Abstract
Objective:
Echinococcus granulosus is the causative agent of cystic echinococcosis in humans and many domestic animals, and remains an important global health problem. The aim of this study was to genotype E. granulosus isolates obtained from humans and animals in the Thrace Region of Turkey.
Material and Methods:
A total of 58 isolates were obtained from patients who underwent surgery at several hospitals and from animals at a slaughterhouse in the province of Edirne. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of ribosomal internal transcribed spacer 1 fragments, and polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) of the partial mitochondrial NADH dehydrogenase subunit 1 (ND1) gene, was used to characterize human and animal E. granulosus isolates. To investigate the genetic characteristics of isolates, deoxyribonucleic acid (DNA) sequencing of the mitochondrial cytochrome c oxidase subunit 1 (CO1) and ND1 genes was performed.
Results:
Fifty-eight E. granulosus isolates, including 42 from human, 13 from cattle and 3 from sheep were, analyzed. The results indicated two distinct genotypes: the G1 (sheep strain) and G7 (pig strain) genotypes. The sheep strain was shown to be the most common genotype of E. granulosus affecting humans, sheep and cattle. Among the concatenated partial CO1 and ND1 sequence data, eight haplotypes of Echinococcus species were identified in the present study.
Conclusion:
This is the first report indicating that the E. granulosus pig strain is present in humans in this region. We suggest that new strategies be designed for E. granulosus control programs in Turkey.
Keywords: Echinococcus granulosus, genotype, PCR-RFLP, PCR-SSCP, CO1, ND1, Edirne, Thrace region, Turkey
Introduction
Echinococcus granulosus, a small cestode, belongs to the Taeniidae family. The adult cestode lives in the intestines of dogs and other carnivores (definitive hosts), while the larval form (metacestode) lives in the internal organs of ungulates and humans (intermediate hosts). The metacestode form causes cystic echinococcosis in intermediate hosts (1, 2). E. granulosus is globally distributed and causes public health problems in many parts of Eurasia, including Turkey, as well as in Africa, Australia and South America (3). Between 2001 and 2005, various hospitals and Health Ministry documents recorded 14,789 and 526 human cases of cystic echinococcosis in Turkey and Thrace region, respectively (4).
To date, considerable phenotypic and genetic variability has been observed within the species E. granulosus, and ten genotypes have been identified from different intermediate and definitive hosts. E. granulosus has been split into E. granulosus sensu stricto (G1–G3), E. equinus (G4), E. ortleppi (G5), and E. canadensis (G6–G10) (5, 6). This variability may be reflected in characteristics which affect the life-cycle pattern, host specificity, development rate, pathogenicity, antigenicity and sensitivity to chemotherapeutic agents, transmission dynamics, epidemiology, and control of echinococcosis (7). In particular, because of variations in the period of egg production in some strains (prepatent period in dogs for G1 genotype is an average of 45 days while this period for G7 genotype is 34 days), determination of genotypes is important in control programs (8–10).
A number of deoxyribonucleic acid (DNA) identification techniques have been employed to better characterize Echinococcus species and genotypes from different intermediate and definitive hosts and in different geographical settings (11). Although there are some molecular studies of E. granulosus isolates originating from different hosts in Turkey (12–18), no mitochondrial DNA sequence data exists for Echinococcus species from the European part of Turkey.
The aim of this study was to characterize the E. granulosus genotypes currently infecting humans and livestock animals in Turkey, using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) analyses, and to estimate the genetic variability within the strains by sequencing the mitochondrial cytochrome c oxidase subunit 1 (CO1) and NADH dehydrogenase subunit 1 (ND1) genes.
Material and Methods
Parasites and isolation of genomic DNA
A total of 58 isolates were obtained from patients who underwent surgery at several hospitals and from animals in a slaughterhouse all located in Edirne, in the Thrace region of Turkey, between January and December 2009. Hydatid fluid and/or germinal layers were examined by light microscopy. Also, all isolates were examined pathologically. Then, parasite materials were stored in 70% ethanol at −20°C until used. Total genomic DNA was extracted from the parasite materials using DNeasy Blood & Tissue Kit (Qiagen, Germany), according to the manufacturer’s protocol.
PCR-RFLP analysis
The PCR-RFLP method was performed according to the Bowles and McManus (19) protocol with slight modifications. Ribosomal DNA-internal transcribed spacer gene 1 (rDNAITS1) was amplified from each sample using the BD1 and 4S primers. For each set of PCRs, positive (EU503084) (20) and negative (no-DNA) controls were included. All PCR products were digested by four-base cutting restriction endonucleases (AluI and RsaI, Bioron, Germany; MspI and HhaI, Fermentas, Lithuania). The restriction fragments were separated by electrophoresis on a 3% Tris-Borate-EDTA (TBE) agarose gel containing ethidium bromide.
