (G) Using ImageJ, cell area was significantly (*p<0.05, n=3, m=12) higher for Schwann cells cultured within collagen–Matrigel (gray squares) over those cultured within collagen (black circles). As expected, there was a significant increase in the overall cell area with increasing seeding density (+p<0.05, n=3, m=12). (H) Normalizing the cell area to cell number provides the average spreading area per cell, which was shown to be significantly (*p<0.05, n=3, m=12) higher for cells seeded within collagen–Matrigel (gray squares) than Schwann cells cultured in collagen (black circles). There was not a significant difference in spreading area per cell between seeding densities when normalized to cell number. (I) High-throughput, low-resolution scanner intensity measurements from RAMP correlate with low-throughput, higher resolution microscopy measurements for samples with varying seeding density, matrix, and duration with few outliers. “Hits” found in the scanned image show comparable correlation to the samples with the highest Schwann cell spreading area. Error bars represent one standard deviation. Scale bar=200 μm. 4×magnification. DAPI, 4′,6-diamidino-2-phenylindol. Color images available online at www.liebertpub.com/tec
Using both RAMP and conventional microscopy, differences in signal intensity were detected between the two hydrogels at the same seeding density (within a given range), suggesting that differences in cell morphology can be detected at a given seeding density. The cell area (actin signal) was normalized to the cell number (DAPI signal) to rapidly estimate the average individual cell area. Cell spreading is limited in collagen-only biomaterials (130±78 μm2·cell−1) compared with Schwann cells cultured within collagen–Matrigel (1571±575 μm2·cell−1; Fig. 2A–F, H). Schwann cells exhibit a round morphology in collagen hydrogels, while a more spread morphology is evident with the addition of Matrigel. Differences in individual cell morphology within the collagen-based biomaterials were first detected by RAMP and confirmed using microscopy.
The RAMP assay is designed to rapidly screen cell response in 3D to a biomaterial library. Its utility, in practice, is dependent on good quantitative agreement with macroscale hydrogels imaged using traditional microscopy. The average spot intensity from RAMP arrays for different biomaterials (e.g., seeding density and matrix composition; Fig. 1) as well as experiment duration (day 3 and 7 data) were rank-ordered with the highest intensity values corresponding to the highest rank order. The average cell area as measured by microscopy was similarly rank-ordered by cell area. The correlation between RAMP and traditional methods (microscopy) was linear with a few outliers (Fig. 2I). Most importantly, there was a good correlation between the rank for the top three hits (identified as GS biomaterials) using the scanner and the microscope. The lower ranked biomaterials (indicated as GI biomaterials) also showed a correlation between the two assays, indicating that RAMP is adequately sensitive in quickly identifying GI biomaterials as well.
Average intensity and cell distribution is independent of well diameter
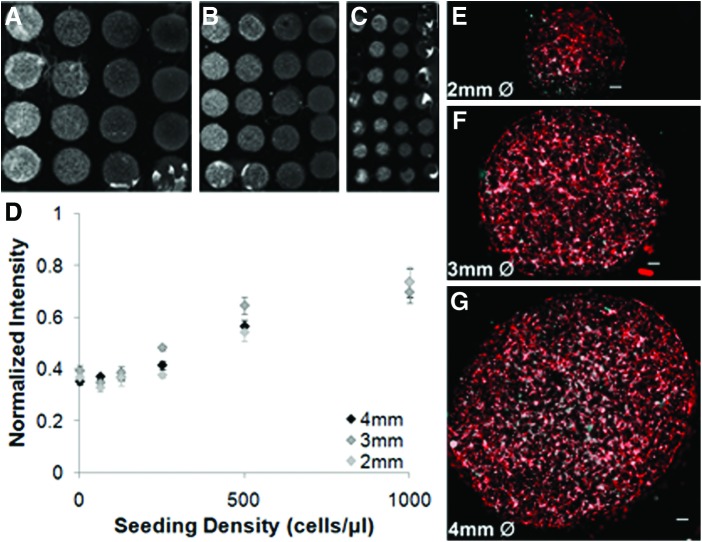
Assay throughput was explored by conducting biomaterial screening experiments over a range of well diameters (2-, 3-, or 4-mm ID). A serial dilution of Schwann cells from 63 to 1000 cells per μL was seeded into arrays of varying diameter. The thickness of each of the biomaterial regardless of well diameter was kept constant to allow a direct comparison between well sizes. Single scans of 2-, 3-, and 4-mm-ID wells exhibited consistent signal within seeding densities ranging from 1000 cells per μL (left) to 125 cells per μL (right) (Fig. 3A–C). Regardless of well diameter, the average intensity increases with initial seeding density over the range of 250–1000 cells per μL (Fig. 3D). The scanned images provide qualitative evidence that the cells were well distributed within each biomaterial. Regardless of the imaging modality, the normalized intensity at a given seeding density was independent of well diameter (Fig. 3D–G).
