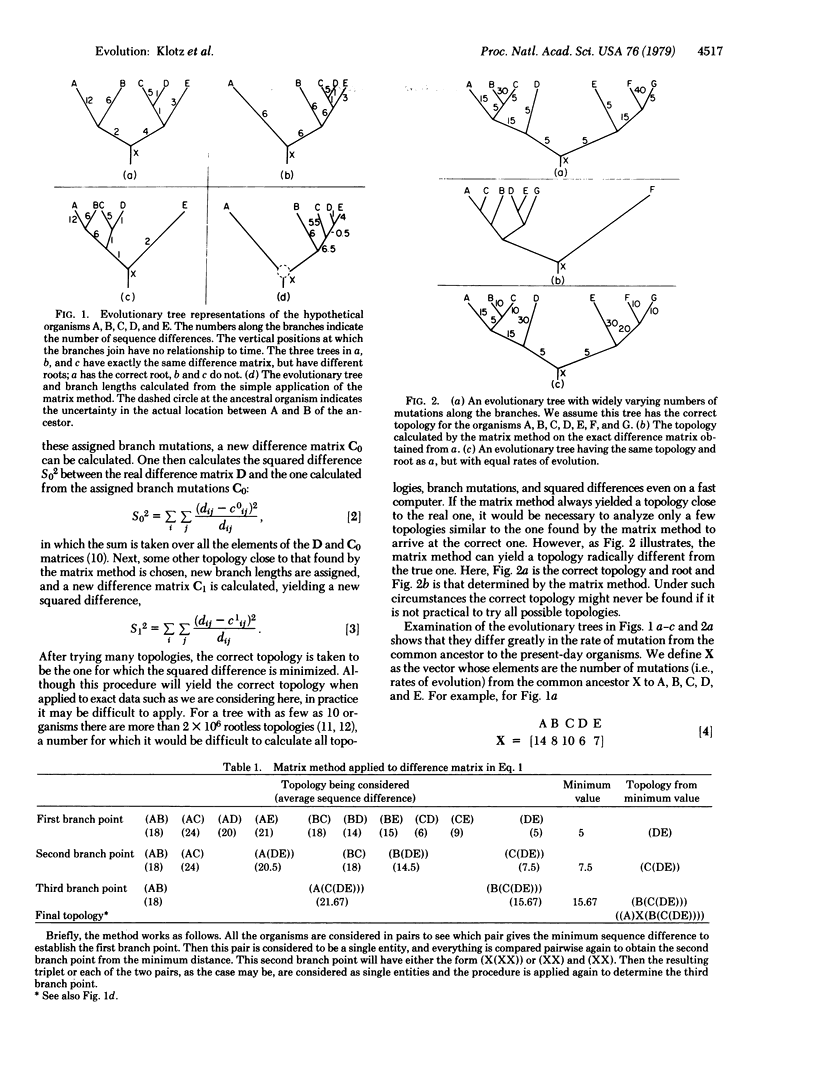
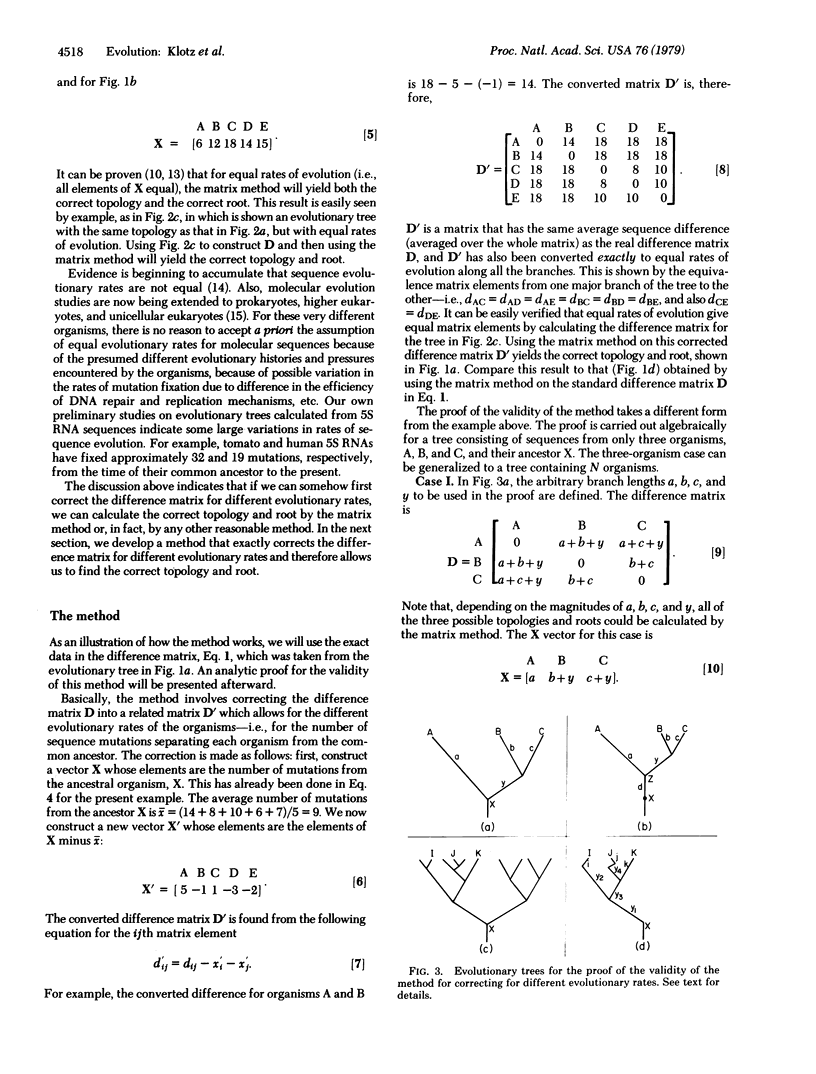
Abstract
Evolutionary trees are usually calculated from comparisons of protein or nucleic acid sequences from present-day organisms by use of algorithms that use only the difference matrix, where the difference matrix is constructed from the sequence differences between pairs of sequences from the organisms. The difference matrix alone cannot define uniquely the correct position of the ancestor of the present-day organisms (root of the tree). Furthermore, methods using the difference matrix alone often fail to give the correct pattern of tree branching (topology) when the different sequences evolve at different rates. Only for equal rates of evolution can the difference matrix (when used with the so-called matrix method) yield exactly the correct topology and root. In this paper we present a method for calculating evolutionary trees from sequence data that uses, along with the difference matrix, the rate of evolution of the various sequences from their common ancestor. It is proven analytically that this method uniquely determines both the correct topology and root in theory for unequal rates of sequence evolution. How one would estimate an ancestral sequence to be used in the method is discussed in particular for the 5S RNA sequences from prokaryotes and eukaryotes and for ferredoxin sequences.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. R., Roberts M., Loeblich A. R., 3rd, Klotz L. C. Characterization of the DNA from the dinoflagellate Crypthecodinium cohnii and implications for nuclear organization. Cell. 1975 Oct;6(2):161–169. doi: 10.1016/0092-8674(75)90006-9. