Abstract
Objective
Short bowel syndrome remains a condition of high morbidity and mortality, and current therapeutic options carry significant side effects. To identify new treatments we focused on postresection changes in microRNAs—short noncoding RNAs, which suppress target genes—and suggest a previously undiscovered role for microRNA-125a (mir-125a) in intestinal adaptation.
Methods
Rats underwent either 80% massive small bowel resection or transection and were harvested after 48 hours. Jejunum was harvested for microRNA microarrays, laser capture microdissection, and RNA and protein analysis. Mir-125a was overexpressed in intestinal epithelium–6 (crypt-derived) cells (IEC-6) and effects on proliferation and apoptosis determined using MTS and flow cytometry. Expression of potential targets of mir-125a in rat jejunum and IEC-6 cells was determined using quantitative real-time polymerase chain reaction (RNA) and Western blotting (protein).
Results
Resection upregulated mir-125a and mir-214 by 2.4-folds and 3.2-folds, respectively. Highest levels of expression were noted in the crypt fraction. Mir-125a overexpression induced apoptosis and resultant growth arrest in IEC-6 cells. The expression of the prosurvival Bcl-2 family member Mcl-1 was downregulated in both mir-125a-overexpressing IEC-6 cells and in jejunum of resected rats, confirming Mcl-1 as a previously undiscovered target of mir-125a.
Conclusions
Upregulation of mir-125a suppresses the prosurvival protein Mcl1, producing the increase in apoptosis known to accompany the proliferative changes characteristic of intestinal adaptation. Our data highlight a potential role for microRNAs as mediators of the adaptive process and may facilitate the development of new therapeutic options for short bowel syndrome.
Short bowel syndrome results from the loss of significant intestinal length in which insufficient water and nutrient absorption by the remnant bowel necessitates dependence on total parenteral nutrition (TPN) to meet nutritional requirements.1 Moreover, TPN is itself associated with side effects such as line sepsis and impairment of liver function,2 whereas other alternatives such as intestinal transplantation are associated with high morbidity and mortality,3–7 making current therapeutic options suboptimal. Understanding the regulatory mechanisms behind the adaptive response of the intestine after massive intestinal loss may reveal new therapeutic options for this condition.
The intestine is able to mount an adaptive response to the loss of a large extent of small bowel as associated with short bowel syndrome.6,7 The events that occur during intestinal adaptation to short bowel syndrome have been well characterized in the rat model of massive small bowel resection.8 Morphological changes include increases in villus height and crypt depth, increasing the intestinal absorptive surface as compensation for the reduced intestinal length to minimize malabsorption.7,9 The adaptive process is associated with increases in apoptosis and proliferation in the intestinal crypts10 and has been noted to occur as early as 48 hours postresection.1,11,12 Recent studies have revealed several key genes that are activated during adaptation, for example, Bcl-2 and epidermal growth factor (EGF) receptor family members, initiating the programs to increase proliferation and the accompanying apoptosis.13–16 However, factors regulating these pathways have not been fully elucidated.
MicroRNAs are short, noncoding RNAs able to silence numerous genes simultaneously. Bioinformatics analysis suggests that up to 30% of mammalian gene transcripts are regulated by microRNAs.17–20 MicroRNAs suppress protein expression by interacting with complementary sequences on the 3′ untranslated region (3′UTR) of target gene messenger RNAs (mRNAs) and either directly inhibiting translation (detected as changes in protein levels without changes in mRNA) or via inducing mRNA cleavage (manifested as reductions in both protein and mRNA levels).21–23 Presence of the target sequence for each microRNA on multiple genes permits simultaneous regulation of protein expression from numerous genes by a single microRNA.17,24,25 The postulated role of microRNAs in “fine-tuning” gene expression has been confirmed in other models of intestinal regulation, such as the circadian rhythmicity seen in many of the intestinal genes and proteins.26–30
Given the involvement of microRNAs in the regulation of many gene expression pathways, it is likely that they are also involved in intestinal adaptation. The ability of microRNAs to regulate gene expression post-transcriptionally would provide additional control and flexibility to this process. However, the role of microRNAs in the adaptive response to small bowel resection has yet to be explored. To address this gap, we examined the changes in microRNA expression generated by massive small bowel resection in rats. We found increased expression of microRNAs 214 and 125a (mir-214 and mir-125a) in the jejunum during adaptation. Overexpression of mir-125a in enterocytes reduced cell growth and cell number because of an increased rate of apoptosis, possibly via suppression of the prosurvival protein Mcl1. The regulation of apoptosis by mir-125a highlights a potential role for microRNAs in the intestinal adaptive process.