FIG. 3.
The average spot intensity for a given cell density is independent of spot diameter (2–4 mm). Schwann-cell-laden collagen was printed in RAMP arrays of varying spot sizes, cultured for 24 h, and stained with phalloidin. Representative scans at (A) 4, (B) 3, and (C) 2 mm are depicted with increasing seeding density (decreasing from left to right). (D) No significant differences in the average intensity were detected due to spot size at a given seeding density (p>0.05). As expected, significant increases in average spot intensity with increasing seeding density were detected in the range of 125–1000 cells per μL (p<0.05, n=3, m=12). (E–G) Microscopy verified uniform dispersal of cells within the biomaterials in all arrays regardless of spot size. Error bars represent standard deviation. Color images available online at www.liebertpub.com/tec
Schwann cell migration is sensitive to presence of Matrigel
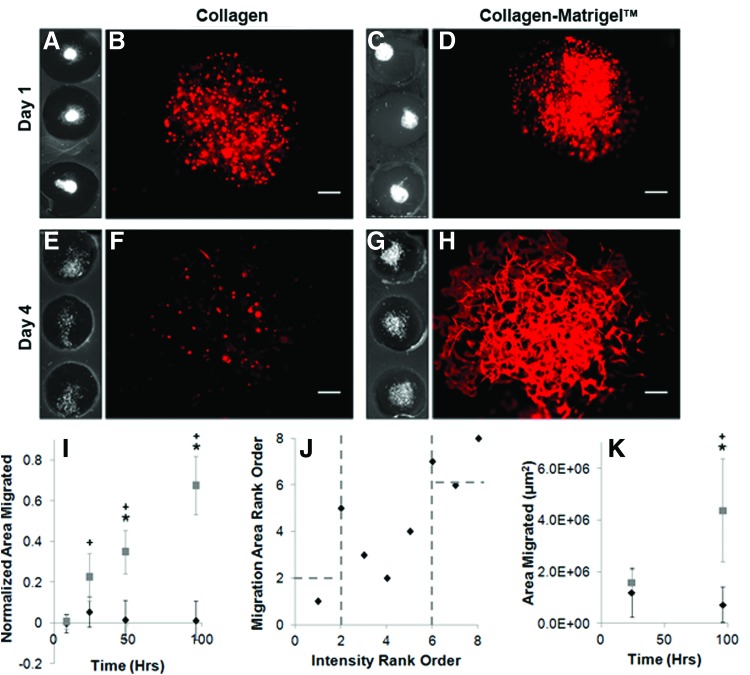
Cell attachment, proliferation, and viability over time are straightforward and necessary metrics to quantify when determining whether a given biomaterial is appropriate. Once basic metrics of cell attachment are satisfied, increasingly complex cell response metrics, such as migration, need to be assessed to identify appropriate biomaterials for tissue repair. In this work, we extended the cell population assay to rapidly assess a more complex cell response, for example, cell migration. Typically, migration assays are performed in a large-scale, multiwell format, which are both effort and resource intensive. Rapid-screening technologies currently do not incorporate assessment of migration despite its importance in tissue repair. Simple modifications to cell placement within the biomaterial allow for the rapid screening of cell migration. To demonstrate this, Schwann cells were injected in the center of an acellular hydrogel and these arrays were scanned after 8 h to visualize cell placement (Fig. 4A, C). By 96 h, migration was quantified and was observed extending beyond the initial injection site within the collagen–Matrigel (Fig. 4G), but migration was not apparent within the collagen biomaterials (Fig. 4E). In collagen, there was an obvious reduction in signal, indicating a loss in cell number. These results were confirmed by microscopy at higher resolution (Fig. 4B, D, F, H). Using RAMP, migration was rapidly assessed over time (8, 24, 48, and 96 h). Schwann cells did not migrate to any significant extent within collagen, but did migrate into collagen–Matrigel after 24 h and continued to migrate at 48 and 96 h, in agreement with our qualitative assessment (Fig. 4I, p<0.05).