PCR-SSCP analysis
Echinococcus granulosus genomic DNA samples were analyzed by PCR-SSCP using the described methods with minor modifications (21). The mitochondrial ND1 region was amplified by PCR using MS1 and MS2 primers (22). For each set of PCRs, positive [G1: EU503084 (20), and G6–G10 complex: GU951511 (18)] and negative (no-DNA) controls were included. Four microliters of each PCR products were mixed with 12 μl of denaturating buffer. After denaturation at 95°C for 10 min and subsequent snap-cooling on a freeze block, 10 μl of individual samples were loaded into the wells. Electrophoresis was carried out on 12% acrylamide-bisacrylamide non-denaturing gels containing 5% glycerol at 200 V for 20 hours at room temperature. The gel was stained using the silver-staining technique modified by previously described methods (23).
Mitochondrial DNA sequencing and phylogenetic analysis
Thirteen amplicons, representing each unique SSCP and RFLP profile, were selected, and fragments of amplicons mitochondrial CO1 and ND1 genes were amplified with primers published in Bowles et al. (24) and Sharbatkhori et al. (22), respectively. Then, PCR products were purified using the High Pure PCR Product Purification Kit (Roche, Germany). Sequencing was performed on the ABI 3130 XL Genetic Analyzer (Applied Biosystems, USA) with the BigDye Cycle Sequencing Kit (Applied Biosystems, USA) using the corresponding PCR primers by Refgen Company (Ankara, Turkey). DNA sequences were compared with partial CO1 and ND1 sequences (6, 24–31) representing all currently known genotypes of E. granulosus, all species of Echinococcus and Taenia saginata (the out-group) (Table 1) which we obtained from previous publications and National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Phylogenetic analysis of partial sequence data from mitochondrial CO1 and ND1 genes was performed by Iontek Company (İstanbul, Turkey). Phylogenetic analysis of concatenated CO1 and ND1 data was conducted by the maximum likelihood (ML) method, employing the MEGA5 (Molecular Evolutionary Genetics Analysis) software.
Table 1.
Echinococcus granulosus CO1 and ND1 sequence data from the present study, together with reference sequences from previous studies used for nucleotide sequence comparison and phylogenetic analysis (GenBank accession numbers)
| Haplotype/Genotype | Host origin (no) | Accession no (CO1) | Accession no (ND1) | References (CO1 and ND1, respectively) |
|---|---|---|---|---|
| Haplotype 1 | Human (1), Cattle (1) | HQ717150 | HQ717151 | The present study |
| Haplotype 2 | Human (1) | HQ717155 | HQ717154 | The present study |
| Haplotype 3 | Human (1) | HQ717150 | HQ717152 | The present study |
| Haplotype 4 | Human (1) | HQ717148 | HQ717153 | The present study |
| Haplotype 5 | Human (1) | HQ717148 | HQ717151 | The present study |
| Haplotype 6 | Human (5) | HQ703429 | HQ717151 | The present study |
| Haplotype 7 | Sheep (1) | HQ717149 | HQ717151 | The present study |
| Haplotype 8 | Cattle (1) | HQ717156 | HQ717151 | The present study |
| Genotype 1 | Sheep | U50464 | AJ237632 | 25, 26 |
| Genotype 2 | Sheep | M84662 | AJ237633 | 24, 26 |
| Genotype 3 | Buffalo | M84663 | AJ237634 | 24, 26 |
| Genotype 4 | Horse | M84664 | AJ237635 | 24, 26 |
| Genotype 5 | Cattle | M84665 | AJ237636 | 24, 26 |
| Genotype 6 | Camel | M84666 | AJ237637 | 24, 26 |
| Genotype 7 | Pig | M84667 | AJ237638 | 24, 26 |
| Genotype 8 | Moose | AB235848 | AJ237643 | 6, 27 |
| Genotype 10 | Reindeer | AF525457 | AF525297 | 28 |
| E. multilocularis* | Human | M84668 | AJ237639 | 24, 26 |
| E. multilocularis** | Rodent | M84669 | AJ237640 | 24, 26 |
| E. shiquicus | Pika | AB208064 | AB208064 | 6 |
| E. vogeli | Rodent | M84670 | AJ237641 | 24, 26 |
| E. oligarthrus | Rodent | M84671 | AJ237642 | 24, 26 |
| E. felidis | Lion | EF558356 | EF558357 | 29 |
| Outgroup: | ||||
| T. saginata | Cattle | AB465239 | AJ239106 | 30, 31 |
CO1: Cytochrome c oxidase subunit 1, ND1: NADH dehydrogenase subunit 1,
Asterisks relate also to Figure 2
Results
PCR-RFLP analysis
ITS1-PCR was carried out on 58 isolates from 42 humans, 13 cattle and 3 sheep. All isolates showed two fragments of approximately 1000 bp and 1100 bp. ITS-PCR products of the 58 isolates were digested by restriction endonucleases, and band patterns were compared with the band patterns of reference isolates. The results demonstrated that 47 isolates had the same band pattern as the reference G1 isolate (pattern I), 10 isolates had pattern II and one isolate had pattern III (Table 2).