METHODS
Animal Studies
All animal study protocols were prospectively approved by the Harvard Medical Area Standing Committee on Animals. Sprague-Dawley rats (16 male rats, 7-week old) were purchased from Harlan World (Indianapolis, IN) and acclimatized to a 12:12–hour light–dark photoperiod for 7 days with ad libitum access to food and water. Surgery was performed at 10 AM to minimize effects of circadian rhythmicity. Rats were subjected to a midline laparotomy followed by either a massive mid-small bowel resection or transection (n = 8 per group). For the resection, the intestine was transected 10 cm distal to the ligament of Treitz and 80% of the small intestine resected before reanastomosis of jejunum to terminal ileum. For the transection group, the transected limbs were reanastomosed without removal of intestine. Intestinal continuity in both groups was restored with interrupted 6-0 polydioxanone sutures (Ethicon, Somerville, NJ). Rats were fed a liquid diet for the first 24 hours postsurgery (Ensure; Ab-bott Laboratories, Abbott Park, IL), then returned to a chow diet for the remaining 24 hours. Rats in the transection group were pair-fed to those in the resection group to ensure matched nutrient intake. Three animals in the resected group died of complications before harvest and were excluded from the study. The remaining animals all seemed healthy and well.
Tissue Harvest
Rats were killed at 48 hours postsurgery at 10 AM and jejunum harvested for RNA and protein determination. Rats were anesthetized with sodium pentobarbital (50 mg/kg; Ovation Pharmaceuticals, Deerfield, IL). A 6-cm length of small intestine proximal to the anastomosis and beginning 1 cm distal to the ligament of Treitz was harvested via midline laparotomy and rinsed with ice-cold saline to remove luminal contents. We chose a segment proximal to the anastomosis to ensure similar regions of the intestine were compared between the resection and transaction groups. The harvested segment was divided along the antimesenteric border, mucosa scraped from the underlying muscle, snap-frozen in liquid nitrogen, and stored at −80°C for subsequent RNA or protein extraction.
MicroRNA Microarrays and MicroRNA Validation by Real-Time Polymerase Chain Reaction
Total RNA from jejunum was extracted using the mirVana kit (Ambion, Austin, TX) and profiled on probes printed in quadruplicate on in situ hybridization arrays (Exiqon, Woburn, MA) against a reference sample consisting of RNA pooled from transected rats. Dye swaps were incorporated in the arrays to correct for dye bias. The limma package in the statistical language R (http://www.r-project.org) was used for background correction, loess normalization, and calculation of t tests and false discovery rates (q values) to assess differential expression.31,32 For microarray data, microRNAs with false discovery rates (q values) less than 0.05 after multiple-testing correction were considered significant. Those microRNAs exhibiting a 2-fold or more difference in expression between the resection and transection groups and conserved across human, mouse, and rat species were selected for further analysis. Raw data showing expression of all profiled microRNAs have been submitted to the Gene Expression Omnibus Web site.33
For quantification of microRNA expression by real-time quantitative polymerase chain reaction (qPCR), selected microRNAs were reverse transcribed from extracted total RNA samples using the stem-loop hybridization–based microRNA reverse-transcription kit and microRNA-specific primers (Taqman microRNA reverse-transcription kit and Taqman microRNA assays; Applied Biosystems, Carlsbad, CA). MicroRNA expression was quantified using the Taqman microRNA PCR primers and Taqman gene expression Master Mix (Applied Biosystems); all reactions were carried out in triplicate. Samples underwent reverse transcription and PCR simultaneously to minimize errors introduced by variations in reaction efficiency.
Construction of the Mir-125a Overexpression Vector
An inducible human mir-125a overexpression vector was constructed in TRIPZ, an shRNAmir vector carrying a tetracycline-inducible promoter (Open Biosystems, Huntsville, AL). The human mir-125a gene along with the adjacent upstream and downstream 200-bp flanking sequences was amplified from human genomic DNA using primers incorporating MluI and ClaI restriction sites (Table 1). The PCR product was ligated into the MluI/ClaI sites of TRIPZ. A nonsilencing short hairpin RNA (shRNA) in TRIPZ was used as a control. Vectors were sequenced to ensure correct insertion sequence fidelity.
TABLE 1.
| Target | Forward Primer | Reverse Primer |
|---|---|---|
| mir-214 | GCGCATCGATGGCACCACTCACTTTACTTTTG | ATACGCGTCGCAAATTATCCATGTTAGCAC |
| mir-125a | ATACGCGTGGGCTCTTTTCTGTCCTTGTC | GCCGATCGATTTTCAGTTGGTGGTCAAATGTC |
| Mcl1 | TGGGTTTGTGGAGTTCTTCC | GTTGGTGGCTGGAGGTTTTA |
| β-actin | GGATCAGCAAGCAGGAGTACGA | AACGCAGCTCAGTAACAGTCCG |
Cell Transfection
Rat crypt-derived intestinal epithelial (IEC-6) cells were purchased from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA) and 0.1% bovine insulin (Sigma, St. Louis, MO). Cells between passages 6 to 8 at 80% confluence were transfected with either the mir-125a overexpression construct or the nonsilencing control using Lipofectamine (Invitrogen). Puromycin selection (2 μg/mL; Invivogen, San Diego, CA) was commenced 48 hours after transfection. MicroRNA overexpression was induced by addition of 2 μg/mL doxycycline (Clontech, Mountain View, CA) to the cell culture media 72 hours before cells were plated for specific assays.
