Abstract
Uromodulin is expressed exclusively in the thick ascending limb and is the most abundant protein excreted in normal urine. Variants in UMOD, which encodes uromodulin, are associated with renal function, and urinary uromodulin levels may be a biomarker for kidney disease. However, the genetic factors regulating uromodulin excretion are unknown. We conducted a meta-analysis of urinary uromodulin levels to identify associated common genetic variants in the general population. We included 10,884 individuals of European descent from three genetic isolates and three urban cohorts. Each study measured uromodulin indexed to creatinine and conducted linear regression analysis of approximately 2.5 million single nucleotide polymorphisms using an additive model. We also tested whether variants in genes expressed in the thick ascending limb associate with uromodulin levels. rs12917707, located near UMOD and previously associated with renal function and CKD, had the strongest association with urinary uromodulin levels (P<0.001). In all cohorts, carriers of a G allele of this variant had higher uromodulin levels than noncarriers did (geometric means 10.24, 14.05, and 17.67 μg/g creatinine for zero, one, or two copies of the G allele). rs12446492 in the adjacent gene PDILT (protein disulfide isomerase-like, testis expressed) also reached genome-wide significance (P<0.001). Regarding genes expressed in the thick ascending limb, variants in KCNJ1, SORL1, and CAB39 associated with urinary uromodulin levels. These data indicate that common variants in the UMOD promoter region may influence urinary uromodulin levels. They also provide insights into uromodulin biology and the association of UMOD variants with renal function.
CKD is a major public health problem affecting 5%–10% of adults worldwide,1,2 and is associated with increased risks of cardiovascular morbidity and mortality.3,4 Although hypertension and diabetes are established risk factors for developing CKD,5,6 they do not fully identify individuals at risk for this condition.7 Therefore, further exploration of novel biomarkers for early detection and development of CKD beyond the current measures of kidney function and damage is necessary.8
Uromodulin (Tamm–Horsfall protein), the most abundant protein excreted in the urine, is increasingly considered as a potential biomarker for CKD.9–11 Uromodulin is exclusively produced in the epithelial cells lining the thick ascending limb (TAL) of the loop of Henle, where it is sorted to the apical plasma membrane and released into the tubular lumen by proteolytic cleavage. Uromodulin is an abundant transcript in the TAL cells, and its rate of secretion in the urine ranges from 20 to 100 mg/d in physiologic conditions.12 Recent genome-wide association studies (GWAS)13–15 and sequencing efforts16 have identified common single-nucleotide polymorphism (SNP) variants in the UMOD gene in association with the eGFR and the risk of developing CKD.14 On the other hand, mutations in UMOD cause dominantly inherited forms of tubulointerstitial kidney disease collectively referred to as uromodulin-associated kidney disease (UAKD).17 These disorders include defective urinary concentrating ability, hyperuricemia, gout, and progression to ESRD between the second and fourth decades of life. Analyses of renal biopsies and in a transgenic mouse model of UAKD revealed that UAKD is coupled to a significant reduction of uromodulin excretion in the urine. Studies in UMOD knockout mice showed that uromodulin may play a role in the transport processes operating in the TAL18,19 and protects against urinary tract infection20 and formation of kidney stones.21
Despite these data, obtained essentially in mouse or cellular models, the biologic function of uromodulin is still undefined and the genetic contribution to common variation in uromodulin production and release remains to be determined. Thus, this study aimed to investigate the genetic association of urinary uromodulin levels in the general population. We performed a meta-analysis of GWAS for urinary uromodulin concentration in six studies including three urban cohorts and three genetic isolates amounting to 10,884 participants of European ancestry. We also used a candidate gene approach to seek whether urinary uromodulin concentrations are associated with variants in genes regulating transport processes in the TAL.
Results
Table 1 shows study sample characteristics for 10,884 participants from all six cohorts (Cohort Lausannoise [CoLaus], Croatia-Korcula, Croatia-Split, Framingham Heart Study, Network Italiano Isolati Genetici (INGI)-Carlantino, and INGI-Val Borbera) with urinary uromodulin levels. Detailed information on samples, assays, genotyping, and imputation platforms are included in Supplemental Tables 1 and 2.
Table 1.
Study sample size characteristics
| Study | Population Type | Sample Size (n) | Women | Age (yr) | BMI (kg/m2) | eGFR Cr (ml/min per 1.73 m2) | HTN | DM | UMOD Indexed to UCr (mg/g Cr) | UMOD Unindexed (μg/ml) |
|---|---|---|---|---|---|---|---|---|---|---|
| CoLaus | Nonisolate urban | 5077 | 53.5 (2716) | 53.3±10.7 | 25.7±4.5 | 90.1± 19.5 | 36.0 (1830) | 4.5 (229) | 18.4 (3.6–46.7) | 25.8 (4.9–73.7) |
| Croatia-Korcula | Isolate | 887 | 63.7 (565) | 56.3±13.9 | 28.0±4.1 | 91.3±29 | 55.9 (496) | 14.2 (126) | 6.4 (1.37–27.98) | 6.9 (2.28–26.85) |
| Croatia-Split | Nonisolate urban | 488 | 56.6 (276) | 49±14.7 | 26.9±4.2 | 95.6±23.8 | 35.2 (172) | 3.2 (16) | 20.6 (3.54–48.82) | 28.6 (5.05–88.68) |
| Framingham Heart Study | Nonisolate urban | 2640 | 53.2 (1404) | 58.4±9.6 | 27.9±5.1 | 87.5±25 | 40.0 (1055) | 9.4 (249) | 9.5 (0.3–27) | 7.42 (0.30–33) |
| INGI-Carlantino | Isolate | 360 | 56.9 (205) | 48.1±20.4 | 26.7±5.9 | 94.8±25.7 | 40.4 (122) | 10.0 (29) | 6.7 (1.44–29.43) | 5.09 (0.97–20.74) |
| INGI-Val Borbera | Isolate | 1432 | 55.7 (797) | 54.1±18.3 | 25.9±4.5 | 88.9±22.4 | 41.4 (593) | 6.6 (95) | 8 (1.54–34.54) | 6.75 (1.52–30.37) |
Data are presented as mean±SD for continuous variables, % (n) for categorical variables, or median (5th percentile, 95th percentile). BMI, body mass index; Cr, creatinine; HTN, hypertension; DM, diabetes mellitus; UCr, urine creatinine.
Heritability measures performed in the Framingham Heart Study showed that uromodulin levels are heritable at 17% (P=0.003), whereas the Croatia-Korcula study showed heritability of 28%.