Table 2.
Characteristics of Echinococcus granulosus human (n=42), sheep (n=3), and cattle (n=13) isolates from the Thrace region, Turkey, 2009
| Host/no | Pattern of PCR-RFLP | Pattern of PCR-SSCP | DNA Sequencing | Final Genotype/Haplotype | |
|---|---|---|---|---|---|
| CO1 | ND1 | ||||
| Human(1) | I | A | G1 | G1 (195 C/T) | G1/H1 |
| Cattle (1) | |||||
| Human (1) | III | B | G7 | G7 (3 A/G) | G7/H2 |
| Human (1) | I | C | G1 | G1 (195 C/T, 288 T/C) | G1/H3 |
| Human (1) | I | A | G1 (56 C/T) | G1 | G1/H4 |
| Human (1) | I | A | G1 (56 C/T) | G1 (195 C/T) | G1/H5 |
| Human (5) | II | A | G1 (108 T/C) | G1 (195 C/T) | G1/H6 |
| Sheep (1) | I | A | G1 (66 C/T, 306 G/A) | G1 (195 C/T) | G1/H7 |
| Cattle (1) | I | A | G1 (66 C/T) | G1 (195 C/T) | G1/H8 |
| Human (27) | I | A | ND | ND | G1 |
| Sheep (2) | |||||
| Cattle (11) | |||||
| Human (5) | II | A | ND | ND | G1 |
| All Isolates | I: 47 | A: 56 | G1: 57 | ||
| II: 10 | B: 1 | G7: 1 | |||
| III: 1 | C: 1 | ||||
PCR-RFLP: Polymerase chain reaction - restriction fragment length polymorphism, PCR-SSCP: Polymerase chain reaction - single stranded conformation polymorphism, CO1: Cytochrome c oxidase subunit 1, ND1: NADH dehydrogenase subunit 1, G: Genotype, C:Cytosine, T: Thymine, G: Guanine, A: Adenine, H: Haplotype, ND: Not Done
PCR-SSCP analysis
ND1-PCR was carried out on 58 isolates, and all isolates showed a fragment of approximately 400 bp. SSCP analyses of all 58 amplicons displayed three unique profiles for partial ND1. These patterns were compared with the domestic sheep strain and E. canadensis patterns. 56 isolates (pattern A) corresponded to the G1 strain pattern. Two human isolates (patterns B and C) produced different patterns from the reference G1 and G6–G10 complex strains (Figure 1, Table 2).
Figure 1.
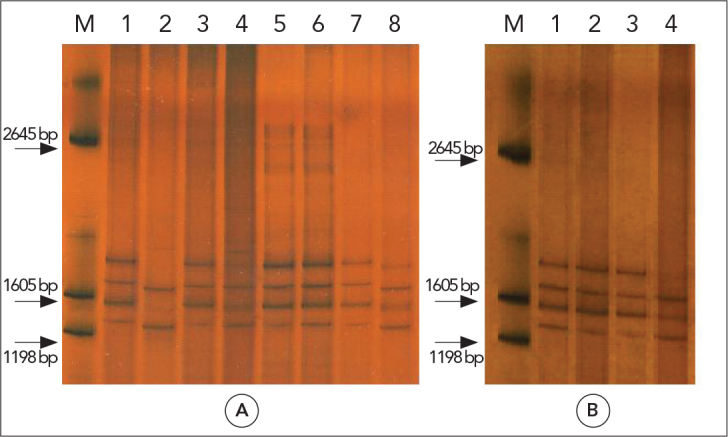
SSCP analysis of mitochondrial ND1 PCR products of Echinococcus granulosus
M: marker (Promega, USA); A: G1 control (lane 1), G6–G10 complex control (lane 2), pattern A (lane 3,4,5,6 and 7) and pattern B (lane 8); B: G1 control (lane 1), pattern A (lane 2 and 3), pattern C (lane 4)
Mitochondrial DNA sequencing and phylogenetic analysis
Thirteen amplicons, representing each unique RFLP and SSCP profile, were selected. ND1 and CO1-PCR were carried out on these isolates, and all isolates showed fragments of approximately 400 bp and 450 bp, respectively. Twelve isolates, including nine human, two cattle and one sheep were determined to be the G1 strain, while one human isolate was determined to be the G7 strain. The obtained sequences were submitted to the GenBank database under their accession numbers (Table 1).
The CO1 region of three of the 13 isolates fully corresponded with the G1 reference sequence. Two isolates possessed the non-synonymous nucleotide substitution 56 C/T (inducing substitution of alanine with valine). One isolate presented a C/T exchange at position 66. Five human isolates possessed the nucleotide substitution 108 T/C. One sheep isolate presented two mutations (66 C/T and 306 G/A). One human isolate produced a sequence which fully corresponded with the G7 reference sequence.