In Vitro Proliferation and Cell Counting
For proliferation assays, cells were seeded in 96-well plates at a density of 1000 cells per well in triplicate and the proliferation index measured at 48 hours using the CellTiter96 Aqueous One Solution Cell Proliferation Assay (MTS assay; Promega, Madison, WI). Cell growth rates were corroborated by counting cells trypsinized 48 hours after seeding at a density of 10,000 cells/mL in triplicate in 6-well dishes. All experiments were performed thrice.
Analyses for Cell Cycle, Apoptosis, and Viability
For cell cycle analysis, cells were trypsinized, counted and fixed in 70% ethanol overnight at −20°C. Fixed cells were centrifuged at 1200 rpm for 10 minutes at 4°C and the pellet suspended in propidium iodide (BD Biosciences, San Jose, CA) for 30 minutes at 37°C in the dark. Samples were processed on a BD FACScan (BD, Franklin Lakes, NJ) and 10,000 events each were analyzed using ModFit LT (Verity, Topsham, ME). For determination of apoptosis and viability, cells were trypsinized, counted, and stained with Annexin V-FITC (BD) and Sytox Blue (Invitrogen), respectively. Samples were processed as earlier and 10,000 events each were analyzed using the BD FACSDiva software.
RNA Extraction, mRNA Reverse Transcription, and Real-Time PCR
Total RNA was extracted from rat jejunum or IEC-6 cells using the mirVana kit (Ambion). Samples were reverse transcribed simultaneously with Superscript III (Invitrogen) and oligo-DT. Real-time PCR was performed as previously described.34 Levels of Mcl1 mRNA were expressed as ratios to the stably expressed β-Actin. Rat-specific primers for rat jejunal samples and rat-derived IEC-6 cells were obtained from Invitrogen (Table 1).
Protein Extraction and Western Blotting
Protein expression of Mcl1 was measured in total lysates from jejunal mucosal scrapings or IEC-6 cell lysates using Western blotting as previously described.34 Protein extracts from cells or tissue (15 and 75 μg, respectively) were resolved on 4% to 12% bistris polyacrylamide gels, transferred to polyvinylidene fluoride membranes, blocked, and then incubated with rabbit anti-Mcl1 (1:200; Santa Cruz Biotechnology, Santa Cruz, CA). Protein expression was visualized by chemiluminescence, quantified using the freely available ImageJ (http://rsbweb.nih.gov/ij/) and normalized to β-Actin (mouse anti–β-Actin, 1:1000; Labvision, Fremont, CA).
Laser Capture Microdissection
To localize mir-125a and mir-214 within intestinal sections, 3 additional Sprague-Dawley rats (Harlan World) were acclimatized as earlier for 7 days, anesthetized with sodium pentobarbital (50 mg/kg; Ovation Pharmaceuticals) and harvested without surgical intervention. A 2-cm section of jejunum from 5 cm distal to the ligament of Treitz was harvested and embedded in optimal cutting temperature compound over dry ice and isopentane. Sections were cut from the fresh frozen specimens at 9-μm thickness and stained with Histogene staining solution (Molecular Devices, Sunnyvale, CA). Laser capture microdissection was used to isolate fractions from crypts (lower half), villi (top half), or smooth muscle (Veritas Microdissection System, Molecular Devices). Total RNA was extracted from each fraction (RNAqueous Kit; Ambion) and subjected to microRNA reverse transcription and real-time PCR as described earlier.
Statistical Analysis
Data are presented as means ± SE. Statistical analysis was performed with the statistical language R and graphical analysis was performed using GraphPad Prism (San Diego, CA). T test was used for comparing 2 groups; analysis of variance was used for comparing more than 2 groups.