Variants Identified through Meta-Analysis of GWAS
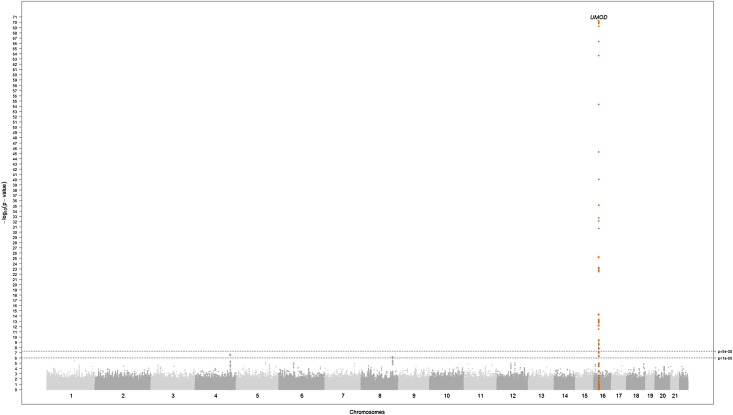
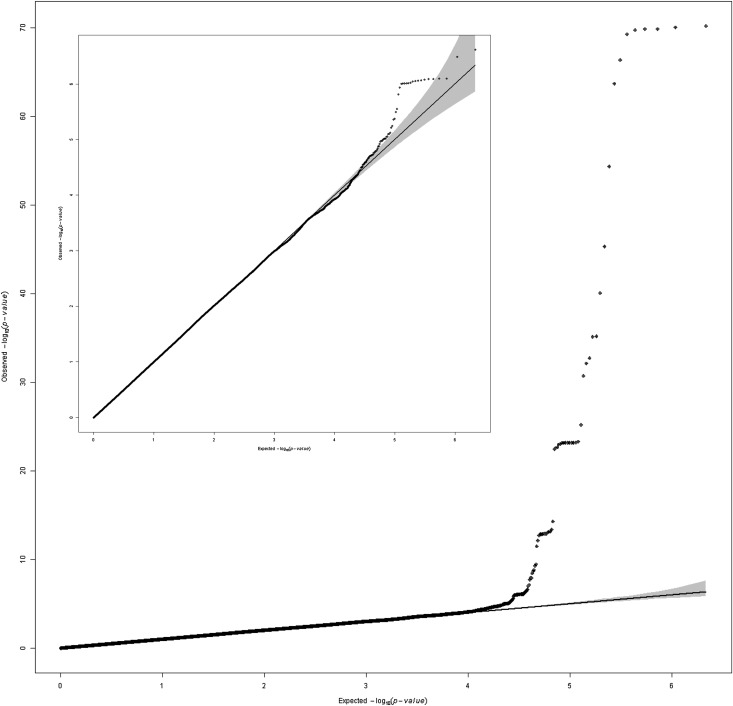
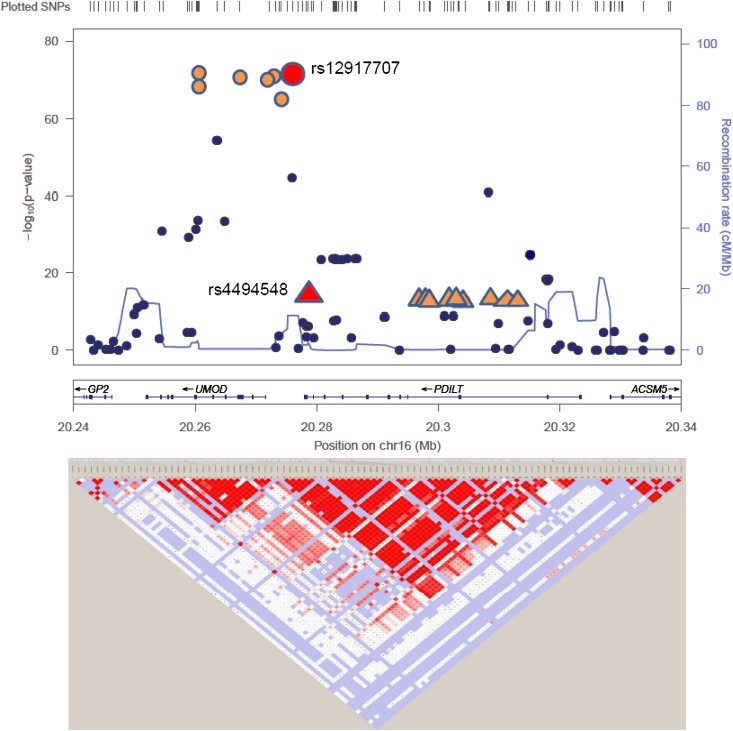
Figure 1 displays P values from the meta-analysis of GWAS of urinary uromodulin indexed to urinary creatinine on a −log10 scale versus their genomic position. A genome-wide significant signal (P<1E-72) was present on chromosome 16 in and near the UMOD gene, followed by two loci, which did not reach genome-wide significance. In addition, an excess of low P values was present in the quantile-quantile plot of the –log10 observed versus expected P values (Figure 2), which was still present after removing a 1 Mb–long region around the UMOD gene on chromosome 16 (Figure 2, inset). The regional association plot of UMOD (Figure 3) shows that the urinary uromodulin association signal spreads over several linkage disequilibrium (LD) blocks.
Figure 1.
The −log10(P value) plot by genomic position (Manhattan plot) for results from the meta-analysis of GWAS of uromodulin indexed to creatinine in six population-based studies. The reported locus (P<1E-72) is highlighted in orange. SNPs with minor allele frequency <5% are excluded. P values are double corrected for genomic control.
Figure 2.
Quantile-quantile plot of results from the meta-analysis of GWAS of uromodulin indexed to creatinine in six population-based studies. The inset graph shows the remaining signals after excluding SNPs in and 1 Mb around the UMOD gene. SNPs with minor allele frequency <5% are excluded from the meta-analysis. P values are double corrected for genomic control.
Figure 3.
Regional association plot of UMOD gene for the analysis indexed to urinary creatinine. The lead GWAS hit (rs12917707) and GCTA hit (rs4494548) are highlighted. The plot is modified from LocusZoom49 and Haploview.50 The small figure shows the R2 between the two SNPs highlighted in red. LD information is based on HapMap CEU.
Table 2 presents results from the inverse-variance meta-analysis for urinary uromodulin indexed to urinary creatinine. The variant with the lowest P value (7.85E-73) was rs12917707 located in the 5′ promoter region of the UMOD gene. Each copy of the G allele was associated with high levels of uromodulin in our analyses (P=6.6E-71) and lower levels of eGFR in CKDGen Consortium participants (P=1.2E-20). This SNP was previously reported in association with eGFR and CKD.13 A second variant (rs12446492, within the PDILT gene located near the UMOD gene) showed similar associations with uromodulin (P=6.6E-26) and eGFR (P=6.9E-07). This variant has low R2 but high D′ values (R2=0.17, D′=0.83) with the lead GWAS SNP, rs12917707. The next two independent variants identified through meta-analysis (rs4533720 on chromosome 4 near the MARCH1 gene and rs6988636 on chromosome 8 near the FAM83A gene) did not reach genome-wide significance (P=2.41E-07 and P=7.97E-07, respectively) and were not associated with eGFR in 67,093 individuals in the CKDGen Consortium (P=0.47 and P=0.58, respectively).15,22 A sensitivity meta-analysis of raw unindexed uromodulin (results in Supplemental Table 3) yielded similar values to the ones indexed to creatinine. The association of eGFR with the four variants restricted to the six cohorts included in this study is not significant.
Table 2.