The ND1 region of one of 13 isolates fully corresponded with the G1 reference sequence. Ten isolates possessed the nucleotide substitution 195 C/T. One isolate presented two mutations (195 C/T and 288 T/C). One human isolate, which was identified as the G7 genotype, had a guanine nucleotide at position 3. All of these nucleotide substitutions were synonymous.
Among the concatenated partial CO1 and ND1 sequence data, eight haplotypes were detected in the present study using reference data (comprising concatenated partial sequences from previous studies representing all recognized E. granulosus genotypes; Table 1). Based on the phylogenetic analyses, haplotype 2 were classified as G6–G10 complex; and the other seven haplotypes (haplotype 1, 3–8) were classified as G1–G3 complex (Figure 2).
Figure 2.
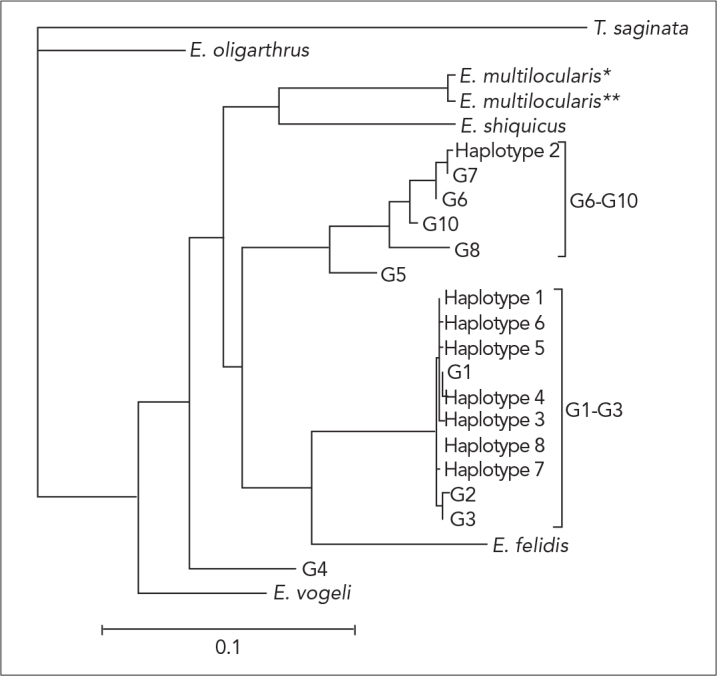
Genetic relationships of haplotypes 1–8, which represent Echinococcus granulosus samples from Turkey (present study), to reference sequences selected from previous studies. The relationships were inferred based on phylogenetic analysis of concatenated CO1 + ND1 sequence data (see Table 1) using maximum likelihood (ML) method
Discussion
Some studies investigating the molecular characterization of E. granulosus isolates from humans, sheep, goats, cattle, camels and dogs in different parts of Turkey have been performed previously. The investigators used different methods to classify E. granulosus isolates from different hosts in Turkey, such as PCR-RFLP of the ITS-1 region of the rRNA gene and sequencing of CO1 (15); comparative sequencing of the CO1 and/or ND1 genes (12–14); specific PCR analysis of the 12S rRNA gene and sequencing of CO1 (16, 18); and specific PCR analysis of the 12S rRNA gene. Another study performed SSCP analysis of the CO1 gene and sequencing of CO1 (17). According to these reports, G1 was the most prevalent strain among human and animal isolates. The G1 genotype has been demonstrated in humans, sheep, goats, cattle, camels, water buffaloes and dogs (12–16); the G3 genotype has been demonstrated in humans, sheep and cattle (12, 13); and the G7 genotype has been demonstrated in humans and sheep (13), whereas G6 has only been identified in humans (18).
The present study showed that G1 is the dominant isolate in Turkey and is the principal agent in human and animal cystic echinococcosis. In addition, the G7 genotype was detected for the second time in human isolates from Turkey. In the previous study, the pig strain G7 was reported in one human isolate and two isolates from sheep (13). Our results are consistent with the data obtained by other authors who used different techniques, such as PCR-RFLP (15, 32–37) and SSCP (17, 22, 38–40) analyses for the genotyping of E. granulosus. In addition, we examined 13 isolates of E. granulosus from different intermediate hosts using sequence analysis of the mitochondrial ND1 and CO1 genes. We identified two E. granulosus genotypes (G1 and G7). Although the human, sheep and cattle isolates shared identical sequences to the sheep strain (G1 genotype), one human isolate produced identical sequences to the pig strain (G7 genotype). Previous analyses (22, 39, 40) revealed that concatenated sequence data was reliable for estimating haplotypic variability within E. granulosus. The present phylogenetic analysis classified 12 isolates (representing haplotype 1, 3–8) as E. granulosus sensu stricto (G1–G3 complex) and one isolate as E. canadensis (G6–G10 complex).