RESULTS
MicroRNAs Are Differentially Expressed After Intestinal Resection
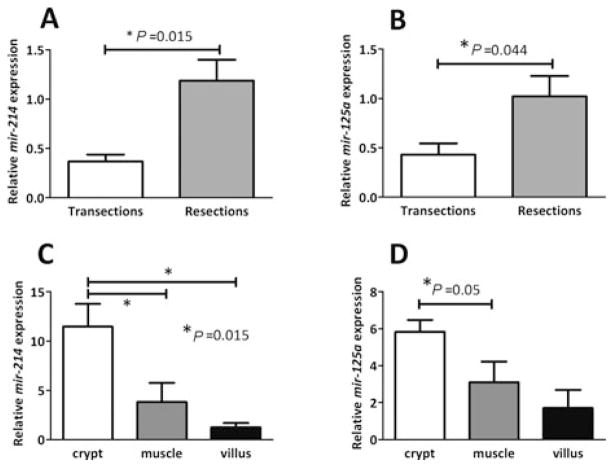
Of the 238 known rat microRNAs tested on in situ hybridization microarrays, expression of 15 was altered by 2 folds or more with statistical significance after massive small bowel resection (ranges, 0.2- to 0.5-fold or 2.0- to 4.4-fold resection versus transection, q < 0.05). Raw data showing expression of all profiled microRNAs in resected rats and transected controls have been submitted to the Gene Expression Omnibus Web site.33 Five microRNAs were not conserved in human and/or mouse and therefore not examined further. The 10 remaining microRNAs conserved in both human and mouse included 7 with increased expression after resection (range, 2.0- to 4.4-fold resection versus transection, q < 0.05) and 3 with decreased expression (range, 0.2- to 0.4-fold, q < 0.05). Expression changes in these 10 microRNAs were further validated by qPCR. Two exhibited statistically significant increases 2 folds or more after resection by qPCR: mir-214 (3.2-fold, Fig. 1A, P = 0.015) and mir-125a (2.4-fold, Fig. 1B, P = 0.044). Of the remaining 8 microRNAs validated, 5 exhibited changes in the same direction as the microarray but the differences did not reach statistical significance. We thus confined our analyses to mir-214, which has been implicated in myogenic differentiation35 among other functions,36–40 and mir-125a, which targets p5341 and the EGF receptor pathway.42–44
FIGURE 1.
Expression of mir-214 and mir-125a in jejunal mucosal scrapings of rats subjected to transection (n = 8) or resection (n = 5) (A, B). RNA was analyzed for mir-214 (A) and mir-125a (B) expression by real-time (RT)-PCR. Relative abundance of mir-125a in crypts versus villi (C, D). Laser capture microdissection of jejunal sections from intact rats (n = 3) was used to isolate fractions of villi (upper half), crypts (lower half), and muscle (circular and longitudinal). Mir-214 (C) and mir-125a (D) expression in each fraction was analyzed as earlier. Thus, both mir-214 and mir-125a seem to be enriched in the crypts, the proliferative compartment of the intestinal mucosa.
Intestinal Crypt-Enriched Expression of Mir-214 and Mir-125a
To ascertain the crypt-villus distribution of mir-214 and mir-125a in the jejunum, we performed laser capture microdissection on mucosal sections in a separate group of intact rats (n = 3), acclimatized as were the experimental rats but not subjected to surgery. Three fractions were compared: villus (upper half), crypt (lower half), and smooth muscle (circular plus longitudinal muscle as a control). Both mir-214 and mir-125a exhibited highest expression in crypts. Mir-214 abundance was 9.2-fold and 3.0-fold greater in crypts than in villi and muscle, respectively (P = 0.015, Fig. 1C). Mir-125a abundance was 3.4-fold greater in crypts than in villi (P = 0.05; Fig. 1D). Although mir-125a was ~2 folds more abundant in crypts versus smooth muscle, the difference did not achieve statistical significance.
Mir-125a Induces Apoptosis in Enterocytes
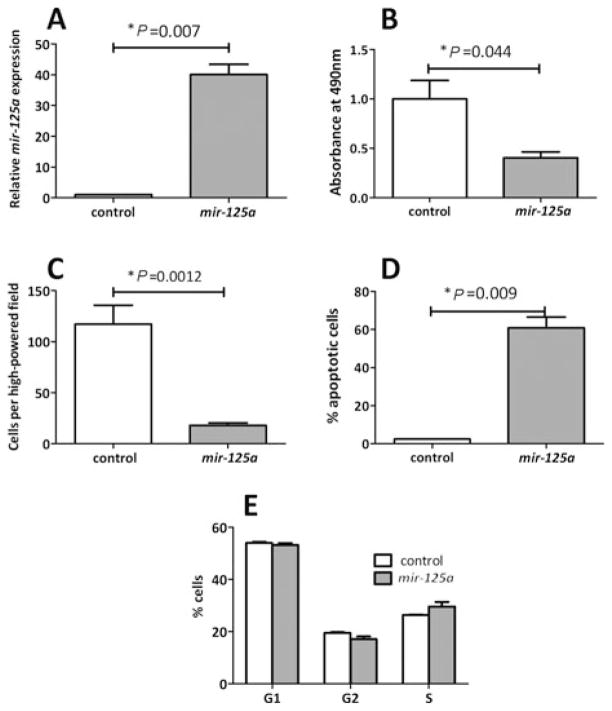
Mir-125a has been previously identified as a tumor suppressor in other cell lines,42,44–46; hence, our next series of experiments aimed to investigate the role of mir-125a in the regulation of proliferation and apoptosis, key features of the adaptive process. To determine the effect of mir-125a on enterocyte apoptosis, mir-125a was overex-pressed in rat-derived IEC-6 crypt cells, chosen because crypts are the main site of proliferation in the adaptive process.9 Stable transfection of IEC-6 cells with the mir-125a expression vector led to a 40-fold increase in mir-125a expression versus the control (P = 0.0007, Fig. 2A). Overexpression of mir-125a significantly reduced proliferation in IEC-6 cells. At 48 hours after plating, the proliferation rate was decreased by 60% versus control cells as measured by the MTS assay (P = 0.044, Fig. 2B) and by 85% as measured by cell counts (P = 0.026, Fig. 2C). Cells overexpressing mir-125a had a significantly higher rate of apoptosis compared with control as revealed by flow cytometry (61% versus 2.4%, P = 0.009, Fig. 2D). In contrast, over-expression of mir-125a did not seem to affect cell cycle progression (Fig. 2E). These findings indicate that the effect of mir-125a on inhibiting cell growth is induced by increasing cellular apoptosis rather than cell cycle progression, and suggest that mir-125a may mediate apoptosis via members of the apoptotic pathway.