FIG. 4.
Schwann cell migration can be rapidly screened over time using a flatbed scanner. Schwann cells were printed into acellular biomaterials and incubated over 4 days in culture. Samples were fixed at 8, 24, 48, and 96 h postprinting. Scanned slides and microscopy images show Schwann cells largely remain localized within an acellular collagen (A, B) and collagen–Matrigel (C, D) biomaterials (8 h). In contrast, there is no migration within the collagen-only biomaterial (E, F) and there is significant migration within collagen–Matrigel (G, H) after 4 days. Higher resolution images (F, H) confirmed these observations. (I) Schwann cell migration was quantified based on the scanned data and plotted against time. The migration area was normalized to the 8-h time points for each matrix type. Area significantly increases with increasing time for the collagen–Matrigel (gray square) biomaterial, but does not change significantly for the collagen (black diamond). A significant difference between the biomaterials at 48 and 96 h was detected, +p<0.05 for matrix biomaterial, *p<0.05 for collagen–Matrigel time sensitivity compared to 8-h sample (n=3, m=12). (J) High-throughput, low-resolution scanner intensity measurements correlate with low-throughput, high-resolution microscopy measurements. Planar scan “hits” that support Schwann cell migration, dependent on matrix and duration, result in an increased Schwann cell migration area based on traditional microscopy, indicating a linear correlation between the two imaging modalities. (K) Macroscale migration assays were used to confirm differences between the biomaterials after 96 h. Microscopy data of the cell area confirmed a significant increase in cell migration after 96 h in collagen–Matrigel compared with collagen (*p<0.05, n=3, m=3) and compared with the 24-h samples (+p<0.05). Error bars represent standard error. Scale bar=200 μm. Color images available online at www.liebertpub.com/tec
Similar to the cell culture arrays, there was a positive correlation in the migrated area (microscopy) and average intensity (scanner) (Fig. 4J). Over 96 h, a similar overall trend was confirmed in a macroscale, 48-well-plate assay using traditional microscopy and quantified with image analysis (Fig. 4K, p<0.05). Thus, RAMP is able to quickly screen higher order metrics (migration) in a resource- and time-efficient manner making it attractive for evaluating a larger experimental space.
Discussion
We report on the development of an assay to extend high-throughput techniques to screen for basic metrics, such as cell spreading, cell viability, and cell proliferation, to include cell migration in a rapid format. The data presented is a proof-of-concept evaluating the sensitivity of cells seeded within a mixture of two common hydrogels: type 1 collagen and Matrigel. First, RAMP sensitivity to cell density was validated at a single time point over a range of seeding densities; however, this assay could be seeded at a single density and increases in cell number could be screened over time to evaluate toxicity to (decrease) or proliferation within (increase) a biomaterial (Fig. 1). To investigate this concept, assay sensitivity to biomaterial composition was demonstrated using two model biomaterials. Collagen is a commonly used biomaterial for neural tissue engineering, supportive of neurite outgrowth but less supportive of Schwann cell spreading and migration. Unlike collagen, the collagen–Matrigel composite biomaterial incorporates many soluble and insoluble factors, including, but not limited to, laminin and type IV collagen—key components of the natural Schwann-cell-produced basal lamina—which support Schwann cell proliferation and migration.13 Differences in cell response to acellular collagen and 4:1 collagen–Matrigel were attributed to biochemical differences and not biomechanical variability as these materials have similar elastic moduli of 6.4 and 6.6 kPa, respectively.13 Utilizing these model biomaterials, we demonstrate RAMP to be a flexible platform that can be used to assess multiple cell metrics, including cell number (Fig. 1), gross cell morphology (Fig. 2), and cell migration (Fig. 4), in response to different biomaterials.
