Abstract
Siglec-G/10 is broadly expressed on B cells, dendritic cells and macrophage subsets. It binds strongly to CD24, a small glycosyl-phosphatidylinositol-anchored sialoprotein, in a sialylation-dependent manner. Targeted mutation of Siglecg dramatically elevates the level of natural IgM antibodies and its producer, B1 B cells. Incorporation of Siglec-G ligands to both T-dependent and T-independent immunogens reduces antibody production and induces B-cell tolerance to subsequent antigen challenges. By interacting with CD24, Siglec-G suppresses inflammatory responses to danger (damage)-associated molecular patterns, such as heat-shock proteins and high mobility group protein 1, but not to Toll-like receptor ligands. By a CD24-independent mechanism, Siglec-G has been shown to associate with Cbl to cause degradation of retinoic acid-inducible gene 1 and reduce production of type I interferon in response to RNA virus infection. The negative regulation by Siglec-G/10 may provide a mechanism for the host to discriminate between infectious nonself and noninfectious self, as envisioned by the late Dr. Charles A. Janeway.
Keywords: DAMP, PAMP, self–nonself, Siglec-G
Introduction
Pathogen-associated molecular patterns (PAMPs), such as bacterial DNA, RNA, lipopolysaccharide from Gram-negative bacterial cell walls and peptidoglycan from Gram-positive bacterial cell walls, interact with pattern recognition receptors to activate immune responses (Janeway 1989; Medzhitov et al. 1997; Poltorak et al. 1998). Toll-like receptors (TLRs), Nod-like receptors and RIG-I-like receptors (RLR) are the dedicated receptors for PAMPs on immune cells (Medzhitov et al. 1997; Inohara et al. 2002; Meylan et al. 2006). The concept of pattern recognition in immunology was invoked as a sensor of microbial infection, allowing the host to discriminate noninfectious self from infectious nonself (Janeway 1989, 1992).
Surprisingly, endogenous molecules released from necrotic cells during infection or tissue injury activate the immune system (Matzinger 1994). The endogenous molecules released from necrotic cells are called danger (damage)-associate molecular patterns (DAMPs). High-mobility group box 1 protein (HMGB1) and heat-shock proteins (HSP70/90) are prototypical DAMPs released from necrotic cells during infection or tissue injury (Apetoh et al. 2007). Under physiological conditions, DAMP-triggered inflammation has been shown to be involved in tissue remodeling (Takahashi et al. 2008). However, under pathological conditions, stimulation by DAMPs may contribute to the pathogenesis of autoimmunity (Urbonaviciute et al. 2008; Andersson and Harris 2010), sepsis (Wang et al. 1999) and cancer (Mittal et al. 2010; Sims et al. 2010).
A critical issue is the switch between physiological and pathological responses. Here, we use Siglec-G as an example to suggest that pattern recognition by some Siglecs may be essential in this switch. Recent advances on the roles for Siglec-G in B-cell biology, including development, response and tolerance, as well as innate response to RNA viruses are also reviewed.
Sialic acids and Siglecs
Sialic acids are a family of nine-carbon sugars found on secreted or cell surface glycopeptides or glycolipids. N-Glycolylneuraminic acid (Neu5Gc) and N-acetylneuraminic acid (Neu5Ac) are the major sialic acids found in mammalian cells. The only difference between the two sialic acid species is an additional oxygen atom in the N-glycolyl group of Neu5Gc (Varki 2007). Human CMP-N-acetylneuraminic acid hydroxylase, which catalyzes the generation of Neu5Gc by the transfer of a single oxygen atom to the acyl group of CMP-Neu5Ac, was genetically mutated during human evolution. Therefore, humans are unable to produce endogenous Neu5Gc (Chou et al. 2002), whereas many model organisms, such as mice, produce Neu5Gc. Sialic acids attach to glycans via α2-3, α2-6 or α2-8 linkages. These linkages are often the critical determinants for recognition by the sialic acids binding proteins which are expressed by mammalian cells or pathogens.
Sialic acids residues are broadly expressed and could act as a marker of self in the immune system, as such residues are absent from most microbes. The frequent observation that host cells can be lysed by immune cytotoxic effector mechanisms after extensive desialylation demonstrates the significant role played by cell surface sialic acids in this recognition process (Pilatte et al. 1990). One of the most striking examples of the importance of sialic acid in self–nonself discrimination is the presence of natural antibodies to sialidase-treated red blood cells (Vaith and Uhlenbruck 1978; Springer 1984; Pilatte et al. 1990), lymphocytes (Rogentine and Plocinik 1974) and thymocytes (Pilatte et al. 1990) in normal human serum. These antibodies are specific for cryptantigens that maybe exposed on the surface of human cells by the action of sialidase. Recently, Meesmann et al. (2010) showed that desialylation acted as an “eat me” signal and caused an enhanced uptake of apoptotic cells.