FIGURE 2.
Proliferation of IEC-6 cells overexpressing either mir-125a or a nonsilencing control RNA (control). A, Mir-125a expression was analyzed by RT-PCR. B, Proliferation index was determined using the MTS assay. C, Cell numbers were assessed by counting cell densities. Apoptosis (D) and cell cycle progression (E) were assessed by fluorescence-activated cell sorting using Annexin V or Sytox Blue, respectively.
Mir-125a Suppresses the Prosurvival Protein Mcl1 in Enterocytes
To identify potential mir-125a targets that might regulate apoptosis in enterocytes, we interrogated TargetScan,47 a microRNA target prediction algorithm, for mir-125a binding sequences in the 3′ untranslated regions of apoptosis-regulating genes. This search identified the prosurvival protein Mcl1 as a potential target of mir-125a in human, mouse, and rat. Overexpression of mir-125a significantly decreased protein levels of Mcl1 in IEC-6 cells to 0.55 that of the nonsilencing control (P = 0.046, Fig. 3A). Suppression of Mcl1 expression by mir-125a seemed to occur at a translational level rather than mRNA cleavage because mRNA levels did not change detectably compared with controls (P = 0.78, Fig. 3B). These findings confirm that Mcl1 is a target of mir-125a and may mediate the proapoptotic effects of mir-125a in IEC-6 cells.
FIGURE 3.
Effect of mir-125a on Mcl1 expression in IEC-6 cells overexpressing mir-125a or control. Mcl1 protein expression was assessed by immunoblotting (A) or Mcl1 mRNA expression by RT-PCR (B). Data are expressed relative to β-actin protein or mRNA, respectively, and indexed to the control values.
Mcl1 is Downregulated After Intestinal Resection
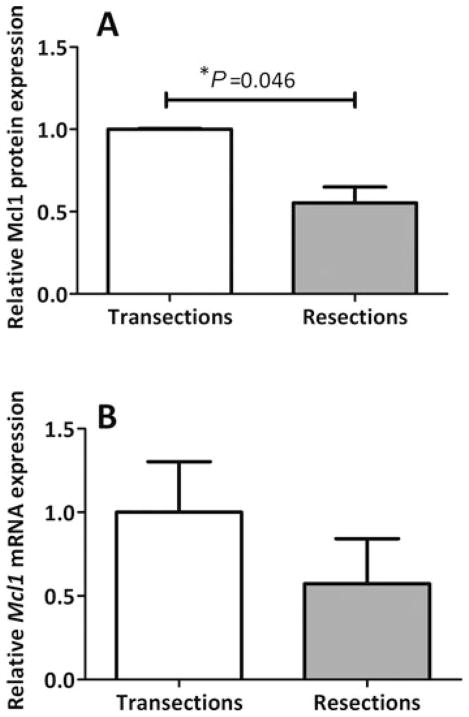
The adaptive response to massive intestinal resection is associated with a marked increase in the rate of crypt apoptosis.10,15,48 Mcl1 has been shown to induce apoptosis in other tissues and, as shown earlier, is a target of mir-125a. To determine whether Mcl1 expression is also altered during intestinal adaptation, we assessed protein and mRNA levels after resection. Mcl1 protein abundance in the jejunum of resected rats was 45% lower than in control rats (P = 0.046, Fig. 4A). Mcl1 mRNA expression trended to be lower after resection versus transection, but this difference did not achieve statistical significance (P = 0.31, Fig. 4B). We take this result to conform to our finding in IEC-6 cells that the inhibition of Mcl1 expression occurred at the translational level.
FIGURE 4.
Effect of bowel resection on Mcl1 expression. Mucosal scrapings from rats subjected to transection (n = 8) or resection (n = 5) were analyzed for Mcl1 protein expression by immunoblotting (A) or Mcl1 mRNA expression by RT-PCR (B). Data are expressed relative to β-actin protein or mRNA, respectively, and indexed to the transection values.