In the present study, analysis of partial mitochondrial CO1 sequence data revealed a C/T change at position 56 (HQ717148), the only mutation in our study that causes an amino acid change. Similar C/T changes were seen in many sequences including Turkey isolates, such as EU178104, JF775379, JF775380, HM598459, EF689726, AB491447 and AJ508019 deposited in the GenBank database (12–15, 41, 42). The C/T change at position 66, which was identified in a cattle isolate (HQ717156), has been previously noted in various parts of the world (HM130578, AB491456, DQ356881) (41, 43, 44). Similar patterns recorded in the study of Vural et al. (12) from eastern Turkey were classified as G3 (EF545563, EU178105) and Snabel et al. (13) from western Turkey were classified as G1/G3 genotype cluster.
The T/C change at position 108, which was identified in our human isolates (HQ703429), was reported in the sheep isolate (AJ508005) in Australia (42). The sequence in which the 66C/T and 306G/A changes existed together was identified in the sheep isolate (HQ717149) and was not found among the other sequences in GenBank. However, a human isolate (AJ508024) with a 306G/A mutation was identified by Obwaller et al. (42).
Analysis of partial mitochondrial ND1 sequence data revealed a C/T change at position 195 in human, sheep and cattle isolates (HQ717151) that has been reported in many isolates from various parts of the world (including HM853645, DQ856470 and AF408688, among others) and from Turkey (13, 45–47). The co-occurrence of the 195 C/T and 288 T/C changes (HQ717152) has been previously noted [FJ796212 (22)]. In our study, the human isolate identified as the G7 genotype had a guanine nucleotide at position 3 (HQ717154), unlike the reference G7 sequence. The 3A/G mutation was not found in the sequences deposited in the GenBank database.
The domestic sheep strain (G1) of E. granulosus has a worldwide geographical distribution. It is found in parts of South America, South and East Europe, North and East Africa, Asia and Australia. The pig strain (G7) is found in some countries in central and east Europe, parts of Russia, and Mexico and Peru (48–50). The pig strain is considered to have low infectivity for human, however, infected human cases with pig strain have been recorded in Poland and Turkey (8, 13, 51).
A study including sheep and goats in Greece, Turkey’s neighbour which has been placed in the North Thracian region, showed G1 and G3 genotypes in sheep, and G7 genotype in all goats (46). In Bulgaria,another neighbour, Breyer at al. (52) detected G1 genotype in all E. granulosus isolates including cattle, sheep, pigs, jackals and a wolf. Casulli et al. (53) investigated the mitochondrial variability of E. granulosus s.s. in the European countries of Bulgaria, Hungary, Romania and Italy. Some isolates were identified as G7 genotype from Hungary and Romania. All other isolates were identified as E. granulosus sensu stricto.
There is a single pig farm in Turkey, however, because the majority of Turkey’s population are Muslims, and pork consumption is very low. In our study, the single human case enfected with pig strain has never left the Thrace region. Although there is no pig farm in the Thrace region, the number of wild boars is not low. For these reasons, we suppose that the pig strain obtained from a patient from Edirne may have come through a wild cycle from the neighbouring countries. However, in order to speak more clearly about this subject, studies which include more patients, livestock, wild boars and definitive hosts from a wide geographical area are required.
Conclusion
Our genotypic data strongly suggest that most of the human and animal isolates are of the G1 genotype. As this study identifies the G7 genotype in human isolates from the Thrace region for the first time, and because the G7 and G1 genotypes have different maturation periods in definitive hosts, we suggest that new strategies be designed for further genetic analysis and E. granulosus control programs in Turkey.
Acknowledgments
This study was financially supported by the Trakya University Scientific Research Fund (TUBAP-2009/61). Ethics approval was granted by the Ethical Committee of Trakya University Medical Faculty.
We thank the nurses and medical doctors of hospitals in Edirne for helping isolate collection; Tammam Sipahi (Department of Biophysics, Faculty of Medicine, Trakya University, Edirne) for excellent scientific support; Sami Simsek (Department of Parasitology, Faculty of Veterinary Medicine, University of Firat, Elazig) for providing positive controls.
Footnotes
Conflict of Interest
No conflict of interest was declared by the authors.