Top loci with P values up to 1E-7 associated with uromodulin indexed to creatinine in the overall population
| SNP ID | Chr | RA | NRA | Effect Size | P Valuea | SEMa | P Valuea | RAF | Nearest Geneb | Imputation Qualityc | eGFR Cr Direction Relative to RA | eGFR Cr P Value | R2 to Top SNP rs12917707 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12917707 | 16 | T | G | −0.32 | 7.85E-73 | 0.02 | 7.85E-73 | 0.18 | UMOD | 0.94 | + | 1.20E-20 | Same SNP |
| rs12446492 | 16 | A | T | −0.15 | 5.60E-27 | 0.01 | 5.60E-27 | 0.45 | PDILT | 0.72 | + | 6.90E-07 | 0.16 |
| rs4533720 | 4 | A | G | 0.07 | 3.50E-7 | 0.01 | 3.50E-7 | 0.53 | MARCH1 | 1 | + | 0.47 | NA |
| rs6988636 | 8 | T | C | −0.13 | 9.47E-7 | 0.03 | 9.47E-7 | 0.92 | FAM83A | 0.99 | − | 0.58 | NA |
Genes nearby were based on RefSeq genes (build 36). Chr, chromosome; RA, reference allele; NRA, nonreference allele; RAF, reference allele frequency; Cr, creatinine; NA, not available.
P value from inverse-variance meta-analysis, corrected for genomic control. Sample size weighted meta-analysis yielded similar results.
The gene closest to the SNP is listed first and is marked bold if the SNP is located within the gene.
Median imputation quality across participating studies.
In the CoLaus cohort (N=5077), stratified analyses for the association of urinary uromodulin indexed to urinary creatinine with rs127917707 by hypertension status (–0.2849±0.0418 [P=9.64E-12] in individuals with hypertension versus −0.3123±0.0318 [P=9.37E-23] in normotensive people; P for difference between strata=0.60) or by age groups (−0.31±0.04 [P=1.58E-17] in people aged≤53 years versus −0.30±0.04 [P=6.88E-17] in younger people; P for between strata difference=0.79) led to similar results. Adding hypertension as a covariate in the model barely affected this same association (−0.2960±0.0354 [P=6.49E-17] including hypertension versus −0.3018±0.0254 [P=1.22E-32] not including hypertension).
Association of rs12917707 Genotypes with Uromodulin Levels
Table 3 presents the association of rs12917707 genotypes with urinary uromodulin levels. The G allele of rs12917707 was consistently associated with higher urinary uromodulin levels in an additive manner. The 2-fold increase in participants harboring the GG genotype was observed in each of the six cohorts investigated, with a meta P value of 2.41E-58.
Table 3.
Association of rs12917707genotypes with urinary uromodulin levels in the six cohorts
| Cohort | UMOD Indexed to UCr (μg/g Cr) | Sample Size (n) | Total Analyzed Sample Size (n) | P Value in Each Cohorta | ||||
|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | TT | TG | GG | |||
| CoLaus | 13.90±8.92 | 18.74±13.08 | 22.80±17.27 | 195 | 1472 | 3410 | 5077 | 2.31E-26 |
| Croatia-Korcula | 4.83±3.63 | 8.07±8.16 | 9.97±10.83 | 12 | 207 | 655 | 874 | 4.97E-03 |
| Croatia-Split | 12.80±4.97 | 17.43±10.03 | 26.09±16.83 | 11 | 123 | 347 | 481 | 9.27E-10 |
| Framingham Heart Study | 5.27 (4.35) | 8.98±7.53 | 12.60±11.88 | 103 | 851 | 1689 | 2643 | 7.37E-25 |
| INGI-Carlantino | 5.67±6.25 | 9.00±8.49 | 11.13±15.2 | 13 | 108 | 220 | 341 | 6.08E-02 |
| INGI- Val Borbera | 6.21±4.31 | 10.27±10.86 | 12.43±11.89 | 26 | 319 | 1028 | 1373 | 1.50E-04 |
Data are presented as mean±SD unless otherwise indicated. UCr, urine creatinine; Cr, creatinine.
The meta P value for the six cohorts is 2.41E-58.
Joint Analyses
Table 4 shows the results from a joint analysis conditioned on the rs12917707 SNP. This analysis revealed another nearby variant in the PDILT gene, rs4494548, in low LD with the conditioned SNP (R2=0.02, D′=0.16), which is associated with uromodulin (meta-analysis, P=3.9E-15; joint conditional analysis, P=2.3E-08). This variant was not significantly associated with eGFR in CKDGen Consortium participants (P=4.3E-04, N=67,093).13,22 Variance analysis using genotype data from the CoLaus study showed that the lead GWAS hit rs12917707 explains 1.2% of the variance of urinary uromodulin levels, with an additional 0.8% variance explained by the second SNP, rs4494548. These results suggest that at least two independent signals for urinary uromodulin exist at the UMOD locus.
Table 4.
Joint conditional analysis for loci associated with urinary uromodulin indexed to creatinine in the overall population
| Chr | SNP | RA | RAF | Meta-Analysisa | Sample Size (n) | GAF | Joint Analysis | R2 to Top SNP rs12917707 | Variance (%)b | eGFR Cr P Value in CKDGen | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect Size | SEM | P Value | Effect | SEM | P Value | |||||||||
| 16 | rs12917707 | T | 0.18 | −0.32 | 0.02 | 7.9E-73 | 10,417 | 0.17 | −0.31 | 0.018 | 3.4E-64 | 1.0 | 1.2 | 1.2E-20 |
| 16 | rs4494548 | A | 0.13 | 0.17 | 0.02 | 3.9E-15 | 10,417 | 0.10 | 0.12 | 0.022 | 2.3E-08 | 0.0 | 0.8 | 4.3E-04 |
Joint analysis of the meta-analysis results for unindexed urinary uromodulin did not yield any additional signal beyond the lead SNP rs12917707. Chr, chromosome; RA, reference allele; RAF, reference allele frequency; GAF, genotype allele frequency; Cr, creatinine.
Values from the inverse-variance meta-analysis.
Linear model using phenotype and genotype data from the CoLaus cohort.
Gene-Based Association Analyses
Results from the Versatile Gene-based Association Study (VEGAS) analysis are included in Supplemental Table 4. This gene-based association analysis identified the region of chromosome 16 containing UMOD, PDILT, GP2, and ACSM5 as being the only region with a statistically significant result (P<2.8×10−6) and identified the top SNP from the meta-analysis (rs12917707, P=7.9E-73) as the main contributor to the finding. No other genes were identified.
Candidate Gene Analyses Results
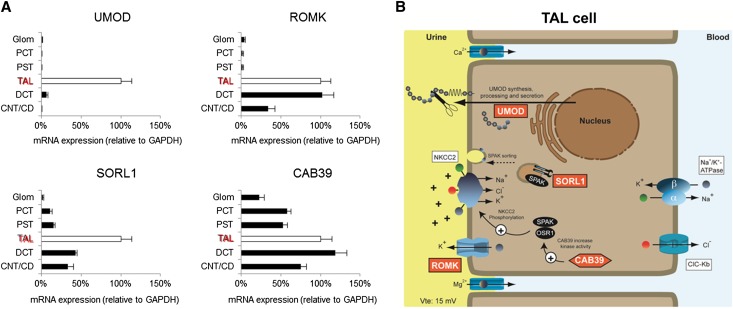
Uromodulin is exclusively synthesized in the epithelial cells lining the TAL (Figure 4). These cells reabsorb approximately 5% of the filtered NaCl and generate a lumen-positive transepithelial voltage. Previous studies suggested that urinary uromodulin concentration reflects the transport activity of the TAL.9,12 The potential association of 24 candidate genes expressed in the TAL and involved in TAL transport processes revealed that SNPs in KCNJ1(ROMK), CAB39, and SORL1 genes were associated (P value lower than the gene-specific threshold) with the level of uromodulin indexed to urinary creatinine (Table 5). Of the other 21 investigated candidate genes, 5 genes were only nominally associated with uromodulin and the rest did not show any association. We calculated the effective number of independent tests of the 1739 SNPs examined in the 24 candidate genes to be 129. When accounting for all candidate SNPs tested, declaring the four most significant SNPs (rs2855800, rs2438298, rs1532763, rs4239217) as true associations would result in a false discovery rate below 75% (Table 5). SNP rs28555800 in KCNJ1 was also associated with eGFR (P=0.02; N=67,093) in previously published CKDGen meta-analysis results.13,22 Expression studies performed on well characterized microdissected tubule segments showed that Kcnj1, Cab39, and Sorl1 were enriched with Umod in the TAL of mouse kidney (Ct values: Kcnj1, 26.43±0.41; Cab39, 30.36±0.44; Sorl1, 28.34±0.40; Umod, 19.71±0.41; n=3 independent experiments performed on tubular fractions pooled from two kidneys), where they are known to be involved in the regulation of NaCl transport activity (Figure 4).