Siglecs are Ig-like type I transmembrane proteins with an IgV-like domain that binds to sialic acids attached to the terminal regions of cell surface glycoconjugates (Varki and Angata 2006; Crocker et al. 2007). They are divided into two groups based on structure. The first group of Siglecs, which include Siglec-1, -2, -4 and -15, are conserved structurally between rodents, humans and other vertebrates, and share ∼25–30% amino acid identity. The second group of Siglecs, which include Siglec-3/CD33 and CD33-related Siglecs, have less conserved structure between humans and other vertebrates but have high homology to CD33 in their extracellular domains (50–85% amino acid identity). The CD33-related Siglecs of humans are Siglec-3, -5, -6, -7, -8, -9, -10, -11, -12, -14 and -16, whereas the CD33-related murine Siglecs are Siglec-3, Siglec-E, -F, -G and -H. With the exceptions of murine Siglec-1, -4, -15 and -H, and human Siglec-1, -4, -14, -15 and -16, all known Siglecs have immunoreceptor tyrosine-based inhibition motifs (ITIM, S/I/V/LxYxxI/V/Lor) or ITIM-like motifs in their intracellular domains. Recognition of ligands by Siglecs results in an induction of accessibility of the cytosolic ITIM or ITIM-like tyrosine to Src family kinases such as Lyn. These kinases phosphorylate cytosolic ITIM tyrosines, which in turn recruit tyrosine phosphatases such as SHP-1 or SHP-2 to attenuate signal transduction. Given the dedicated function of Siglecs in recognizing sialic acid-containing structures, it is of interest to determine if they may be involved in recognition of sialic acid-based self-markers as outlined above. In the context of adaptive immunity, Siglecs recognizing sialylated glycans as self-ligands also play a major role in the tolerance of B cells to self-antigens (O'Keefe et al. 1996; Ferry et al. 2005; Nitschke 2009; Pillai et al. 2009; Duong et al. 2010; Jellusova, Duber et al. 2010, Jellusova, Wellmann et al. 2010; Poe and Tedder 2012). More comprehensive perspectives of Siglecs and their functions can be found in these excellent reviews (Crocker et al. 2001, 2007; Crocker 2005; Crocker and Redelinghuys 2008; von Gunten and Bochner 2008; O'Reilly and Paulson 2009; Pillai et al. 2012).
Siglec-G/10 expression
Siglec-G is a member of the CD33-related Siglec family in the mouse. The Siglecg gene is found in a cluster with genes of the related proteins Siglec-E, CD33 and Siglec-F on mouse chromosome 7 (Angata et al. 2001). Siglec-G has five extracellular Ig domains, a transmembrane region and an intracellular tail with three tyrosine-based motifs, among them one ITIM and one Grb-2-binding motif (Hoffmann et al. 2007). Using the expression of GFP to report transcription of Siglec-G in GFP-knock-in mice revealed that Siglec-G is widely expressed in multiple lineages (Ding et al. 2007). All the major known B-cell subsets, B-1a, B-1b, FOB, MZB and Pre-Pro/Pro B cells expressed high levels of Siglecg. However, significant levels were also detected in DC, myeloid cells, and to a lesser extent, T cells. This observation is consistent with the report by Hoffmann et al. (2007) which measured Siglecg transcript levels by reverse-transcriptase polymerase chain reaction and confirmed with an intracellular tail-specific antibody. Recently, Pfrengle et al. (2013) investigated cell surface expression of Siglec-G on murine leukocytes by using a mAb toward the extracellular portion of Siglec-G. They found that among splenic leukocytes, Siglec-G is expressed at highest levels on B cells and, to a lesser extent, on dendritic cells (DCs) and a subset of macrophages and neutrophils. Significant Siglec-G expression appears early on in B-cell development, as early as pre-pro and immature B cells. All three studies showed that the Siglecg locus is transcribed in essentially all the major hematopoietic cell types analyzed, although B cells, as a group, appear to have the highest levels. Siglec-10 (human homolog of mouse Siglec-G) was detected on all B cells and also subsets of human leukocytes including eosinophils, monocytes and a minor population of natural killer cells, by using antibody staining (Munday et al. 2001). High levels of mRNA expression were observed in peripheral blood leukocytes, spleen and liver as demonstrated by Northern blot analysis (Li et al. 2001; Whitney et al. 2001).
An important, but less investigated, issue is how Siglec-G is regulated under pathological conditions. A recent study demonstrated that Siglec-G is elevated in peritoneal cells after infection by RNA viruses (Chen et al. 2013).