References
- 1.Soulsby EJL. Helminths, arthropods and of domesticated animals. 7th ed. London: Bailliere Tindall; 1982. pp. 92–136. [Google Scholar]
- 2.Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of Echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev. 2004;17:107–35. doi: 10.1128/CMR.17.1.107-135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckert J, Schantz PM, Gasser RB, Torgerson PR, Bessonov AS, Movsessian SO, et al. Geographic distribution and prevalence. In: Eckert J, Gemmell MA, Meslin F-X, Pawlowski ZS, editors. WHO/OIE manual on Echinococcosis in humans and animals: a public health problem of global concern. Paris: World organisation for animal health; 2001. pp. 101–43. [Google Scholar]
- 4.Yazar S, Ozkan AT, Hökelek M, Polat E, Yilmaz H, Ozbilge H, et al. Cystic echinococcosis in Turkey from 2001–2005. Turkiye Parazitol Derg. 2008;32:208–20. [PubMed] [Google Scholar]
- 5.Thompson RCA. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. 2008;119:439–46. doi: 10.1016/j.exppara.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Nakao M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology. 2007;134:713–22. doi: 10.1017/S0031182006001934. [DOI] [PubMed] [Google Scholar]
- 7.Thompson RCA, McManus DP. Aetiology: parasites and life-cycles. In: Eckert J, Gemmell MA, Meslin F-X, Pawlowski ZS, editors. WHO/OIE manual on Echinococcosis in humans and animals: a public health problem of global concern. Paris: World organisation for animal health; 2001. pp. 1–19. [Google Scholar]
- 8.Thompson RCA, Lymbery AJ. The nature, extent and significance of variation within the genus Echinococcus. Adv Parasitol. 1988;27:209–58. doi: 10.1016/s0065-308x(08)60356-5. [DOI] [PubMed] [Google Scholar]
- 9.Thompson RCA. Biology and systematics of Echinococcus. In: Thompson RCA, Lymbery AJ, editors. Echinococcus and hydatid disease. Wallingford: CAB International; 1995. pp. 1–50. [Google Scholar]
- 10.Zhang L, Eslami A, Hosseini SH, McManus DP. Indication of the presence of two distinct strains of Echinococcus granulosus in Iran by mitochondrial DNA markers. Am J Trop Med Hyg. 1998;59:171–4. doi: 10.4269/ajtmh.1998.59.171. [DOI] [PubMed] [Google Scholar]
- 11.McManus DP. Molecular discrimination of taeniid cestodes. Parasitol Int. 2006;55(Suppl):S31–7. doi: 10.1016/j.parint.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Vural G, Baca AU, Gauci CG, Bagci O, Gicik Y, Lightowlers MW. Variability in the Echinococcus granulosus cytochrome c oxidase 1 mitochondrial gene sequence from livestock in Turkey and a re-appraisal of the G1-3 genotype cluster. Vet Parasitol. 2008;154:347–50. doi: 10.1016/j.vetpar.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Snabel V, Altintas N, D’Amelio S, Nakao M, Romig T, Yolasigmaz A, et al. Cystic echinococcosis in Turkey: genetic variability and first record of the pig strain (G7) in the country. Parasitol Res. 2009;105:145–54. doi: 10.1007/s00436-009-1376-2. [DOI] [PubMed] [Google Scholar]
- 14.Beyhan YE, Umur S. Molecular characterization and prevalence of cystic echinococcosis in slaughtered water buffaloes in Turkey. Vet Parasitol. 2011;181:174–9. doi: 10.1016/j.vetpar.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 15.Utuk AE, Simsek S, Koroglu E, McManus DP. Molecular genetic characterization of different isolates of Echinococcus granulosus in east and southeast regions of Turkey. Acta Trop. 2008;107:192–4. doi: 10.1016/j.actatropica.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 16.Simsek S, Balkaya I, Koroglu E. Epidemiological survey and molecular characterization of Echinococcus granulosus in cattle in an endemic area of eastern Turkey. Vet Parasitol. 2010;172:347–9. doi: 10.1016/j.vetpar.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Simsek S, Balkaya I, Ciftci AT, Utuk AE. Molecular discrimination of sheep and cattle isolates of Echinococcus granulosus by SSCP and conventional PCR in Turkey. Vet Parasitol. 2011;178:367–9. doi: 10.1016/j.vetpar.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 18.Simsek S, Kaplan M, Ozercan IH. A comprehensive molecular survey of Echinococcus granulosus in formalin-fixed paraffin-embedded tissues in human isolates in Turkey. Parasitol Res. 2011;109:411–6. doi: 10.1007/s00436-011-2269-8. [DOI] [PubMed] [Google Scholar]
- 19.Bowles J, McManus DP. Rapid discrimination of Echinococcus species and strains using a polymerase chain reaction-based RFLP method. Mol Biochem Parasitol. 1993;57:231–40. doi: 10.1016/0166-6851(93)90199-8. [DOI] [PubMed] [Google Scholar]