Figure 4.
Candidate gene analysis and nephron segmentation. (A) Expression levels of UMOD, ROMK, SORL1, and CAB39 in microdissected mouse tubule segments (relative to TAL taken as 100%, white bars). Expression levels are relative to reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are means±SEM of at least three independent experiments obtained for pooled tubules from two mouse kidneys. (B) Model for the intersection of ROMK, SORL1, and CAB39 with uromodulin in cells lining the TAL.
Table 5.
Candidate gene analysis for uromodulin indexed to creatinine in the overall population
| Gene Name | Chr | SNPs Found in Gene (n) | LD Blocks in Gene at R2=0.2 (n) | Gene-Specific Threshold | SNPs in Gene with P Value Below Threshold (n) | SNP ID with Lowest P Value in Gene | Lowest P Value in Gene | FDR | eGFR Cr P Value in CKDGen | Gene Category |
|---|---|---|---|---|---|---|---|---|---|---|
| KCNJ1(ROMK)a | 11 | 40 | 5 | 0.010 | 1 | rs2855800 | 0.001 | 0.1914 | 0.023 | Ion transport |
| CAB39a | 2 | 43 | 6 | 0.008 | 1 | rs2438298 | 0.002 | 0.2962 | 0.065 | Regulator, ion transport |
| SORL1a | 11 | 120 | 9 | 0.006 | 3 | rs1532763 | 0.004 | 0.4381 | 0.29 | Regulator, ion transport |
| HNF1B | 17 | 36 | 9 | 0.006 | 0 | rs4239217 | 0.01 | 0.7178 | 0.38 | Transcription factor |
| REN | 1 | 21 | 6 | 0.008 | 0 | rs11571093 | 0.02 | >0.75 | 0.64 | Regulator, ion transport |
| STK39 | 2 | 362 | 15 | 0.003 | 0 | rs2203703 | 0.02 | >0.75 | 0.19 | Regulator, ion transport |
| CLDN14 | 21 | 165 | 12 | 0.004 | 0 | rs4816538 | 0.03 | >0.75 | 0.9 | Paracellular, ion transport |
| CLDN16 | 3 | 56 | 6 | 0.008 | 0 | rs17445530 | 0.03 | >0.75 | 0.69 | Paracellular, ion transport |
| CNNM2 | 10 | 137 | 6 | 0.008 | 0 | rs12769080 | 0.06 | >0.75 | 0.1 | Ion transport |
| SLC9A3 | 5 | 27 | 7 | 0.007 | 0 | rs1035908 | 0.07 | >0.75 | 0.94 | Ion transport |
| CLCNKB | 1 | 27 | 5 | 0.010 | 0 | rs2007471 | 0.07 | >0.75 | 0.21 | Ion transport |
| SLC9A4 | 2 | 168 | 7 | 0.007 | 0 | rs17027258 | 0.07 | >0.75 | 0.45 | Ion transport |
| NFAT5 | 16 | 75 | 6 | 0.008 | 0 | rs1064825 | 0.08 | >0.75 | 0.017 | Transcription factor |
| SLC9A2 | 2 | 81 | 12 | 0.004 | 0 | rs13011360 | 0.11 | >0.75 | 0.49 | Ion transport |
| AGT | 1 | 48 | 5 | 0.010 | 0 | rs3789664 | 0.11 | >0.75 | 0.52 | Regulator, ion transport |
| WNK1 | 12 | 104 | 8 | 0.006 | 0 | rs7137188 | 0.13 | >0.75 | 0.15 | Regulator, ion transport |
| CLCNKA | 1 | 34 | 3 | 0.017 | 0 | rs1889785 | 0.15 | >0.75 | 0.34 | Ion transport |
| CASR | 3 | 75 | 8 | 0.006 | 0 | rs3804593 | 0.15 | >0.75 | 0.65 | Regulator, ion transport |
| OXSR1 | 3 | 19 | 5 | 0.010 | 0 | rs4955408 | 0.22 | >0.75 | 0.75 | Regulator, ion transport |
| SLC12A1 | 15 | 44 | 1 | 0.050 | 0 | rs1843144 | 0.22 | >0.75 | 0.38 | Ion transport |
| CLDN19 | 1 | 8 | 3 | 0.017 | 0 | rs7548008 | 0.31 | >0.75 | 0.73 | Paracellular, ion transport |
| BSND | 1 | 20 | 2 | 0.025 | 0 | rs12134118 | 0.31 | >0.75 | 0.55 | Ion transport |
| WNK4 | 17 | 1 | 1 | 0.050 | 0 | rs873084 | 0.34 | >0.75 | 0.2 | Regulator, ion transport |
| HIF1A | 14 | 28 | 2 | 0.025 | 0 | rs7143164 | 0.43 | >0.75 | 0.19 | Transcription factor |
The table is sorted after lowest P value in the gene. Significance was defined as P value lower than a gene-specific threshold, calculated as 0.05 divided by the number of found LD blocks at R2=0.2 in that gene. Chr, chromosome; FDR, false discovery rate; Cr, creatinine.
Three loci with at least one SNP with a P value below the threshold in the gene. The FDR values <0.75 are provided as an alternative threshold (see the Concise Methods).
Discussion
Our study provides the first meta-analysis of uromodulin levels in urine, including six population-based cohorts amounting to >10,000 individuals of European descent. Our analyses document a robust locus with two independent signals around the UMOD gene (rs12917707 and rs4494548), with rs12917707 included in a LD block encompassing the UMOD promoter. We show that the G allele of the rs12917707 variant, associated with higher risk of CKD in previous studies and with lower eGFR in this study, is associated with up to 2-fold higher levels of uromodulin in urine. This finding is consistent across all cohorts, in both genetic isolates and nonisolated populations. rs4494548 is also associated with urinary uromodulin levels, even upon adjustment by rs12917707. rs4494548 is an intronic SNP of the PDILT gene, which is known to be transiently expressed during urinary tract development and is associated with male infertility.23 Finally, we used a candidate gene approach to show that SNPs in three genes (ROMK, CAB39, and SORL1) known to mediate or regulate transport processes in TAL cells are associated with urinary uromodulin levels. Taken together, our findings suggest that common SNP variants identified through GWAS and conditional analysis may account for variation in urinary uromodulin and eGFR.