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967 May;19(3 Pt 1):233–257. [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck R. V., Dayhoff M. O. Evolution of the structure of ferredoxin based on living relics of primitive amino Acid sequences. Science. 1966 Apr 15;152(3720):363–366. doi: 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M., Ishii K., Matsuda H. Comments of Holmquist's theory for paleogenetics: the effect of multiple hits on nucleotide differences between homologous DNA's. J Mol Evol. 1975 Aug 5;5(3):249–254. doi: 10.1007/BF01741245. [DOI] [PubMed] [Google Scholar]
- Lauer G. D., Klotz L. C. Determination of the molecular weight of Saccharomyces cerevisiae nuclear DNA. J Mol Biol. 1975 Jun 25;95(2):309–326. doi: 10.1016/0022-2836(75)90397-6. [DOI] [PubMed] [Google Scholar]
- Lauer G. D., Roberts T. M., Klotz L. C. Determination of the nuclear DNA content of Saccharomyces cerevisiae and implications for the organization of DNA in yeast chromosomes. J Mol Biol. 1977 Aug 25;114(4):507–526. doi: 10.1016/0022-2836(77)90175-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R. M., Loeblich L. A., Klotz L. C., Loeblich A. R., 3rd DNA organization of Methanobacterium thermoautotrophicum. Science. 1979 Jun 8;204(4397):1082–1084. doi: 10.1126/science.377486. [DOI] [PubMed] [Google Scholar]
- Moore G. W., Goodman M., Barnabas J. An iterative approach from the standpoint of the additive hypothesis to the dendrogram problem posed by molecular data sets. J Theor Biol. 1973 Mar;38(3):423–457. doi: 10.1016/0022-5193(73)90251-8. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Klotz L. C., Loeblich A. R., 3rd Characterization of a blue-green algal genome. J Mol Biol. 1977 Feb 25;110(2):341–361. doi: 10.1016/s0022-2836(77)80076-4. [DOI] [PubMed] [Google Scholar]