- 20.Simsek S, Eroksuz Y. Occurrence and molecular characterization of Echinococcus granulosus in Turkish mouflon (Ovis gmelinii anatolica) Acta Trop. 2009;109:167–9. doi: 10.1016/j.actatropica.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Gasser RB, Zhu X, McManus DP. Display of sequence variation in PCR-amplified mitochondrial DNA regions of Echinococcus by single-strand conformation polymorphism. Acta Trop. 1998;71:107–15. doi: 10.1016/s0001-706x(98)00052-7. [DOI] [PubMed] [Google Scholar]
- 22.Sharbatkhori M, Mirhendi H, Jex AR, Pangasa A, Campbell BE, Kia EB, et al. Genetic categorization of Echinococcus granulosus from humans and herbivorous hosts in Iran using an integrated mutation scanning-phylogenetic approach. Electrophoresis. 2009;30:2648–55. doi: 10.1002/elps.200900145. [DOI] [PubMed] [Google Scholar]
- 23.Byun SO, Fang Q, Zhou H, Hickford JG. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal Biochem. 2009;385:174–5. doi: 10.1016/j.ab.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54:165–73. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto M, Bessho Y, Kmiya M, Kurosawa T, Horii T. Phylogenetic relationships within Taenia taeniaeformis variants and other taeniid cestodes inferred from the nucleotide sequence of the cytochrome c oxidase subunit I gene. Parasitol Res. 1995;81:451–8. doi: 10.1007/BF00931785. [DOI] [PubMed] [Google Scholar]
- 26.Bowles J, McManus DP. NADH dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus. Int J Parasitol. 1993;23:969–72. doi: 10.1016/0020-7519(93)90065-7. [DOI] [PubMed] [Google Scholar]
- 27.Bowles J, Blair D, McManus DP. Molecular genetic characterization of the cervid strain (‘northern form’) of Echinococcus granulosus. Parasitology. 1994;109:215–21. doi: 10.1017/s0031182000076332. [DOI] [PubMed] [Google Scholar]
- 28.Lavikainen A, Lehtinen MJ, Meri T, Hirvela-Koski V, Meri S. Molecular genetic characterization of the Fennoscandian cervid strain, a new genotypic group (G10) of Echinococcus granulosus. Parasitology. 2003;127:207–15. doi: 10.1017/s0031182003003780. [DOI] [PubMed] [Google Scholar]
- 29.Hüttner M, Nakao M, Wassermann T, Siefert L, Boomker JD, Dinkel A, et al. Genetic characterization and phylogenetic position of Echinococcus felidis (Cestoda: Taeniidae) from the African lion. Int J Parasitol. 2008;38:861–8. doi: 10.1016/j.ijpara.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto M, Nakao M, Blair D, Anantaphruti MT, Waikagul J, Ito A. Evidence of hybridization of between Taenia saginata and Taenia asiatica. Parasitol Int. 2010;59:70–4. doi: 10.1016/j.parint.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Gasser RB, Zhu X, McManus DP. NADH dehydrogenase subunit 1 and cytochrome c oxidase subunit 1 sequences compared for members of the genus Taenia (Cestoda) Int J Parasitol. 1999;29:1965–70. doi: 10.1016/s0020-7519(99)00153-8. [DOI] [PubMed] [Google Scholar]
- 32.Harandi MF, Hobbs RP, Adams PJ, Mobedi I, Morgan-Ryan UM, Thompson RCA. Molecular and morphological characterization of Echinococcus granulosus of human and animal origin in Iran. Parasitology. 2002;125:367–73. doi: 10.1017/s0031182002002172. [DOI] [PubMed] [Google Scholar]
- 33.Wachira TM, Bowles J, Zeyhle E, McManus DP. Molecular examination of the sympatry and distribution of sheep and camel strains of Echinococcus granulosus in Kenya. Am J Trop Med Hyg. 1993;48:473–9. doi: 10.4269/ajtmh.1993.48.473. [DOI] [PubMed] [Google Scholar]
- 34.Bowles J, van Knapen F, McManus D. Cattle strain of Echinococcus granulosus and human infection. The Lancet. 1992;339:1358. doi: 10.1016/0140-6736(92)92003-x. [DOI] [PubMed] [Google Scholar]
- 35.Scott JC, Stefaniak J, Pawlowski ZS, McManus DP. Molecular genetic analysis of human cystic hydatid cases from Poland: identification of a new genotypic group (G9) of Echinococcus granulosus. Parasitology. 1997;114:37–43. doi: 10.1017/s0031182096008062. [DOI] [PubMed] [Google Scholar]
- 36.Sharbatkhori M, Mirhendi H, Harandi MF, Rezaeian M, Mohebali M, Eshraghian M, et al. Echinococcus granulosus genotypes in livestock in Iran indicating high frequency of G1 genotype in camels. Exp Parasitol. 2010;124:373–9. doi: 10.1016/j.exppara.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 37.M’rad S, Filisetti D, Quidni M, Mekki M, Belguith M, Nouri A, et al. Molecular evidence of ovine (G1) and camel (G6) strains of Echinococcus granulosus in Tunisia and putative role of cattle in human contamination. Vet Parasitol. 2005;129:267–72. doi: 10.1016/j.vetpar.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Gasser RB, Zhu X, McManus DP. Screening for different genotypes of Echinococcus granulosus within China and Argentina by single-strand conformation polymorphism (SSCP) analysis. Trans R Soc Trop Med Hyg. 1999;93:329–34. doi: 10.1016/s0035-9203(99)90043-3. [DOI] [PubMed] [Google Scholar]