Siglec-G/10 in adaptive and innate immunity
Siglec-G/10 in B-cell development
Deletion of Siglecg selectively expands the B1a B-cell lineage, including the frequency of the B1-cell progenitor in the bone marrow and the number of B1a cells in the peritoneal cavity (Ding et al. 2007; Hoffmann et al. 2007). The expansion of B1a B cells in the peritoneum is a cell-intrinsic effect and is correlated with enhanced activation of NFκB (Ding et al. 2007). Lower level of spontaneous apoptosis and prolonged life span of Siglecg−/− B1a B cells might contribute to the expansion of B1a B cells in Siglecg−/− mice. The lower apoptosis could result from higher expression levels of the transcription factor NFATc1 in Siglec-G-deficient B1a cells (Jellusova, Duber, et al. 2010). Intriguingly, steroid- and xenobiotic receptor (SXR)-deficient mice also showed expansion of the B1a B-cell lineage (Casey et al. 2011). Notably, the expression of Siglec-G and PTPN6 were downregulated in SXR−/− mice (Casey et al. 2011). Peritoneal B1a B cells are severely reduced in IgHEµGFP/Eµ-GFP mice (Brenner et al. 2011), but the number of B1a B cells returned to normal levels in Siglecg−/− IgHEµGFP/Eµ-GFP double knockout mice. These data demonstrate that by regulating BCR signaling strength, Siglec-G directly impacts the development of the distinct peritoneal B-cell subset (Brenner et al. 2011).
Siglec-G/10 in B-cell tolerance
Sialylated glycans are ubiquitously expressed in nearly all animal tissues, but largely absent from most microbes, with the exception of some pathogenic strains. This difference in sialylated glycan expression between pathogen and host has suggested that B-cell-restricted Siglecs may play an important role in B-cell self–nonself discrimination. Both CD22 and Sigelc-G are able to recognize synthetic T-independent type 2 (TI-2) antigens harboring sialic acid motifs (Duong et al. 2010). NeuGcα2-6Galβ1-4GlcNac (NeuGc) conjugated to polyacrylamide (PA), the synthetic sialylated T-independent type 2 antigens (TI-2 Ag), bound well to B cells from wild-type mice. However, PA-NeuGc binding was reduced on B cells from either Cd22−/− or Siglecg−/− mice, and was undetectable on B cells from Cd22−/−Siglecg−/− mice (Duong et al. 2010), indicating CD22 and Siglec-G cooperatively mediate the binding of native sialylated ligands. To test whether self TI-2 antigens could induce B-cell tolerance via CD22 and Siglec-G, the hapten nitrophenol (NP) was conjugated to synthetic TI-2 antigens. These conjugates included NP-PA, NP-PA-NeuGc and NP-PA-bNeuGc, which carried an additional modification by the molecule biphenyl-acetyl which appears to selectively augment CD22-binding activity. NP-specific IgM and IgG3 responses were robust for NP-PA, but negligible for NP-PA-bNeuGc, and significantly reduced for NP-PA-NeuGc when the wild-type mice were immunized with these synthetic TI-2 antigens. Siglecg−/− mice generated similar, robust IgM and IgG3 anti-NP response to both NP-PA and NP-PA-NeuGc. In wild-type and Cd22−/− mice, however, IgM and IgG3 responses to NP-PA-NeuGc immunization were reduced relative to NP-PA, suggesting that Siglec-G played a dominant inhibitory role in controlling the immune response to this synthetic NeuGc ligand (Duong et al. 2010).
Interestingly, for both T-dependent and T-independent antigens, incorporation of the NeuGc ligand not only reduced the primary response but also prevented the immune response to subsequent antigen challenge. Therefore, NeuGc coadministration leads to B-cell immune tolerance. Since Siglecg deletion is necessary and sufficient to abrogate tolerance induction, Siglec-G plays a critical role for B-cell tolerance induced by coadministration of Siglec ligands (Pfrengle et al. 2013).
Siglec-G/10 and diseases
Tissue injury
During tissue injury, intracellular components are released. The molecules capable of triggering inflammation are called DAMPs. The DAMPs trigger the innate immune response through TLR and NLR (Dostert et al. 2008). HMGB1 and the heat-shock proteins HSP70/90 are the most intensively-studied DAMPs, and are capable of inducing significant inflammatory responses (Millar et al. 2003; Apetoh et al. 2007). The function of Siglecs in the host response to tissue injuries was discovered during study of the function of CD24, which is a natural ligand for CD33-like Siglecs on host cells (Chen et al. 2009). Subsequently, other pathogen-associated (Carlin et al. 2009) and host cell-associated ligands (Bandala-Sanchez et al. 2013; McMillan et al. 2013) for CD33-like (or CD33 family) Siglecs were identified.