DISCUSSION
The adaptive response of the jejunum to massive small bowel resection is associated with increases in crypt cell proliferation, enterocyte migration, and apoptosis,1,11,12,49 and entails changes in the expression of numerous protein-encoding genes. We undertook this study with the supposition that expression changes would also occur in microRNA genes as part of intestinal adaptation. We examined microRNA abundance 48 hours after surgery, a time chosen to correspond to the onset of adaptive changes and thereby allow us to identify microRNAs involved in initiating the adaptive response.49 We found that the mucosal abundance of mir-214 and mir-125a increased 3.2 folds and 2.4 folds, respectively. This is the first report to our knowledge that demonstrates altered microRNA expression triggered by small bowel resection. As with other physiological processes, microRNAs would be expected to provide additional control over the adaptive process.
A total of 15 microRNAs exhibited alterations 2 or more folds by microarray analysis after small bowel resection. Of these, 2 met the criteria of conservation in humans and mice and of a statistically significant 2-fold or more difference on qPCR validation. Conservation in humans (and mice) would indicate relevance to human biology that might translate into therapeutic use. The 2-fold threshold is in line with other studies profiling differential microRNA expression.50,51 Although only 2 microRNAs subjected to validation met our criteria in this experimental paradigm, others may exhibit substantial changes with different time courses after bowel resection. We fully expect that microRNAs other than mir-214 and mir-125a will also be found that contribute to intestinal adaptation in either human or rodents. However, as discussed earlier, we chose to assess changes at 48 hours postresection under the premise that microRNAs involved in initiating the adaptive process would be detected at this time.
Although several studies have characterized expression patterns and functions for mir-125a or mir-214, ours is the first to detect them in the intestine. In particular, our finding that both are more abundant in intestinal crypts than villi may be pertinent to their functions in this tissue. Changes in the abundances of mir-214 and mir-125a have been associated with cellular proliferation and differentiation. MicroRNA mir-214 is dysregulated in cervical and pancreatic cancers36,38 and contributes to the differentiation of myocytes and neurons.35,52 Increased mir-214 expression in the adapting intestine would be consistent with the increases in proliferation and/or the number of cells undergoing differentiation.
We obtained further information on the potential role of mir-125a in intestinal adaptation. Specifically, our data provide evidence that this microRNA contributes to the increased apoptosis induced in intestinal crypts after bowel resection.10,15,48 First, mir-125a overexpression in IEC-6 cells dramatically increased the rate of apoptosis. The lack of effect of mir-125a on cell cycle progression indicated that the reduced proliferation was due largely to increased apoptosis rather than decreased cell division. Second, mir-125a overexpression in IEC-6 cells reduced expression of Mcl1, an antiapoptotic member of the Bcl2 family.53–55 Finally, Mcl1 expression was reduced in the remnant bowel after resection. Our results suggest that one role of mir-125a in intestinal adaptation is to activate the apoptotic pathway, in part by suppressing expression of the antiapoptotic Mcl1. Increased apoptosis may be necessary for tissue remodeling to increase the mucosal surface area of the remnant intestine and seems to occur as early as 48 hours postresection.12 Further studies examining the effect of apoptosis-inducing agents such as chemotherapy or irradiation on mir-125a expression may help unravel the role of this microRNA as a mediator of apoptosis.
Suppression of Mcl1 expression seems to be a previously unidentified regulatory pathway contributing to intestinal adaptation. Earlier studies have demonstrated that the rate of apoptosis, which peaks within 3 days of intestinal resection, is governed by the ratio of the proapoptotic Bcl-family member Bax to the prosurvival Bcl-family member Bcl-w.15 Bax in particular is an essential mediator of resection-induced apoptosis.15,56 Activation of Bax induces a conformational change, which results in mitochondrial fragmentation in turn releasing mediators of caspase activation.57 The expression of Bax and Bcl-w in the adapting intestine is influenced by the EGF signaling pathway56, which decreases the ratio of Bax to Bcl-w thereby suppressing apoptosis. Of relevance to our studies, previous publications have documented interactions between EGF and mir-125a in several malignancies.43,44,58 For example, EGF has been shown to repress mir-125a transcription in ovarian cells.58 Further studies are necessary to investigate the relationship between activation of this signaling pathway and the apoptosis-inducing effects of mir-125a in the intestine.
A recent study examining the impact of global microRNA ablation in mouse intestine and colon by disrupting Dicer, encoding the key microRNA processing enzyme,59 has interesting parallels to our study. These investigators observed growth retardation, water retention, and impaired fat absorption. Morphological changes were also observed, including increased lymphocyte infiltration and decreased numbers of goblet cells. Of greater interest, Dicer-deficient mice showed increased rates of crypt apoptosis compared with controls, accompanied by increased rates of proliferation and epithelial cell migration. Investigation of the adaptive response in Dicer-deficient mice would allow confirmation of the role of microRNAs in this process.
Our data therefore highlight a potential role for microR-NAs in mediation of the adaptive response to massive small bowel resection and identify a suppressive effect of the microRNA mir-125a on the prosurvival protein Mcl1. Further studies are necessary to better delineate the role of Mcl1 in the adaptive response and the targets of mir-214. MicroRNAs are known to simultaneously regulate multiple targets, and more detailed studies are necessary to identify any other potential targets of mir-125a in the adapting intestine. Indeed, mir-125a can regulate the abundance of p53,41 which could impact expansion of the stem cell pool.60 Our study paves the way for further investigation of the role of microRNAs in intestinal adaptation and the future development of new therapeutic modalities for conditions of intestinal failure such as short bowel syndrome.