No differences were observed in RAMP assays ranging from 2- to 4-mm ID, indicating that RAMP can be scaled below the 2-mm (4 μL) threshold to increase throughput without a reduction in accuracy, as demonstrated in Figure 3. In this work, the number of sample replicates for each condition (m) is 8–12 within each of the three trial runs; thus, there were 288 (cell population arrays) or 96 (cell migration arrays) individual samples in each trial array run. RAMP is only limited by the size of the PDMS gasket, pHEMA-coated glass, and well diameter used. Here, only two biomaterials were assayed so a smaller glass substrate was used to accommodate the samples, but a larger setup could be easily used to accommodate more samples. Alternatively, reducing the sample size to a diameter of 2 mm or less would increase the throughput of RAMP when larger biomaterial libraries are screened. We do, however, expect that there is a lower limit where a decreasing well diameter will eventually decrease the signal area below detection limits of the scanner detector. Three factors—printing parameters (accuracy and reproducibility), cell type, and dye choice—dictate the lower limits of the assay. The printing accuracy within the well is an important factor to obtain reproducible results. For example, in the 2-mm wells (Fig. 3), off-centered printing results in uneven gelation along the well sides. Poor printing results in nonuniform distribution of cells/biomaterials within the well and these samples were discarded from further analysis. This hurdle can be overcome through the addition of an automated seeding device, or by simply using a thinner PDMS gasket with manual printing that makes it easier to maintain an even deposition within the well. A second factor is cell type and the inherent size and morphology of the cell type used in the screen. For example, in this work, Schwann cells have the capacity to spread significantly within a GS biomaterial, which allows for scanner sensitivity to a more graded cell response to different materials and a larger disparity between GS and GI biomaterials. Primary Schwann cells can be printed with a high purity, so the use of phalloidin to label actin within all cells is an efficient stain to visualize and quantify cell spreading. In contrast, neurons isolated from tissue have proliferating glial cells, and the use of a neuro-specific marker would be more appropriate. RAMP is a comparative assay that would require some preliminary experiments to determine the dynamic range for each cell type and dye combination. It is not necessary to optimize cell response to each biomaterial, rather the ability to compare all novel biomaterials with a control biomaterial (positive and negative) on-chip is important for the rapid identification of relative changes of GS or GI biomaterial candidates that can be verified using traditional microscopy.
High-throughput assessment of cell metric with the flatbed scanner correlated linearly with traditional, low-throughput inverted microscopy with few outliers, as demonstrated through rank-order analysis (Figs. 2I and 4J). Further, data acquired by conventional microscopy were comparable to those collected from confocal microscopy images despite the 3D biomaterials being 1-mm thick. Presumably due to even seeding of cells within the transparent biomaterial and the small z-dimension of the constructs, confocal microscopy did not provide different outcomes relative to the conventional microscopy. An outlier of particular interest in the basic cell metric arrays was the collagen samples seeded at 1000 cells per μL indicated as a GS hit via the scanner, but as a GI hit using microscopy. This disparity may be due to the high concentration of cells that diffract the light throughout the biomaterial, leading to a false positive. It is therefore proposed that an optimal seeding density would be <1000 cells per μL for this cell type to achieve consistent scanner data and to allow for growth of the cell population without reaching saturation. By first pinpointing an optimal seeding density for a given cell type, RAMP can assess basic cell characteristics thus providing a promising technique to quickly screen a library of biomaterials. Biomaterials identified as GS or GI can be further screened based on higher order cellular metrics/functions, such as migration, to efficiently predict cell response within a novel composite biomaterial. A second screen serves to confirm or reject false positives and false negatives based upon the candidates (GS or GI) identified in the first screen.
RAMP is a fluorescence-based assay and therefore has an inherent limitation in that the 3D biomaterials of interest must be optically transparent within the emission spectrum of the dyes; however, simple modifications to RAMP would overcome this limitation. Nontransparent biomaterials may be used if cell response is evaluated indirectly. Developing a deeper well (thick PDMS sheets) or a gasket overlay postprinting would confine the medium for each individual sample instead of submerging the entire array within a common medium. This partitioning would allow for the collection of medium from each sample to be used for the numerous subsequent assays (viability, proliferation, or metabolism) compatible with high-throughput arrays. Assessing general metabolism (Alamar Blue) or examination of specific metabolites in cells could be used to rank-order biomaterials in a two-step fashion.