At doses that are significantly higher than what is normally used for pain relief, acetaminophen causes release of HMGB1 from hepatocytes and limited inflammation in the liver (Scaffidi et al. 2002). Interestingly, mice with targeted mutation of CD24 are extremely sensitive to liver injury and are more susceptible to develop “cytokine storm,” producing higher level of cytokines than the control wild-type mice (Chen et al. 2009). Using mass spectrometry, we identified several CD24-associated DAMPs, including HMGB1, Hsp70/90 and nucleolin. The functional significance of HMGB1 in CD24-regulated tissue injuries was confirmed by neutralization with a high-affinity anti-HMGB1 antibody.
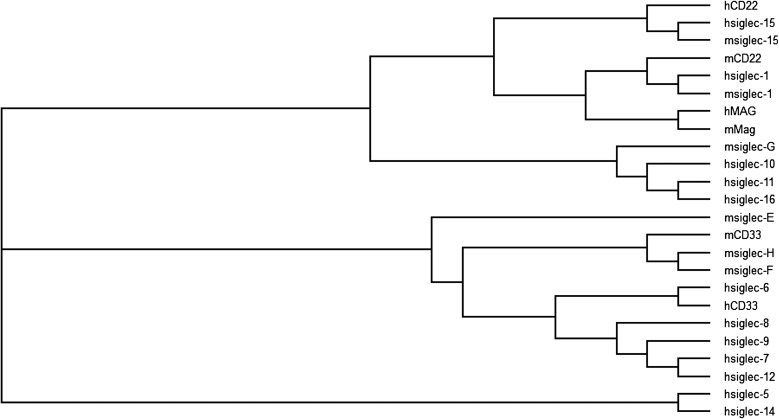
CD24 is a small glycosyl-phosphatidyl-inositol-anchored molecule with broad expression in hematopoietic, neuronal and epithelial cells. Both native CD24 expressed in the brain and recombinant CD24 are heavily sialylated. To test whether CD24 interacts with Siglecs, we tested if recombinant human Siglec-Fc fusion proteins can capture CD24 expressed on spleen cells. We found that CD24 can bind to Siglec-10 but not to Siglec-5, -7 and -11 (Chen et al. 2009). The association of CD24 with Siglec-10 (human homolog of mouse Siglec-G) depends on the critical arginine residue in the V-set domain of Siglec-10 that is involved in sialic acid recognition. Phylogenetic analysis of the sialic acid binding Ig domain of the entire siglec family was performed by the ClustalW (Thompson et al. 1994) and TreeView (Page 1996) programs. The results revealed a close association between Siglec-G and Siglec-10, -11 and -16 (Figure 1), indicating that Sigelc-G and Siglec-10 have similar ligand-binding activity. Analysis of the glycan array data in CFG (US consortium for Functional Glycomics) for Siglec-G (Bochner et al. 2005) and Siglec-10 (Blixt et al. 2008) indicated that Siglec-G binds to ligands carrying α2-3- or α2-6-linkages with similar affinity, while Siglec-10 also binds to the ligands carrying α2-3- or α2-6-linkages, but prefers α2-6-linkages. Indeed, these ligand specificities for the Siglec-G and Siglec-10 were also confirmed by published reports: Siglec-G recognizes Sia ligands carrying both α2-3- and α2-6-linkage with similar affinity (Duong et al. 2010); Siglec-10 recognizes Sia ligands carrying α2-3- or α2-6-linkage, but prefers α2-6-linkage (Chen et al. 2011). In addition, treatment of recombinant CD24 with sialidase abrogated interactions between recombinant Siglec-10 and CD24 and resialylation of CD24 with either 2,3 or 2,6 sialyltransferase restored CD24-Siglec-10 interaction (Chen et al. 2011). Importantly, targeted mutation of CD24 significantly reduced Siglec-10 binding to mouse spleen cells, which suggests that CD24 is likely the dominant ligand for Siglec-10/G on the hematopoietic cells (Chen et al. 2009).
Fig. 1.
Phylogenetic analysis of the Siglec family. The sialic acid binding Ig V-set domain was aligned to create the phylogenetic tree using ClustalW (Thompson et al. 1994) and TreeView (Page 1996).
Siglec-G/10 forms a complex with HMGB1 through CD24. The significance of the CD24-Siglec-G/10 interaction is demonstrated by the fact that mice with targeted mutation of Siglecg phenocopies Cd24−/− mice in the acetaminophen-induced liver injury model. The negative regulation is reflected at the level of DCs, as Cd24−/− and Siglecg−/− DC exhibit significantly enhanced responses to HMGB1 (Chen et al. 2009).