Previous GWAS have shown that promoter variants in the UMOD gene in high LD with our top rs12917707 SNP are associated with eGFR and CKD. A first GWAS with nearly 20,000 individuals from population-based studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium identified rs12917707, whereby each copy of the minor allele T was associated with higher levels of eGFR and a lower risk of CKD.24 On the basis of these findings, investigators then measured urinary uromodulin in several nested case-control studies. Among 200 individuals in the Framingham Heart Study, the C allele at rs4293393 (in perfect LD with rs12917707, R2=1) was associated with lower urinary uromodulin levels, and these levels predicted higher eGFR at 10 years.15 In light of these intriguing case-control studies, we sought to measure urinary uromodulin in much larger sample sizes, and to apply genome-wide association to uncover loci in association with urinary uromodulin levels. Our study advances knowledge in this area in several important ways. First, we evaluated the association between genetic variants and urinary uromodulin at a genome-wide level, evidencing two robust independent signals around the UMOD gene. We confirm the strong and consistent association of the promoter variant with an up to 2-fold increase in urinary excretion of uromodulin. That association is detected in each cohort analyzed, either from urban or genetic isolate origin, and (in CoLaus) is not altered by adjustment for hypertension nor upon stratification for hypertension status and for age. Of note, large differences in the median value of urinary uromodulin have been observed across cohorts even when using the same sensitive assay, and they are not related to storage conditions. We do not think that this variation influenced our meta-analysis results, because we first performed study-specific analyses that served to internally calibrate the data. Large variations in the daily physiologic excretion of uromodulin have been reported, probably reflecting the complex secretory pathway of the protein and its potential interactions with multiple urinary components.9 Second, we used a joint conditional analysis to extract other independent signals at the same locus that account for a higher variation of the observed signal. Third, we leveraged our existing urinary uromodulin GWAS data set to interrogate genomic regions suggested through gene expression and functional studies in order to better understand the link between uromodulin and ion transport in the TAL segment.
The potential mechanisms of the association of SNPs (including the top rs12917707) in the promoter of UMOD include potential cis-acting effects that could modulate the transcription of uromodulin by the TAL cells, and thus its excretion in the urine. The dose-dependent effect, which is consistently observed in all cohorts, supports the latter hypothesis, in line with the recent results of Trudu et al. showing an effect of UMOD promoter variants on gene expression.25 The fact that age does not modify the association of urinary uromodulin with rs12917707 may suggest that elevated urinary uromodulin could lead to progressive renal damage. Indeed, evidence obtained in a transgenic mouse model suggest that such deleterious effects of uromodulin would take a long time and it could reflect multiple hits.25 Alternatively, increased urinary uromodulin levels may be secondary to renal damage caused by mechanisms unrelated to uromodulin. This second hypothesis would be in line with a protective effect of urinary uromodulin.9 The mechanism of the SNPs in PDILT is not predictable, although it is unlikely that the association is driven by LD with the UMOD promoter variants.
Our candidate gene analysis revealed that variants in KCNJ1/ROMK, SORL1, and CAB39 are associated with the level of uromodulin in urine. In TAL cells, uromodulin is trafficked to the apical plasma membrane and released into the tubule lumen via proteolytic cleavage. This cleavage is necessary for the polymerization of uromodulin in the urine.9,26 The cells lining the TAL reabsorb approximately 25% of the filtered NaCl, a process that involves the apical, bumetanide-sensitive Na+/K+/2Cl−cotransporter NKCC2, organized in parallel with the renal outer medullary K+ (ROMK) channel, which is essential to recycle K+. Na+ and Cl− are transported across the basolateral membrane via the Na+/K+-ATPase and the Cl− channel ClC-Kb, respectively. These transport processes are highly regulated: the Ste20- and SPS1-related proline and alanine-rich kinase (SPAK) and oxidative stress-responsive kinase (OSR1) activate NKCC2 by N-terminal phosphorylation.27 In turn, SPAK and OSR1 require regulatory cofactors, such as sorting protein-related receptor with A-type repeats 1 (SORL1) and calcium-binding protein 39 (CAB39). CAB39 is a scaffolding protein that interacts with SPAK and OSR1, increasing kinase activation.28 SORL1 was previously shown to mediate SPAK membrane sorting, allowing the phosphorylation of NKCC2 by SPAK.29 The fact that uromodulin expression is limited to the TAL segment in mice and humans30–32 and the enrichment of ROMK, SORL1, and CAB39 in that very segment emphasize the potential link between transport processes and uromodulin secretion in the cells lining the TAL. Furthermore, a functional link between ROMK and uromodulin has already been documented in vitro and in mouse models.18 Future studies will address the biologic mechanisms that underlie these genetic and functional associations.
Our study combines the advantages of the first and large GWAS on urinary uromodulin, performed using a robust assay on various types of well documented cohorts, with an interrogation of candidate genes operating in the very tubular segment that is releasing uromodulin in the urine. Limitations of this study include the availability of data only for individuals of European descent, restricting the generalizability of our findings. The lack of external replication is balanced by the genome-wide significance of the association of rs12917707 in the CoLaus cohort, with replication of this main hit in five independent cohorts. Conversely, the subgenome-wide findings around the FAM83A and MACH1 genes should be interpreted with caution given the absence of replication. In addition, we used common genetic variants (minor allele frequency >5%) and it is possible that there may be other low frequency variants that could additionally explain this association. Future GWAS conducted in larger cohorts and using variants derived from exome sequencing could reveal other genetic associations with urinary uromodulin concentrations.
In conclusion, a meta-analysis of urinary uromodulin concentrations revealed the association of common variants in the promoter region of the UMOD gene, together with a second independent signal located within the flanking gene PDILT. Each copy of the G allele at rs12917707 (risk allele for CKD) was associated with higher urinary uromodulin concentrations. Variants in the KCNJ1, CAB39, and SORL1 genes regulating transport processes in the TAL cells producing uromodulin were also associated with its urinary concentration. These results advance our understanding of the biology of uromodulin and substantiate the association of UMOD variants with renal function.
Concise Methods
Urinary uromodulin and creatinine levels were measured in 10,884 individuals of European descent from six cohorts in both urban and isolate communities (CoLaus, Croatia-Korcula, Croatia-Split, Framingham Heart Study, INGI-Carlantino, and INGI-Val Borbera). All studies performed genome-wide associations for both urinary uromodulin indexed to urinary creatinine and raw urinary uromodulin only, which were then meta-analyzed. Participants provided written informed consent.
Study Samples
The CoLaus study is a population-based study including >6000 people aged 35–75 years from the city of Lausanne, Switzerland, as previously described.33 All participants were of European descent, defined as having both parents and grandparents born in a restricted list of countries. A baseline examination, including collection of morning spot urine, a fasting venous blood sample, as well as a detailed health questionnaire, was conducted by trained health care workers between 2003 and 2006. The study was approved by the ethical committee of Lausanne University Hospital.
The Croatia-Korcula study is a family-based, cross-sectional study in the isolated island of Korcula, Croatia, that included 965 examinees aged 18–95 years. Urine samples were collected in 2007, along with clinical and biochemical measures and lifestyle and health questionnaires.