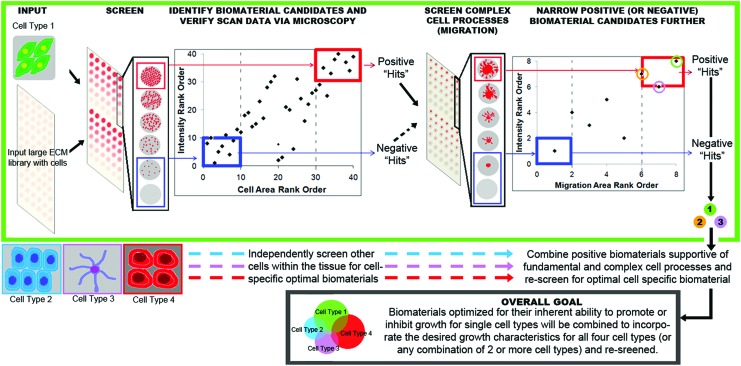
To capitalize on the rapid assessment offered by investigating basic cell metrics and the highly informative functional analysis of the migration cell array, we propose this platform to progress in a two-step manner. First, the cell number and morphology can be rapidly assessed over a wide array of candidate hydrogels to identify relative GS or GI materials. In the second phase, RAMP is used to evaluate cell migration as confirmation of previously identified GS or GI biomaterials, providing a greater level of confidence. Figure 5 outlines the general approach for employing RAMP in the design and discovery of lead candidate biomaterials. “Hits” discovered from screening basic cell metrics for a single cell type could be rank ordered and a subpopulation of materials could be advanced for further screening using the migration assay.
FIG. 5.
RAMP, a novel high-throughput assay, can be used to identify tissue-specific biomaterials for capable of supporting complex cellular processes required for injury repair. This 3D assay utilizes any single cell type embedded within a material; in this case, the model material was collagen embedded with Schwann cells, which is then cultured for several days, at which point the samples are fixed, stained for cytoskeleton structures, and scanned on a flatbed scanner as part of the screening process. Traditional microscopy is used to verify the positive “hits” and negative “hits” for further screening based on cell morphology and number, although work presented here has shown a strong correlation between imaging modalities for the “hits.” These potential biomaterial candidates are then rescreened with enhanced sensitivity via a migration assay. Other cell types can be simultaneously screened in separate, but parallel arrays to discover biomaterials supportive of simple cell processes, such as viability, are then rescreened for higher order processes required for tissue repair (e.g., neurite outgrowth in neurons, differentiation of stem cells, migration, etc.). A composite biomaterial could be developed from the identified “hits” that would be supportive of necessary resident cells as well as potentially inhibitive of the undesired cell types. The overlap of positive biomaterial “hits” across cell types can be employed for the development of a composite tissue-specific biomaterial. Orthogonal biomaterials across different cell types could similarly be used to arrange supportive and inhibitive cues for developing a highly optimized tissue-specific biomaterial. Color images available online at www.liebertpub.com/tec
RAMP data can be processed in parallel evaluating all resident cells simultaneously to quickly identify a library of candidate biomaterials. However, due to its modular nature, RAMP can be applied in series to limit the experimental space. For example, in the peripheral nerve, Schwann cells are abundant and can proliferate in vitro for easy screening, unlike the nonproliferating motor neurons. Therefore, by first identifying the GS materials for the motor-neuron-derived Schwann cells, one can subsequently rapidly identify a common material for motor neurons and motor-derived Schwann cells in a resource-efficient manner. Neurite outgrowth will be a key indicator of neuronal preference for a biomaterial and work is currently underway to validate sufficient sensitivity of RAMP to detect the thin neurite projections. Preliminary 2D culture of sensory neurons on laminin-coated arrays has shown that the fluorescent scanner technology used in RAMP is sensitive enough to detect neurite outgrowth (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tec). These results should be translatable to the 3D biomaterials utilized in RAMP and further work will investigate this supposition and begin to utilize RAMP to screen biomaterial libraries.
Individual cell-specific biomaterials can be combined to generate a tissue-specific biomaterial, supportive of key resident cells necessary for tissue regeneration. Tissue-specific biomaterials would be designed based on supportive biomaterials common to two or more cell types within the target tissue. The Venn diagram in Figure 5 represents a potential situation of overlapping cell-specific biomaterial “hits” for 4 cell types. In this situation supportive biomaterials for cell types 1 and 3 overlap with all the cell types, whereas there may not be any materials found to be GS for both cell types 2 and 4. In this scenario, composite biomaterials could be rationally designed weighing the GS properties identified supportive of two or more resident cell types. For example, in the peripheral nerve, Schwann cell migration and repopulation is a key element of repair, but neurons are necessary to propagate the signal. Ideally, a single biomaterial would be identified that would be supportive of all of these cell types. Clearly, this is not neural engineering specific, but can be applied to any multicellular tissue construct.