Sepsis
Sepsis is generally defined as a systemic inflammatory syndrome in response to infection. By using the cecal ligation and puncture (CLP) model, which mimics the pathophysiological changes in human sepsis (Rittirsch et al. 2009), we found that sialidases from bacteria can remove sialic acid residues from CD24, and abrogate CD24-Siglec-10(G) interactions, and thus enhance the inflammatory process (Chen et al. 2011). Mice deficient in either CD24 or Siglec-G exhibit dramatically increased mortality and production of inflammatory cytokines after CLP treatment. These observations were also confirmed by administration of the sialidase-producing Streptococcus pneumonia D39 to mice, which induced a stronger inflammatory response than a mutant lacking its two sialidase genes (Chen et al. 2011). A combination of two sialidase inhibitors, Neu5Ac2en and Neu5Gc2en, was able to protect wild-type mice, but not CD24 knockout or Siglecg knockout mice, from sepsis, and inhibited the production of inflammatory cytokines and subsequent morbidity (Chen et al. 2011). These results suggest that the protection of the sialidase inhibitors in bacteria-induced sepsis depends on the integrity of the CD24-Siglec-10(G) pathway.
Immune evasion by RNA virus
TLR3, 7, 8 and 9 detect microbial DNA and RNA in the endosome, whereas retinoic acid-inducible gene 1 (RIG-I) and MDA5, members of RLR, sense viral RNAs in the cytoplasm (Kato et al. 2011). Recently, Chen et al. reported that Siglec-G plays a vital role in negatively regulating the RIG-I-mediated antiviral innate immune response. Siglec-G expression was significantly and rapidly induced in peritoneal macrophages after infection with vesicular stomatitis virus (VSV), but not by infection with DNA virus herpes simplex virus I, intracellular bacteria Listeria monocytogenes or extracellular Gram-negative bacteria Escherichia coli. SiRNA knockdown of Siglecg in macrophages increased the VSV-induced IFN-β expression (Chen et al. 2013). More Siglecg−/− mice survived after challenge with VSV, indicating that Siglecg−/− mice are more resistant to VSV infection. Siglecg−/− mice produced higher levels of IFN-β than wild-type control mice, but not TNF-α or IL-6 (Chen et al. 2013). That Siglec-G selectively suppresses production of IFN-β and inhibits antiviral innate responses against viral infection is achieved by selectively enhancing the phosphorylation of IRF3 and IKK but not p65, p38, ERK and JNK (Chen et al. 2013). Siglec-G negatively regulates RNA-virus-induced IRF3 activation by promoting c-Cbl-mediated ubiquitination and degradation of RIG-I via SHP2 to regulate RNA-virus-induced type I IFN production (Chen et al. 2013). Since elevation of Siglecg caused reduced type I interferon production, it was suggested that this elevation contributes to viral evasion of host resistance.
Perspective
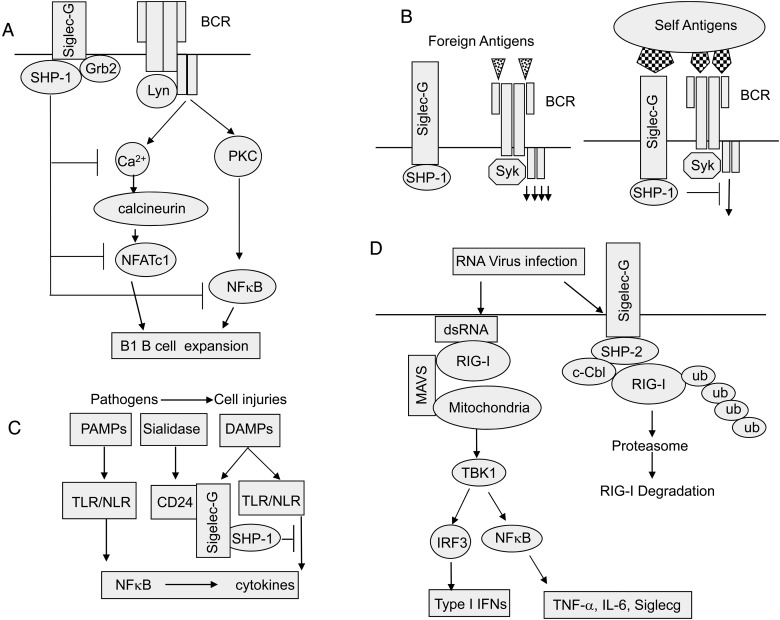
Taken together, accumulating data demonstrate that Siglec-G/10 plays an important role in self–nonself discrimination of the immune system and may be involved in evasion of host immunity by RNA viruses (Figure 2). These data provide several insights with implications for the biology of all Siglec family members.