The Croatia-Split study is a population-based, cross-sectional study in the Dalmatian City of Split, Croatia, that included 1012 examinees aged 18–95 years. Urine samples were collected in 2009.
The Framingham Heart Study is a community-based multigeneration family study, with three generations (1971, original cohort; 1984, offspring cohort; 2002, third generation). The design of this study was previously described.34 The sample used here is a subset of 2640 participants from the offspring cohort at examination 6 (1995–1998), with urinary uromodulin and eGFR levels measured. Participants self-reported their European descent ancestry.
The INGI-Carlantino study is a population-based, cross-sectional study in a village situated in the southeastern part of the Apennines in a hilly area of the Puglia region. We used a subset of 360 individuals (from a total sample size of 1478) who had genotyping information and urine samples available.
The INGI-Val Borbera project was initiated in 2005, involving inhabitants of the geographically isolated Borbera Valley of Northwest Italy, in Piedmont. Information on a large set of phenotypes and biologic samples was obtained from 1803 inhabitants aged between 18 and 102 years.
Uromodulin and Creatinine Measurements
All spot urine samples were aliquoted immediately after collection and were frozen and stored at −80°C before analysis. In the Framingham Heart Study, the Rules-Based Medicine array (Rules-Based Medicine, Inc., Austin, TX) measured urinary uromodulin via immunoassay with a bead Luminex platform. This assay has coefficient of variations of 5.0% at a mean concentration of 37 µg/ml and 11.4% at a mean concentration of 9.4 µg/ml. The assay range is between 6.2 and 135 ng/ml. All other studies measured urinary uromodulin concentration by a well established ELISA35 based on a sheep anti-human uromodulin antibody (K90071C; Meridian Life Science, Memphis, TN) as the capture antibody, a mouse monoclonal anti-human Tamm–Horsfall protein antibody (CL 1032A; Cedarlane Laboratories, Burlington, NC) as the primary antibody, and a goat anti-mouse IgG (H+L) horseradish peroxidase–conjugated protein (172.1011; Bio-Rad Laboratories, Inc., Hercules, CA) as a secondary antibody. Human uromodulin (AG 733, stock solution: 100 µg/ml; EMD Millipore, Temecula, CA) was used to establish the standard curve. The ELISA for human uromodulin showed a sensitivity of 2.8 ng/ml and a linearity of 1.0. The interassay and intraassay variability were determined at 3.28% and 5.46%, respectively. The assay had a detection range between 3.9 and 500 ng/ml (Supplemental Table 1). Uromodulin was indexed to creatinine to compensate for variations in urine concentrations. Uromodulin levels below the detection threshold were set to the lower limit of the urinary uromodulin assay.
Genotyping Platforms and Imputation
Genotyping and imputation were conducted as described in Supplemental Table 2. Genotyped SNPs that passed quality control procedures were then imputed to approximately 2.5 million HapMap CEU SNPs.
Heritability and Proportion of Variance Explained
Heritability of urinary uromodulin levels was assessed in the Framingham Heart Study using SOLAR software (version 1.4),36 with age, sex, and age by sex as covariates, and in the Croatia-Korcula study using an estimate of heritability derived from analysis of the polygenic model in GenABEL software,37 with age and sex as covariates. The proportion of urinary uromodulin variance explained by the reported SNPs was calculated on the basis of the meta-analysis results and the SD of uromodulin levels in the CoLaus cohort using GCTA software (version 1.04).38
Statistical Analyses
Each participating study performed a GWAS of approximately 2.5 million SNPs on both urinary uromodulin indexed to urinary creatinine and raw urinary uromodulin (unindexed) using linear regression and an additive genetic model, adjusting for age and sex and principal components where applicable. Data from the Framingham Heart Study, Croatia-Korcula, Croatia-Split, INGI-Val Borbera, and INGI-Carlantino cohorts were adjusted for family relatedness using the kinship matrices and for population stratification using the first three principal components. These adjustments account for all known and cryptic relationships within these cohorts. The results were then meta-analyzed with METAL software (version 2011-03-05)39 using both sample size and inverse-variance weighted fixed-effects models. The statistical significance threshold was set to 5E-8 for genome-wide significance. P values were corrected for genomic control both at the study level and after meta-analysis. The λ value was around 1.0. SNPs with a minor allele frequency <5% or not present in at least 50% of the studies were dropped from the meta-analysis results. For each locus, independent SNPs with the lowest P value and in low linkage disequilibrium (R2<0.2) were reported.
Stratified Analyses for the Lead SNP
UMOD variants have been associated with hypertension,40 and the association of renal function with UMOD variants was modified by age.14 We therefore conducted stratified analyses for the lead SNP (rs12917707) by hypertension status (defined as being on antihypertensive treatment or having systolic/diastolic BP≥140/90 mmHg) and by age group (below and above the median age of 53 years) in CoLaus, the largest cohort available in our study (n=5077), to explore a potential effect modification by hypertension or age. We compared effect sizes across strata using a two-sample test 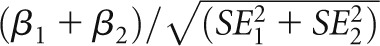 .41 We also conducted sensitivity analyses adding hypertension status as a covariate in the analysis.
.41 We also conducted sensitivity analyses adding hypertension status as a covariate in the analysis.
Associations with Renal Function
We tested the association with kidney function in the six population-based studies using eGFR, determined by the simplified Modification of Diet in Renal Disease study equation. We defined CKD as eGFR<60 ml/min per 1.73 m2. We also tested the association of SNPs most significantly associated with indexed urinary uromodulin with eGFR by querying these in publicly available meta-analysis results of 67,093 individuals of European ancestry from the CKDGen Consortium.13
Gene-Based Association Analyses
The SNP association P values from the meta-analysis of urinary uromodulin were analyzed using VEGAS,42 a program for performing gene-based tests for association using the summary statistics from genetic association studies. The gene-based approach reduces the multiple-testing problem of GWAS by only considering statistical tests for 17,787 genes giving a Bonferroni-corrected threshold of P<2.8E-6.
Joint Analyses
To explore the presence of multiple independent signals at the UMOD locus, we ran a joint step-wise model selection (massoc-slct function) with GCTA38 using the meta-analysis association results for the uromodulin to creatinine ratio. The CoLaus cohort was used as reference to estimate the LD between SNPs. The threshold of significance was set to the default parameters, 5E-8. The resulting estimates are equivalent to that of a multivariate linear model fitting. The LD expressed in R2 between these SNPs, by definition <1, is also indicated in the results file. To quantify the level of variance explained by the different significant SNPs, the adjusted R2 was estimated in the CoLaus population using the LM function in R software.43
Candidate Gene Analyses
We compiled a list of 24 candidate genes for urinary uromodulin levels on the basis of expression profiles showing enrichment in the TAL, and functional data supporting a role in transport processes operating in the cells lining the TAL.44–46 Each gene was queried in the meta-analysis results, by extracting all variants within that gene (including the promoter and the 5′ region around it). For each gene, we accounted for region-specific multiple testing using a gene-specific threshold (0.05 divided by the number of found LD blocks at R2=0.2). SNPs with a median imputation quality <60% and minor allele frequency <5% were excluded before this analysis. Variants that had a P value lower than gene-specific thresholds were declared significant. To assess the overall significance of SNPs in the 24 candidate genes, we estimated the effective number of tests for the 1739 tested SNPs using the method by Gao et al.47 The SNP-by-SNP pairwise correlation matrix was calculated based on the CoLaus data. We then adjusted the Benjamini–Hochberg step-down procedure to compute the false discovery rate for the best SNP in each gene.48
Microdissection and Isolation of Tubular Segments
Tubular segments and glomeruli from mouse kidney were microdissected using a previously described protocol.32 The collagenase-digested segments were sieved and selected on the basis of morphology characteristics. Pooled segments (from two kidneys for each sample) were frozen in liquid nitrogen and kept at −80°C until RNA extraction.