A tissue-specific biomaterial generated from a single component is preferred; however, a composite material could be developed to support cell populations that do not share an affinity for a single biomaterial. For example, in the peripheral nerve, there are motor neurons and sensory neurons as well as motor Schwann cells and sensory Schwann cells that are phenotypically distinct from one another.21 Motor neurons preferentially extend along motor pathways that contain motor Schwann cells,21,22 suggesting that Schwann cells and neurons in the motor nerves exhibit similarities in substrate affinity, as do Schwann cells and neurons in the sensory nerves. Overlap in biomaterial preference between sensory nerve cells could be used to develop scaffolds for mixed nerves, while motor neuron tracts are developed in parallel. In vivo, these tracts would run in parallel down the spinal cord, but there are areas where they would bifurcate. These characteristics could be well captured using a tissue-specific biomaterial that utilizes distinct cell-specific biomaterials patterned to replicate in vivo properties. This strategy would not only overcome the differences in substrate affinity, but would also increase the control over cell populations that are segregated in vivo.
RAMP has been developed to rapidly assess cell response to 3D biomaterials in an effort to identify cell- and tissue-specific biomaterial candidates for in vivo applications. The possible utility of RAMP extends beyond the practice investigated here. Screening biomaterials that promote stem cell self-renewal would be beneficial for enhancing ex vivo expansion of stem cells. For example, clinical trials using stem cells for neurodegenerative diseases are currently underway and require the use of large numbers of stem cells for each patient.23,24 Current protocols are neither time nor cost effective, but RAMP could be implemented to identify biomaterials the stem cells could be cultured on or within that would increase their rate of ex vivo self-renewal and proliferation for subsequent use as a cell-mediated therapeutic. Additionally, there is increasing interest in developing induced pluripotent stem cells to study disease models in vitro.25 Cells are highly influenced by their 3D microenvironment and therefore an appropriate 3D biomaterial to study these cells, as well as develop a 3D model of disease, would be necessary for each disease of interest. RAMP could be implemented to expedite the development of a disease model that could then be used to study treatment options in vitro. The utility of RAMP is not limited to practices explored in this work and can be used on innumerable applications for biomaterial discovery.
This work describes a novel platform (RAMP) to identify GS and GI biomaterials in a two-step assay. First, by quantitatively measuring the changes in cell number and spreading, and, second, by screening previously identified candidates for cell migration in a rapid manner. To our knowledge, this is the first rapid assessment of migration in a high-throughput assay. The preliminary evaluation of the RAMP assay discussed in this study establishes it as a novel, effective, and powerful tool to address the challenges of biomaterial design and discovery. The strength of the RAMP assay lies in the synergy between rational and combinatorial design strategies. Neither is sufficiently adequate nor practical in identifying promising candidates alone. However, a combination of the two strategies—rational selection of matrix proteins and growth factors that are physiologically relevant to a given tissue/organ, and a combinatorial screening of all possible combinations of these in a rapid and high-throughput manner—can facilitate the discovery of compositionally optimized biomaterials. In addition to compositional changes, a library of biomechanically diverse hydrogels can be evaluated using RAMP either in conjunction or independently with compositional changes. Although not explicitly studied here, manipulation of the biomechanical properties of promising biomaterial candidates would allow for further fine tuning of a biomaterial, as the biochemical and mechanobiological responses are known to work in concert to control the cell function.
The results obtained from RAMP are in agreement with conventional, low-throughput methods using macroscale assays analyzed by microscopy and image analysis, indicating sufficient sensitivity for rapid screening of a biomaterial library. Due to the modular nature of this assay, several cell types could be screened in parallel to identify a biomaterial that is GS for all cells or a tissue-specific biomaterial.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation—CBET-1067208 and was performed within the Center for Biotechnology and Interdisciplinary Studies. Experimental data were analyzed using instrumentation within the Molecular Biology CORE and Microscopy CORE facilities.
Disclosure Statement
The authors of this work have no conflicts to disclose.