Fig. 2.
Siglec-G in adaptive and innate immunity. (A) Regulation of B1a B-cell proliferation and BCR signaling by Siglec-G. Siglec-G dampens the calcium signal on B1 cells and inhibits the activity of the transcription factor NFATc1 and NFκB by recruiting the ITIM-binding protein SHP-1 or Grb2. (B) Regulation of B-cell activation and tolerance. Self-antigens are expressed on cells with abundance of Siglec-G ligands and thus likely engage both BCR and Siglec-G. In contrast, foreign antigens are presented to B cells in the absence of Siglec-G ligands. (C) Siglec-G represses host response to DAMPs unless its ligand CD24 is desialylated by bacterial sialidase. (D) Siglec-G suppresses type I IFN production by promoting c-Cbl-mediated ubiquitination and proteasomal degradation of RIG-I.
First, by regulating BCR signaling, Siglec-G regulates both expansion of B1a B cells and tolerance of B cells. It is of note that the role for Siglec-G in B-cell tolerance has only been demonstrated when antigen is administrated with artificial Siglec ligands. Further studies will be needed to determine whether Siglec-G plays a role in B-cell tolerance to endogenous antigens. Since many cell types express high-level Siglec-G ligands, it is of great interest to determine whether these natural ligands may regulate self-tolerance. In this context, it should be noted that CD24, the major Siglec-G/10 ligand, has been shown to be involved in clonal deletion of CD4T cells by self-antigen (Carl et al. 2008). A role for Siglec-10/G in T-cell regulation has also been suggested recently (Bandala-Sanchez et al. 2013).
Second, by selectively affecting innate immune response to DAMPs but not PAMPs, Siglec-G may play a unique role in allowing the innate immune system to sense infection vs. tissue injuries. This provides a plausible mechanism for the host to discriminate between infectious nonself from noninfectious self as suggested by Janeway ∼25 years ago (Janeway 1989, 1992).
Third, although Siglec-G/10 does not directly sense PAMPs, infection can potentially disarm the negative regulation through desialylation of its natural ligands. Many pathogens, including viruses and bacteria, encode sialidases, some of which have been shown to be virulence factors. Disarming sialoside-based negative regulation may allow to host to mount stronger inflammation to tissue injury in the context of infection. In this regard, it is worth noting that although sensitivity to sialidases has long been a gold standard for validating Siglec interactions, the importance of sialidases in Siglec function is only beginning to be appreciated. Meanwhile, these sialidases may be valuable therapeutic targets for taming inflammation.
Finally, identification of natural ligands for Siglecs has helped to shed light on their biological significance. Traditionally, Siglec ligands are identified through synthetic glycans. While these analyses were instrumental in establishing the specificity of Siglecs and yield valuable probes for Siglec biology, recent identification of natural ligands either on the host cells or on pathogens provide a foundation for a better understanding of Siglec function. Additional emphases on natural ligands may be warranted.
Abbreviations
CLP, cecal ligation and puncture; DAMPs, danger (damage)-associate molecular patterns; DC, dendritic cell; HMGB1, high-mobility group box 1 protein; HSP70/90, heat-shock proteins 70/90; ITIM, immunoreceptor tyrosine-based inhibition motif; Neu5Ac, N-acetylneuraminic acid; Neu5Gc, N-glycolylneuraminic acid; NeuGc, NeuGcα2-6Galβ1-4GlcNac; NP, nitrophenol; PA, polyacrylamide; PAMPs, pathogen-associated molecular patterns; RIG-I, retinoic acid-inducible gene 1; RLR, RIG-I-like receptors; TI-2 Ag, T-independent type 2 antigens; TLRs, Toll-like receptors; VSV, vesicular stomatitis virus.
Conflict of interest statement
None declared.
Funding
The work was supported by startup package to Dr Guo-Yun Chen from Children's National Medical Center and by National Institutes of Health (AI105727 to G.Y.C., AI64350 to Y. L., AG036690 to P.Z. and F32AI096730 to N.K.B.).
Acknowledgement
We thank our collaborators, particularly Drs Jie Tang, Xi Chen and Samantha King, for the studies described herein.