Acknowledgments
The authors thank the excellent technical assistance of Jan Rounds. AB, SWA, DBR, and AT designed the studies; AB and ATS conducted the research; AB, PJP, and JMD analyzed the data; AB, DBR, and AT wrote the article. All authors read and approved the final manuscript.
Footnotes
Transcript profiling: Raw microarray data were submitted to Gene Expression Omnibus, submission GSE26542. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xtcnlmqkqmiourq&acc=GSE26542, to access the data.)
Copyright license statement: Data in this manuscript were presented as platform talks at the Society of University Surgeons 2009 and the Society for Academic and Research Surgery 2010, and published only in abstract form (<400 words) in the corresponding supplementary issues of the Journal of the American College of Surgeons and the British Journal of Surgery. The authors attest that this manuscript is not currently under consideration by any other journal.
Disclosures: Supported by grants National Institutes of Health: 5 R01 DK047326 (SWA), American Diabetic Association: 7–05-RA-121 (DBR), Harvard Clinical Nutrition Research Center: P30-DK040561 (AT), Nutricia Research Foundation (AB), and Berkeley Fellowship (ATS).
References
- 1.Vanderhoof JA, Langnas AN. Short-bowel syndrome in children and adults. Gastroenterology. 1997;113:1767–1778. doi: 10.1053/gast.1997.v113.pm9352883. [DOI] [PubMed] [Google Scholar]
- 2.Moukarzel AA, Haddad I, Ament ME, et al. 230 patient years of experience with home long-term parenteral nutrition in childhood: natural history and life of central venous catheters. J Pediatr Surg. 1994;29:1323–1327. doi: 10.1016/0022-3468(94)90107-4. [DOI] [PubMed] [Google Scholar]
- 3.Todo S, Tzakis A, Reyes J, et al. Intestinal transplantation: 4-year experience. Transplant Proc. 1995;27:1355–1356. [PMC free article] [PubMed] [Google Scholar]
- 4.Todo S, Reyes J, Furukawa H, et al. Outcome analysis of 71 clinical intestinal transplantations. Ann Surg. 1995;222:270–280. doi: 10.1097/00000658-199509000-00006. discussion 280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu-Elmagd K, Todo S, Tzakis A, et al. Three years clinical experience with intestinal transplantation. J Am Coll Surg. 1994;179:385–400. [PMC free article] [PubMed] [Google Scholar]
- 6.Drozdowski L, Thomson AB. Intestinal mucosal adaptation. World J Gastroenterol. 2006;12:4614–4627. doi: 10.3748/wjg.v12.i29.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson RC. Intestinal adaptation (first of two parts). Structural, functional, and cytokinetic changes. N Engl J Med. 1978;298:1393–1402. doi: 10.1056/NEJM197806222982505. [DOI] [PubMed] [Google Scholar]
- 8.Wolvekamp MC, Heineman E, Taylor RG, et al. Towards understanding the process of intestinal adaptation. Dig Dis. 1996;14:59–72. doi: 10.1159/000171539. [DOI] [PubMed] [Google Scholar]
- 9.Dowling RH, Booth CC. Structural and functional changes following small intestinal resection in the rat. Clin Sci. 1967;32:139–149. [PubMed] [Google Scholar]
- 10.Wakeman D, Longshore SW, McMellen ME, et al. Extent of small bowel resection does not influence the magnitude of intestinal adaptation in the mouse. J Pediatr Surg. 2010;45:1274–1279. doi: 10.1016/j.jpedsurg.2010.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmrath MA, Erwin CR, Shin CE, et al. Enterocyte apoptosis is increased following small bowel resection. J Gastrointest Surg. 1998;2:44–49. doi: 10.1016/s1091-255x(98)80102-9. [DOI] [PubMed] [Google Scholar]
- 12.Hanson WR, Osborne JW, Sharp JG. Compensation by the residual intestine after intestinal resection in the rat. II. Influence of postoperative time interval. Gastroenterology. 1977;72:701–705. [PubMed] [Google Scholar]
- 13.Helmrath MA, Shin CE, Erwin CR, et al. The EGF\EGF-receptor axis modulates enterocyte apoptosis during intestinal adaptation. J Surg Res. 1998;77:17–22. doi: 10.1006/jsre.1998.5362. [DOI] [PubMed] [Google Scholar]
- 14.Stern LE, Erwin CR, O’Brien DP, et al. Epidermal growth factor is critical for intestinal adaptation following small bowel resection. Microsc Res Tech. 2000;51:138–148. doi: 10.1002/1097-0029(20001015)51:2<138::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 15.Stern LE, Huang F, Kemp CJ, et al. Bax is required for increased enterocyte apoptosis after massive small bowel resection. Surgery. 2000;128:165–170. doi: 10.1067/msy.2000.107370. [DOI] [PubMed] [Google Scholar]