SYBR Green Real-Time Quantitative PCR
The isolated fractions were characterized for their enrichment in mRNA expression of Nphs2 (podocin, glomerulus), Slc38a3 (SNAT3, proximal convoluted tubule), Slc5a2 (SGLT2, proximal straight tubule), Umod (uromodulin, TAL), Slc12a3 (NCC, distal convoluted tubule), and Aqp2 (AQP2, collecting ducts). Total RNA was extracted from isolated segments using the RNAqueousR kit (Applied Biosystems, Inc., Foster City, CA), subjected to DNase treatment, and reverse transcribed by using the iScript cDNA Synthesis Kit (Bio-Rad, Munich, Germany). Quantitative RT-PCR analyses were performed in duplo using 100 nM of both sense and anti-sense primers (Supplementary Table 5) using iQ SYBR Green Supermix (Bio-Rad) and an iCycler IQ System (Bio-Rad). All amplicons showed expected sizes and the dissociation curves showed one melting peak. Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used routinely as a reference gene, because preliminary experiments showed no differences with other reporter genes (Cyclophilin, Hprt1, Actb, 36b4). The relative changes in target gene/GAPDH mRNA ratio were determined by the formula: 2 −ΔΔct.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors acknowledge Viviane Beaujean and Sebastien Druart (UCL Medical School, Brussels, Belgium) for establishing the uromodulin ELISA. The authors wish to specially thank the recruitment teams, the administrative teams, and the study participants in the various cohorts.
This research was conducted in part using data and resources from the Framingham Heart Study of the National Institutes of Health National Heart Lung and Blood Institute (NHLBI) and Boston University School of Medicine. This work was partially supported by the NHLBI Framingham Heart Study (Contract N01-HC-25195). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The CoLaus study received financial contributions from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (33CSCO-122661, 3200BO-111361/2, 3100AO-116323/1, and 310000-112552). The computations for CoLaus imputation were performed in part at the Vital-IT center for high-performance computing of the Swiss Institute of Bioinformatics. M.B. is supported by the Swiss School of Public Health Plus. The Croatia-Korcula and Croatia-Split studies were funded by grants from the United Kingdom Medical Research Council, the European Commission Framework EUROSPAN 6 Project (Contract LSHG-CT-2006-018947), and the Republic of Croatia Ministry of Science, Education, and Sports (Grant 108-1080315-0302 to I.R.). The SNP genotyping for the Croatia-Korcula cohort was performed by Helmholtz Zentrum München (Neuherberg, Germany). The SNP genotyping for the Croatia-Split cohort was performed by AROS Applied Biotechnology (Aarhus, Denmark). The INGI-Carlantino study was funded by Regione FVG (L.26.2008). The INGI-Val Borbera study was supported by funds from Compagnia di San Paolo (Torino, Italy), Fondazione Cariplo, and the Ministry of Health (Ricerca Finalizzata 2008 to D.T.). Other funding sources for this study include the European Commission Seventh Framework Programme (FP7/2007-2013 under Grant 246539 of the Marie Curie Actions Programme and Grant 305608 of the EURenOmics project), Action de Recherche Concertée (ARC10/15-029, Communauté Française de Belgique), Fonds de la Recherche Scientifique and Fonds de la Recherche Scientifique Medicale, Belgium Federal Government Inter-University Attraction Pole, Gebert Rüf Stiftung (Project GRS-038/12), and the Swiss National Science Foundation (Grant 310030-146490 and the NCCR Kidney. CH program).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013070781/-/DCSupplemental.
References
- 1.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS: Prevalence of chronic kidney disease in the United States. JAMA 298: 2038–2047, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Zhang QL, Rothenbacher D: Prevalence of chronic kidney disease in population-based studies: Systematic review. BMC Public Health 8: 117, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsushita K, van der Velde M, Astor BC, Woodward M, Levey AS, de Jong PE, Coresh J, Gansevoort RT, Chronic Kidney Disease Prognosis Consortium : Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: A collaborative meta-analysis. Lancet 375: 2073–2081, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarnak MJ, Levey AS, Schoolwerth AC, Coresh J, Culleton B, Hamm LL, McCullough PA, Kasiske BL, Kelepouris E, Klag MJ, Parfrey P, Pfeffer M, Raij L, Spinosa DJ, Wilson PW, American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention : Kidney disease as a risk factor for development of cardiovascular disease: A statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation 108: 2154–2169, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Shlipak MG, Fried LF, Cushman M, Manolio TA, Peterson D, Stehman-Breen C, Bleyer A, Newman A, Siscovick D, Psaty B: Cardiovascular mortality risk in chronic kidney disease: Comparison of traditional and novel risk factors. JAMA 293: 1737–1745, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, Levy D: Predictors of new-onset kidney disease in a community-based population. JAMA 291: 844–850, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Parikh NI, Hwang SJ, Larson MG, Meigs JB, Levy D, Fox CS: Cardiovascular disease risk factors in chronic kidney disease: Overall burden and rates of treatment and control. Arch Intern Med 166: 1884–1891, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Fassett RG, Venuthurupalli SK, Gobe GC, Coombes JS, Cooper MA, Hoy WE: Biomarkers in chronic kidney disease: A review. Kidney Int 80: 806–821, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Rampoldi L, Scolari F, Amoroso A, Ghiggeri G, Devuyst O: The rediscovery of uromodulin (Tamm-Horsfall protein): From tubulointerstitial nephropathy to chronic kidney disease. Kidney Int 80: 338–347, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Tamm I, Horsfall FL, Jr: Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc Soc Exp Biol Med 74: 106–108, 1950 [PubMed] [Google Scholar]
- 11.Shlipak MG, Day EC: Biomarkers for incident CKD: A new framework for interpreting the literature. Nat Rev Nephrol 9: 478–483, 2013 [DOI] [PubMed] [Google Scholar]
- 12.Serafini-Cessi F, Malagolini N, Cavallone D: Tamm-Horsfall glycoprotein: Biology and clinical relevance. Am J Kidney Dis 42: 658–676, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS: New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O’Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O’Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa M, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson A, Tönjes A, Dehghan A, Chouraki V, Holliday EG, Sorice R, Kutalik Z, Lehtimäki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Kollerits B, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu FB, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Koenig W, Illig T, Döring A, Wichmann HE, Kolcic I, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nöthlings U, Jacobs G, Biffar R, Endlich K, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Völker U, Völzke H, Kovacs P, Stumvoll M, Mägi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Ruggiero D, Bergmann S, Kähönen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Giulianini F, Krämer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Metzger M, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki I, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman JC, Hayward C, Ridker P, Parsa A, Bochud M, Heid IM, Goessling W, Chasman DI, Kao WH, Fox CS, CARDIoGRAM Consortium. ICBP Consortium. CARe Consortium. Wellcome Trust Case Control Consortium 2 (WTCCC2) : Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 8: e1002584, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köttgen