- 39.Abushhewa MH, Abushhiwa MHS, Nolan MJ, Jex AR, Campbell BE, Jabbar A, et al. Gentic classification of Echinococcus granulosus cysts from humans, cattle and camels in Libya using mutation scanning-based analysis of mitochpndrial loci. Mol Cell Probes. 2010;24:346–51. doi: 10.1016/j.mcp.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Jabbar A, Narankhajid M, Nolan MJ, Jex AR, Campbell BE, Gasser RB. A first insight into the genotypes of Echinococcus granulosus from humans in Mongolia. Mol Cell Probes. 2011;25:49–54. doi: 10.1016/j.mcp.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Nakao M, Li T, Han X, Ma X, Xiao N, Qiu J, et al. Genetic polymorphisms of Echinococcus tapeworms in China as determined by mitochondrial and nuclear DNA sequences. Int J Parasitol. 2010;40:379–85. doi: 10.1016/j.ijpara.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obwaller A, Schneider R, Walochnik J, Gollackner B, Deutz A, Janitschke K, et al. Echinococcus granulosus strain differentiation based on sequence heterogeneity in mitochondrial genes of cytochrome c oxidase-1 and NADH dehydrogenase-1. Parasitology. 2004;128:569–75. doi: 10.1017/s0031182004004871. [DOI] [PubMed] [Google Scholar]
- 43.Pour AA, Hosseini SH, Shayan P. Comparative genotyping of Echinococcus granulosus infecting buffalo in Iran using cox1 gene. Parasitol Res. 2011;108:1229–34. doi: 10.1007/s00436-010-2170-x. [DOI] [PubMed] [Google Scholar]
- 44.Bart JM, Abdukader M, Zhang YL, Lin RY, Wang YH, Nakao M, et al. Genotyping of human cystic echinococcosis in Xinjiang, PR China. Parasitology. 2006;133:571–9. doi: 10.1017/S0031182006000734. [DOI] [PubMed] [Google Scholar]
- 45.Sharbatkhori M, Fasihi Harandi M, Mirhendi H, Hajialilo E, Kia EB. Sequence analysis of cox1 and nad1 genes in Echinococcus granulosus G3 genotype in camels (Camelus dromedarius) from central Iran. Parasitol Res. 2011;108:521–7. doi: 10.1007/s00436-010-2092-7. [DOI] [PubMed] [Google Scholar]
- 46.Varcasia A, Canu S, Kogkos A, Pipia AP, Scala A, Garippa G, et al. Molecular characterization of Echinococcus granulosus in sheep and goats of Peloponnesus, Greece. Parasitol Res. 2007;101:1135–9. doi: 10.1007/s00436-007-0568-x. [DOI] [PubMed] [Google Scholar]
- 47.Bardonnet K, Benchikh-Elfegoun MC, Bart JM, Harraga S, Hannache N, Haddad S, et al. Cystic echinococcosis in Algeria: cattle act as reservoirs of a sheep strain and may contribute to human contamination. Vet Parasitol. 2003;116:35–44. doi: 10.1016/s0304-4017(03)00255-3. [DOI] [PubMed] [Google Scholar]
- 48.Eckert J, Thompson RCA. Intraspecific variation of Echinococcus granulosus and related species with emphasis on their infectivity to humans. Acta Trop. 1997;64:19–34. doi: 10.1016/s0001-706x(96)00635-3. [DOI] [PubMed] [Google Scholar]
- 49.Villabos N, Gonzalez LM, Morales J, de Aluja AS, Jimenez MI, Blanco MA, et al. Molecular identification of Echinococcus granulosus genotypes (G1 and G7) isolated from pigs in Mexico. Vet Parasitol. 2007;147:185–9. doi: 10.1016/j.vetpar.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 50.Moro PL, Nakao M, Ito A, Schantz PM, Cavero C, Cabrera L. Molecular identification of Echinococcus isolates from Peru. Parasitol Int. 2009;58:184–6. doi: 10.1016/j.parint.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Pawlowski Z, Stefaniak J. The pig strain of Echinococcus granulosus in humans: a neglected issue? Trends Parasitol. 2003;19:439. doi: 10.1016/s1471-4922(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 52.Breyer I, Georgieva D, Kurdova R, Gottstein B. Echinococcus granulosus strain typing in Bulgaria: the G1 genotype is predominant in intermediate and definitive wild hosts. Parasitol Res. 2004;93:127–30. doi: 10.1007/s00436-004-1116-6. [DOI] [PubMed] [Google Scholar]
- 53.Casulli A, Interisano M, Sreter T, Chitimia L, Kirkova Z, La Rose G, et al. Genetic variability of Echinococcus granulosus sensu stricto in Europe inferred by mitochondrial DNA sequences. Infect Genet Evol. 2012;12:377–83. doi: 10.1016/j.meegid.2011.12.014. [DOI] [PubMed] [Google Scholar]