References
- 1.Finkelstein E.A., Corso P.S., Miller T.R., et al. The Incidence and Economic Burden of Injuries in the United States, 1st edition. Oxford: Oxford University, 2006 [Google Scholar]
- 2.Falsey J.R., Renil M., Park S., Li S., and Lam K.S.Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug Chem 12,346, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Flaim C.J., Chien S., and Bhatia S.N.An extracellular matrix microarray for probing cellular differentiation. Nat Methods 2,119, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Lee M.Y., Kumar R.A., Sukumaran S.M., Hogg M.G., Clark D.S., and Dordick J.S.Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad Sci U S A 105,59, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimetta E., Cagnin S., Volpatti A., Lanfranchi G., and Elvassore N.Dynamic culture of droplet-confined cell arrays. Biotechnol Prog 26,220, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Anderson D.G., Putnam D., Lavik E.B., Mahmood T.A., and Langer R.Biomaterial microarrays: rapid, microscale screening of polymer-cell interaction. Biomaterials 26,4892, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Smalley K.S., Lioni M., and Herlyn M.Life isn't flat: taking cancer biology to the next dimension. In Vitro Cell Dev Biol Anim 42,242, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Anderson D.G., Levenberg S., and Langer R.Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol 22,863, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Yliperttula M., Chung B.G., Navaladi A., Manbachi A., and Urtti A.High-throughput screening of cell responses to biomaterials. Eur J Pharm Sci 35,151, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Chen A.A., Underhill G.H., and Bhatia S.N.Multiplexed, high-throughput analysis of 3D microtissue suspensions. Integr Biol (Cambridge) 2,517, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt C.E., and Leach J.B.Neural tissue engineering: strategies for repair and regeneration. Annu Rev Biomed Eng 5,293, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Rosner B.I., Hang T., and Tranquillo R.T.Schwann cell behavior in three-dimensional collagen gels: evidence for differential mechano-transduction and the influence of TGF-beta 1 in morphological polarization and differentiation. Exp Neurol 195,81, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Dewitt D.D., Kaszuba S.N., Thompson D.M., and Stegemann J.P.Collagen I-matrigel scaffolds for enhanced Schwann cell survival and control of three-dimensional cell morphology. Tissue Eng Part A 15,2785, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Hurtado A., Moon L.D., Maquet V., Blits B., Jerome R., and Oudega M.Poly (D,L-lactic acid) macroporous guidance scaffolds seeded with Schwann cells genetically modified to secrete a bi-functional neurotrophin implanted in the completely transected adult rat thoracic spinal cord. Biomaterials 27,430, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Assouline J.G., and Pantazis N.J.Localization of the nerve growth factor receptor on fetal human Schwann cells in culture. Exp Cell Res 182,499, 1989 [DOI] [PubMed] [Google Scholar]
- 16.Seggio A.M., Narayanaswamy A., Roysam B., and Thompson D.M.Self-aligned Schwann cell monolayers demonstrate an inherent ability to direct neurite outgrowth. J Neural Eng 7,046001, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Koppes A.N., Seggio A.M., and Thompson D.M.Neurite outgrowth is significantly increased by the simultaneous presentation of Schwann cells and moderate exogenous electric fields. J Neural Eng 8,046023, 2011 [DOI] [PubMed] [Google Scholar]
- 18.Morrissey T.K., Kleitman N., and Bunge R.P.Isolation and functional characterization of Schwann cells derived from adult peripheral nerve. J Neurosci 11,2433, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folkman J., and Moscona A.Role of cell shape in growth control. Nature 273,345, 1978 [DOI] [PubMed] [Google Scholar]
- 20.Chernousov M.A., and Carey D.J.Schwann cell extracellular matrix molecules and their receptors. Histol Histopathol 15,593, 2000 [DOI] [PubMed] [Google Scholar]
- 21.Jesuraj N.J., Nguyen P.K., Wood M.D., Moore A.M., Borschel G.H., Mackinnon S.E., et al. Differential gene expression in motor and sensory Schwann cells in the rat femoral nerve. J Neurosci Res 90,96, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoke A., and Brushart T.Introduction to special issue: challenges and opportunities for regeneration in the peripheral nervous system. Exp Neurol 223,1, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glass J.D., Boulis N.M., Johe K., Rutkove S.B., Federici T., Polak M., et al. Lumbar intraspinal injection of neural stem cells in patients with amyotrophic lateral sclerosis: results of a phase I trial in 12 patients. Stem Cells 30,1144, 2012 [DOI] [PubMed] [Google Scholar]
- 24.O'Connor D.M., and Boulis N.M.Cellular and molecular approaches to motor neuron therapy in amyotrophic lateral sclerosis and spinal muscular atrophy. Neurosci Lett 527,78, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., et al. Disease-specific induced pluripotent stem cells. Cell 134,877, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.