- 16.Stern LE, Falcone RA, Jr, Kemp CJ, et al. Effect of massive small bowel resection on the Bax/Bcl-w ratio and enterocyte apoptosis. J Gastrointest Surg. 2000;4:93–100. doi: 10.1016/s1091-255x(00)80038-4. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 19.Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 20.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 23.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 24.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 25.Stark A, Brennecke J, Bushati N, et al. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Yang M, Lee JE, Padgett RW, et al. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng H-Y, Papp JW, Varlamova O, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S, Witmer PD, Lumayag S, et al. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 29.Gatfield D, Le Martelot G, Vejnar CE, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balakrishnan A, Stearns AT, Park PJ, et al. MicroRNA mir-16 is anti-proliferative in enterocytes and exhibits diurnal rhythmicity in intestinal crypts. Exp Cell Res. 2010;316:3512–3521. doi: 10.1016/j.yexcr.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 33.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D990. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balakrishnan A, Stearns AT, Rounds J, et al. Diurnal rhythmicity in glucose uptake is mediated by temporal periodicity in the expression of the sodium-glucose cotransporter (SGLT1) Surgery. 2008;143:813–818. doi: 10.1016/j.surg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Luo XJ, Xiong AW, et al. MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J Biol Chem. 2010;285:26599–26607. doi: 10.1074/jbc.M110.115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Chen S, Luan X, et al. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life. 2009;61:1075–1082. doi: 10.1002/iub.252. [DOI] [PubMed] [Google Scholar]
- 37.Calissano M, Latchman DS. Cell-specific regulation of the pro-survival Brn-3b transcription factor by microRNAs. Mol Cell Neurosci. 2010;45:317–323. doi: 10.1016/j.mcn.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 38.Zhang XJ, Ye H, Zeng CW, et al. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y, Du X, Lonnerdal B. miR-214 regulates lactoferrin expression and pro-apoptotic function in mammary epithelial cells. J Nutr. 2010;140:1552–1556. doi: 10.3945/jn.110.124289. [DOI] [PubMed] [Google Scholar]
- 40.Okada H, Kohanbash G, Lotze MT. MicroRNAs in immune regulation—opportunities for cancer immunotherapy. Int J Biochem Cell Biol. 2010;42:1256–1261. doi: 10.1016/j.biocel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Gao JS, Tang X, et al. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583:3725–3730. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott GK, Goga A, Bhaumik D, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 44.Wang G, Mao W, Zheng S, et al. Epidermal growth factor receptor–regulated miR-125a-5p—a metastatic inhibitor of lung cancer. Febs J. 2009;276:5571–5578. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 46.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 47.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juno RJ, Knott AW, Profitt SA, et al. Preventing enterocyte apoptosis after massive small bowel resection does not enhance adaptation of the intestinal mucosa. J Pediatr Surg. 2004;39:907–911. doi: 10.1016/j.jpedsurg.2004.02.007. discussion 907–911. [DOI] [PubMed] [Google Scholar]
- 49.Hines OJ, Bilchik AJ, Zinner MJ, et al. Adaptation of the Na+/glucose co-transporter following intestinal resection. J Surg Res. 1994;57:22–27. doi: 10.1006/jsre.1994.1103. [DOI] [PubMed] [Google Scholar]
- 50.Lionetti M, Biasiolo M, Agnelli L, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–e26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 51.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 52.Calissano M, Diss JK, Latchman DS. Post-transcriptional regulation of the Brn-3b transcription factor in differentiating neuroblastoma cells. FEBS Lett. 2007;581:2490–2496. doi: 10.1016/j.febslet.2007.04.066. [DOI] [PubMed] [Google Scholar]
- 53.Zhou P, Qian L, Kozopas KM, et al. Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood. 1997;89:630–643. [PubMed] [Google Scholar]
- 54.Reynolds JE, Yang T, Qian L, et al. Mcl-1, a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res. 1994;54:6348–6352. [PubMed] [Google Scholar]
- 55.Craig RW. MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation and tumorigenesis. Leukemia. 2002;16:444–454. doi: 10.1038/sj.leu.2402416. [DOI] [PubMed] [Google Scholar]
- 56.Bernal NP, Stehr W, Coyle R, et al. Epidermal growth factor receptor signaling regulates Bax and Bcl-w expression and apoptotic responses during intestinal adaptation in mice. Gastroenterology. 2006;130:412–423. doi: 10.1053/j.gastro.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 57.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 58.Cowden Dahl KD, Dahl R, Kruichak JN, et al. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–1215. doi: 10.1593/neo.09942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664. 1664e1. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao T, Xu Y. p53 and stem cells: new developments and new concerns. Trends Cell Biol. 2010;20:170–175. doi: 10.1016/j.tcb.2009.12.004. [DOI] [PubMed] [Google Scholar]