References
- Andersson U, Harris HE. The role of HMGB1 in the pathogenesis of rheumatic disease. Biochim Biophys Acta. 2010;1799:141–148. doi: 10.1016/j.bbagrm.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Angata T, Hingorani R, Varki NM, Varki A. Cloning and characterization of a novel mouse Siglec, mSiglec-F: Differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem. 2001;276:45128–45136. doi: 10.1074/jbc.M108573200. [DOI] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Bandala-Sanchez E, Zhang Y, Reinwald S, Dromey JA, Lee BH, Qian J, Bohmer RM, Harrison LC. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat Immunol. 2013;14:741–748. doi: 10.1038/ni.2610. [DOI] [PubMed] [Google Scholar]
- Blixt O, Han S, Liao L, Zeng Y, Hoffmann J, Futakawa S, Paulson JC. Sialoside analogue arrays for rapid identification of high affinity siglec ligands. J Am Chem Soc. 2008;130:6680–6681. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem. 2005;280:4307–4312. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- Brenner S, Drewel D, Steinbart T, Weisel F, Hartel E, Potzsch S, Welzel H, Brandl A, Yu P, Mudde GC, et al. A hypomorphic IgH-chain allele affects development of B-cell subsets and favours receptor editing. EMBO J. 2011;30:2705–2718. doi: 10.1038/emboj.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl JW, Jr, Liu JQ, Joshi PS, El-Omrani HY, Yin L, Zheng X, Whitacre CC, Liu Y, Bai XF. Autoreactive T cells escape clonal deletion in the thymus by a CD24-dependent pathway. J Immunol. 2008;181:320–328. doi: 10.4049/jimmunol.181.1.320. [DOI] [PubMed] [Google Scholar]
- Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113:3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Nelson EL, Turco GM, Janes MR, Fruman DA, Blumberg B. B-1 cell lymphoma in mice lacking the steroid and xenobiotic receptor, SXR. Mol Endocrinol. 2011;25:933–943. doi: 10.1210/me.2010-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Chen X, King S, Cavassani KA, Cheng J, Zheng X, Cao H, Yu H, Qu J, Fang D, et al. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 2011;29:428–435. doi: 10.1038/nbt.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Han C, Xie B, Hu X, Yu Q, Shi L, Wang Q, Li D, Wang J, Zheng P, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152:467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323:1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, Paabo S, Satta Y, Takahata N, Varki A. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci USA. 2002;99:11736–11741. doi: 10.1073/pnas.182257399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR. Siglecs in innate immunity. Curr Opin Pharmacol. 2005;5:431–437. doi: 10.1016/j.coph.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Redelinghuys P. Siglecs as positive and negative regulators of the immune system. Biochem Soc Trans. 2008;36:1467–1471. doi: 10.1042/BST0361467. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Varki A, Wellcome T, Biocentre T, Sciences L. Siglecs in the immune system. Immunology. 2001;103:137–145. doi: 10.1046/j.0019-2805.2001.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Liu Y, Wang Y, Park BK, Wang C-Y, Zheng P, Liu Y. Siglecg limits the size of B1a B cell lineage by down-regulating NFkappaB activation. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000997. e997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, Ota M, Kubitz M, Bovin N, Paulson JC, et al. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. 2010;207:173–187. doi: 10.1084/jem.20091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry H, Crockford TL, Silver K, Rust N, Goodnow CC, Cornall RJ. Analysis of Lyn/CD22 double-deficient B cells in vivo demonstrates Lyn- and CD22-independent pathways affecting BCR regulation and B cell survival. Eur J Immunol. 2005;35:3655–3663. doi: 10.1002/eji.200535247. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Kerr S, Jellusova J, Zhang J, Weisel F, Wellmann U, Winkler TH, Kneitz B, Crocker PR, Nitschke L. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat Immunol. 2007;8:695–704. doi: 10.1038/ni1480. [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Nunez G. Nods: A family of cytosolic proteins that regulate the host response to pathogens. Curr Opin Microbiol. 2002;5:76–80. doi: 10.1016/s1369-5274(02)00289-8. [DOI] [PubMed] [Google Scholar]
- Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Janeway CA., Jr The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- Jellusova J, Duber S, Guckel E, Binder CJ, Weiss S, Voll R, Nitschke L. Siglec-G regulates B1 cell survival and selection. J Immunol. 2010;185:3277–3284. doi: 10.4049/jimmunol.1001792. [DOI] [PubMed] [Google Scholar]