A, Hwang SJ, Larson MG, Van Eyk JE, Fu Q, Benjamin EJ, Dehghan A, Glazer NL, Kao WH, Harris TB, Gudnason V, Shlipak MG, Yang Q, Coresh J, Levy D, Fox CS: Uromodulin levels associate with a common UMOD variant and risk for incident CKD. J Am Soc Nephrol 21: 337–344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köttgen A, Yang Q, Shimmin LC, Tin A, Schaeffer C, Coresh J, Liu X, Rampoldi L, Hwang SJ, Boerwinkle E, Hixson JE, Kao WH, Fox CS: Association of estimated glomerular filtration rate and urinary uromodulin concentrations with rare variants identified by UMOD gene region sequencing. PLoS ONE 7: e38311, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, Shirts B, Xu L, Zhu H, Barmada MM, Bleyer AJ: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renigunta A, Renigunta V, Saritas T, Decher N, Mutig K, Waldegger S: Tamm-Horsfall glycoprotein interacts with renal outer medullary potassium channel ROMK2 and regulates its function. J Biol Chem 286: 2224–2235, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutig K, Kahl T, Saritas T, Godes M, Persson P, Bates J, Raffi H, Rampoldi L, Uchida S, Hille C, Dosche C, Kumar S, Castañeda-Bueno M, Gamba G, Bachmann S: Activation of the bumetanide-sensitive Na+,K+,2Cl-cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J Biol Chem 286: 30200–30210, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bates JM, Raffi HM, Prasadan K, Mascarenhas R, Laszik Z, Maeda N, Hultgren SJ, Kumar S: Tamm-Horsfall protein knockout mice are more prone to urinary tract infection: Rapid communication. Kidney Int 65: 791–797, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Mo L, Liaw L, Evan AP, Sommer AJ, Lieske JC, Wu XR: Renal calcinosis and stone formation in mice lacking osteopontin, Tamm-Horsfall protein, or both. Am J Physiol Renal Physiol 293: F1935–F1943, 2007 [DOI] [PubMed] [Google Scholar]
- 22.CKDGen Consortium: CKDGen meta-analysis data, 2011. Available at: http://www.nhlbi.nih.gov/research/intramural/researchers/pi/fox-caroline/ckdgen-meta-analysis-data/ckdgen-meta-analysis-data.html Accessed July 1, 2013
- 23.Tokuhiro K, Ikawa M, Benham AM, Okabe M: Protein disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility. Proc Natl Acad Sci U S A 109: 3850–3855, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Paré G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS: Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41: 712–717, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L, Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team : Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 19: 1655–1660, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaeffer C, Santambrogio S, Perucca S, Casari G, Rampoldi L: Analysis of uromodulin polymerization provides new insights into the mechanisms regulating ZP domain-mediated protein assembly. Mol Biol Cell 20: 589–599, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercier-Zuber A, O’Shaughnessy KM: Role of SPAK and OSR1 signalling in the regulation of NaCl cotransporters. Curr Opin Nephrol Hypertens 20: 534–540, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Ponce-Coria J, Gagnon KB, Delpire E: Calcium-binding protein 39 facilitates molecular interaction between Ste20p proline alanine-rich kinase and oxidative stress response 1 monomers. Am J Physiol Cell Physiol 303: C1198–C1205, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiche J, Theilig F, Rafiqi FH, Carlo AS, Militz D, Mutig K, Todiras M, Christensen EI, Ellison DH, Bader M, Nykjaer A, Bachmann S, Alessi D, Willnow TE: SORLA/SORL1 functionally interacts with SPAK to control renal activation of Na(+)-K(+)-Cl(-)cotransporter 2. Mol Cell Biol 30: 3027–3037, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahan K, Devuyst O, Smaers M, Vertommen D, Loute G, Poux JM, Viron B, Jacquot C, Gagnadoux MF, Chauveau D, Büchler M, Cochat P, Cosyns JP, Mougenot B, Rider MH, Antignac C, Verellen-Dumoulin C, Pirson Y: A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol 14: 2883–2893, 2003 [DOI] [PubMed] [Google Scholar]
- 31.Bernascone I, Janas S, Ikehata M, Trudu M, Corbelli A, Schaeffer C, Rastaldi MP, Devuyst O, Rampoldi L: A transgenic mouse model for uromodulin-associated kidney diseases shows specific tubulo-interstitial damage, urinary concentrating defect and renal failure. Hum Mol Genet 19: 2998–3010, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Glaudemans B, Terryn S, Gölz N, Brunati M, Cattaneo A, Bachi A, Al-Qusairi L, Ziegler U, Staub O, Rampoldi L, Devuyst O: A primary culture system of mouse thick ascending limb cells with preserved function and uromodulin processing. Pflugers Arch 466: 343–356, 2014 [DOI] [PubMed] [Google Scholar]
- 33.Firmann M, Mayor V, Vidal PM, Bochud M, Pécoud A, Hayoz D, Paccaud F, Preisig M, Song KS, Yuan X, Danoff TM, Stirnadel HA, Waterworth D, Mooser V, Waeber G, Vollenweider P: The CoLaus study: A population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord 8: 6, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP: The Framingham Offspring Study. Design and preliminary data. Prev Med 4: 518–525, 1975 [DOI] [PubMed] [Google Scholar]
- 35.Youhanna S, Weber J, Beaujean V, Glaudemans B, Sobek J, Devuyst O: Determination of uromodulin in human urine: Influence of storage and processing. Nephrol Dial Transplant 29: 136–145, 2014 [DOI] [PubMed] [Google Scholar]
- 36.Almasy L, Blangero J: Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62: 1198–1211, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM: GenABEL: An R library for genome-wide association analysis. Bioinformatics 23: 1294–1296, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Lee SH, Goddard ME, Visscher PM: GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjögren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JM, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Völzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJ, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P, Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF, Global BPgen Consortium : Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet 6: e1001177, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen A: Comparing regression coefficients across subsamples. Sociol Methods Res 12: 77–94, 1983 [Google Scholar]
- 42.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S, AMFS Investigators : A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87: 139–145, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Development Core Team : R: A Language and Environment for Statistical Computing, Vienna, Austria, R Foundation for Statistical Computing, 2011 [Google Scholar]
- 44.Hoorn EJ, Nelson JH, McCormick JA, Ellison DH: The WNK kinase network regulating sodium, potassium, and blood pressure. J Am Soc Nephrol 22: 605–614, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chabardès-Garonne D, Mejéan A, Aude JC, Cheval L, Di Stefano A, Gaillard MC, Imbert-Teboul M, Wittner M, Balian C, Anthouard V, Robert C, Ségurens B, Wincker P, Weissenbach J, Doucet A, Elalouf JM: A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci U S A 100: 13710–13715, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheval L, Pierrat F, Rajerison R, Piquemal D, Doucet A: Of mice and men: Divergence of gene expression patterns in kidney. PLoS ONE 7: e46876, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Starmer J, Martin ER: A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol 32: 361–369, 2008 [DOI] [PubMed] [Google Scholar]
- 48.Li J, Ji L: Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95: 221–227, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ: LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrett JC, Fry B, Maller J, Daly MJ: Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.