- Jellusova J, Wellmann U, Amann K, Winkler TH, Nitschke L. CD22 x Siglec-G double-deficient mice have massively increased B1 cell numbers and develop systemic autoimmunity. J Immunol. 2010;184:3618–3627. doi: 10.4049/jimmunol.0902711. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: Cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang W, Wan T, Zhang J, Chen T, Yu Y, Wang J, Cao X. Cloning and characterization of Siglec-10, a novel sialic acid binding member of the Ig superfamily, from human dendritic cells. J Biol Chem. 2001;276:28106–28112. doi: 10.1074/jbc.M100467200. [DOI] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- McMillan SJ, Sharma RS, McKenzie EJ, Richards HE, Zhang J, Prescott A, Crocker PR. Siglec-E is a negative regulator of acute pulmonary neutrophil inflammation and suppresses CD11b beta2-integrin-dependent signaling. Blood. 2013;121:2084–2094. doi: 10.1182/blood-2012-08-449983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Meesmann HM, Fehr E-M, Kierschke S, Herrmann M, Bilyy R, Heyder P, Blank N, Krienke S, Lorenz H-M, Schiller M. Decrease of sialic acid residues as an eat-me signal on the surface of apoptotic lymphocytes. J Cell Sci. 2010;123:3347–3356. doi: 10.1242/jcs.066696. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Millar DG, Garza KM, Odermatt B, Elford AR, Ono N, Li Z, Ohashi PS. Hsp70 promotes antigen-presenting cell function and converts T-cell tolerance to autoimmunity in vivo. Nat Med. 2003;9:1469–1476. doi: 10.1038/nm962. [DOI] [PubMed] [Google Scholar]
- Mittal D, Saccheri F, Venereau E, Pusterla T, Bianchi ME, Rescigno M. TLR4-mediated skin carcinogenesis is dependent on immune and radioresistant cells. EMBO J. 2010;29:2242–2252. doi: 10.1038/emboj.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday J, Kerr S, Ni J, Cornish AL, Zhang JQ, Nicoll G, Floyd H, Mattei MG, Moore P, Liu D, et al. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem J. 2001;355:489–497. doi: 10.1042/0264-6021:3550489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke L. CD22 and Siglec-G: B-cell inhibitory receptors with distinct functions. Immunol Rev. 2009;230:128–143. doi: 10.1111/j.1600-065X.2009.00801.x. [DOI] [PubMed] [Google Scholar]
- O'Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science. 1996;274:798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- O'Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–248. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pfrengle F, Macauley MS, Kawasaki N, Paulson JC. Copresentation of antigen and ligands of Siglec-G induces B cell tolerance independent of CD22. J Immunol. 2013;191:1724–1731. doi: 10.4049/jimmunol.1300921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilatte Y, Tisserand EM, Greffard A, Bignon J, Lambre CR. Anticarbohydrate autoantibodies to sialidase-treated erythrocytes and thymocytes in serum from patients with pulmonary sarcoidosis. Am J Med. 1990;88:486–492. doi: 10.1016/0002-9343(90)90427-f. [DOI] [PubMed] [Google Scholar]
- Pillai S, Cariappa A, Pirnie SP. Esterases and autoimmunity: The sialic acid acetylesterase pathway and the regulation of peripheral B cell tolerance. Trends Immunol. 2009;30:488–493. doi: 10.1016/j.it.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, Netravali IA, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe JC, Tedder TF. CD22 and Siglec-G in B cell function and tolerance. Trends Immunol. 2012;33:413–420. doi: 10.1016/j.it.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogentine GN, Jr, Plocinik BA. Carbohydrate inhibition studies of the naturally occurring human antibody to neuraminidase-treated human lymphocytes. J Immunol. 1974;113:848–858. [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- Springer GF. T and Tn, general carcinoma autoantigens. Science. 1984;224:1198–1206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Fukushima S, Yamahara K, Yashiro K, Shintani Y, Coppen SR, Salem HK, Brouilette SW, Yacoub MH, Suzuki K. Modulated inflammation by injection of high-mobility group box 1 recovers post-infarction chronically failing heart. Circulation. 2008;118:S106–S114. doi: 10.1161/CIRCULATIONAHA.107.757443. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, Bianchi ME, Kirschning C, Wagner H, Manfredi AA, et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: Implications for the pathogenesis of SLE. J Exp Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaith P, Uhlenbruck G. The Thomsen agglutination phenomenon: A discovery revisited 50 years later. Z Immunitatsforsch Immunobiol. 1978;154:1–15. [PubMed] [Google Scholar]
- Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- Varki A, Angata T. Siglecs – the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- von Gunten S, Bochner BS. Basic and clinical immunology of Siglecs. Ann N Y Acad Sci. 2008;1143:61–82. doi: 10.1196/annals.1443.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- Whitney G, Wang S, Chang H, Cheng KY, Lu P, Zhou XD, Yang WP, McKinnon M, Longphre M. A new siglec family member, siglec-10, is expressed in cells of the immune system and has signaling properties similar to CD33. Eur J Biochem. 2001;268:6083–6096. doi: 10.1046/j.0014-2956.2001.02543.x. [DOI] [PubMed] [Google Scholar]