Abstract
Interindividual variability in response to antiplatelet therapy results in higher platelet reactivity as well as higher rates of cardiovascular events. Despite substantial effort, the genetic and nongenetic determinants of antiplatelet variability remain poorly understood. Emerging pharmacometabolomic paradigms that integrate systems approaches such as pharmacogenomics have the potential to unveil novel biology regarding disease pathogenesis, reveal the effect of drugs on pathways, and allow better understanding of response variability. Such approaches offer great potential for personalized antiplatelet treatment.
ANTIPLATELET THERAPY AND THE ROLE OF ASPIRIN
Aspirin (acetylsalicylic acid) monotherapy contributes significantly to long-term cardiovascular disease prophylaxis. In addition, aspirin therapy, in combination with other antiplatelet medication, is the standard of care for preventing cardiovascular outcomes in patients with coronary artery disease who are undergoing percutaneous coronary intervention. Despite the general effectiveness of aspirin, substantial interindividual variability, either in on-treatment platelet reactivity or in the occurrence of recurrent cardiovascular events, exists, resulting in a subset of patients who do not respond adequately to treatment. The determinants influencing antiplatelet response variability are multifactorial and include factors such as genetic predisposition, existing comorbidities, and other modifying clinical variables. Although a number of these factors are known, the majority remain unidentified.
Aspirin is the most commonly used medication worldwide and is indicated in several clinical settings having a diverse range of therapeutic properties, including antiplatelet, anti-inflammatory, antipyretic, and analgesic effects. Aspirin is a mainstay in antiplatelet therapy, usually prescribed in combination with clopidogrel, prasugrel, or ticagrelor; it inhibits platelet reactivity by acetylating cyclooxygenase 1, thereby inhibiting the formation of thromboxane A2, a potent platelet agonist, from arachidonic acid. However, given the wide interindividual variability observed in aspirin response despite proper thromboxane A2 reduction, it is likely that other, as of yet unidentified, metabolic and biochemical pathways exist that contribute to aspirin’s mechanism of action. Furthermore, other pathways unrelated to thromboxane A2 can compensate for cyclooxygenase 1 suppression, and this capacity may vary between patients. Therefore, identifying these noncyclooxygenase 1–mediated mechanisms is critical in order to better understand response variability and to tailor antiplatelet regimens to improve cardiovascular disease risk reduction.
PHARMACOMETABOLOMIC PARADIGMS IN CLINICAL RESEARCH
The interaction between modifiable and nonmodifiable cardiovascular risk factors and the underlying molecular and biochemical complexities that result in poor response to aspirin treatment have inhibited the ability of clinicians to prescribe the most effective and personalized antiplatelet therapy. Although strides have been made in several individual areas of study, including genetics, pharmacology, and hematology, it has become increasingly apparent that research efforts need to move beyond traditional approaches and utilize multidisciplinary and integrative systems biology study designs to bridge the gap between genotype, phenotype and disease manifestation and/or recurrence.
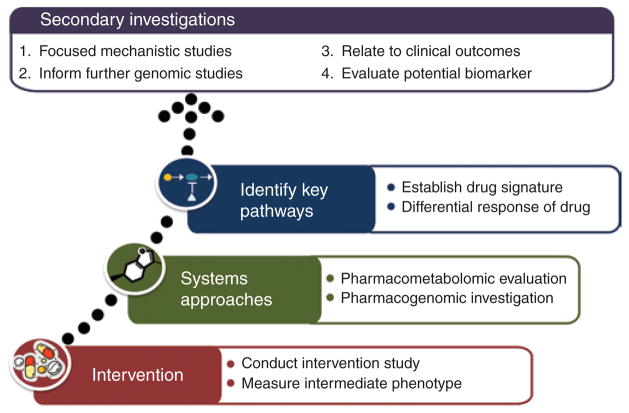
Pharmacometabolomics is a rapidly developing field that leverages a systems pharmacology approach to probe molecular pathways implicated in variability of drug response in order to more comprehensively understand the metabolic changes associated with conditions of interest and to ultimately identify novel biomarkers that can be used for response prediction. More specifically, the use of baseline metabolic profiles and/or the changes in the metabolome induced by pharmacotherapy can aid in the establishment of signatures of drug exposure and elucidate the determinants of drug responsiveness. The true power of this type of inquiry comes from harnessing these observations to complementary studies (Figure 1). For example, by integrating pharmacometabolomics approaches with intermediate clinical phenotypes (e.g., platelet aggregation in the case of antiplatelet therapy), it is possible to design methods-based paradigms that incorporate systematic evaluation of drug exposure and responsiveness. The use of intermediate phenotypes such as platelet aggregation not only allows for comparison of baseline and posttreatment metabolic profiles in good and poor drug responders but also facilitates more mechanistic-driven approaches that can be further probed, validated, and studied ex vivo. Furthermore, by integrating proven pharmacogenomics and functional genetic methodologies, novel information regarding the genetic determinants of drug response and mechanistic causality can be elucidated using informed pathway analysis and detailed functional studies. Indeed, members of the Pharmacometabolomics Research Network have shown the potential of such paradigms by illustrating their use in unveiling novel biology regarding several commonly used medications.1,2
Figure 1.
Example paradigm for pharmacometabolomic investigation.
PHARMACOMETABOLOMICS OF ASPIRIN RESPONSE VARIABILITY
We used pharmacometabolomic approaches to gain insights regarding aspirin response variability in participants of the Heredity and Phenotype Intervention Heart Study.3 To date, we have profiled 165 healthy volunteers who completed a 2-week aspirin intervention (81 mg/day) using a nontargeted gas chromatography–mass spectrometry method, which covers a broad spectrum of the metabolome, and a liquid chromatography— mass spectrometry method targeted for the measurement of metabolites containing an amine functional group. A total of 165 metabolites were measured, among which 49 were significantly affected by aspirin intervention (Table 1; P < 0.05 and Q < 0.05 using Wilcoxon signed-rank test). As expected, metabolites of aspirin, including salicylic acid and 2-hydroxyhippuric acid, were significantly increased after aspirin exposure. Of note, however, metabolites from several other pathways, including purines, fatty acids, glycerol metabolites, amino acids, and carbohydrate- related metabolites, were also significantly altered upon aspirin exposure (Table 1).
Table 1.
Profiled metabolites that are altered upon aspirin exposure
| Pathway | Metabolite | ||
|---|---|---|---|
| Aspirin and xenobiotic metabolism | ↑ 2,183% Salicylic acida | ↑ 7% Glucuronic acida | ↓ 7% 2-Hydroxybutanoic acida |
| ↑ 160% 2-Hydroxyhippuric acida | ↑ 26% Benzylalcohola | — | |
| Purine pathway | ↑ 77% Inosinea | ↓ 5% Hypoxanthinea | — |
| ↑ 8% Guanosinea | ↓ 9% Xanthinea | — | |
| Fatty acids | ↓ 28% Oleic acida | ↓ 20% Azelaic acida | ↓ 15% Lauric acida |
| ↓ 31% Palmitoleic acida | ↑ 11% Threonic acida | ↓ 32% Elaidic acida | |
| ↓ 15% Myristic acida | ↓ 9% Isolinoleic acida | ↓ 14% Capric acida | |
| ↓ 19% Linoleic acida | ↓ 10% Palmitic acida | ↓ 9% Methylhexadecanoic acida | |
| Glycerol metabolism | ↓ 14% 1-Monooleina | ↓ 13% Glycerola | ↑ 14% Glycerol-3-galactosidea |
| Amino acids and related metabolites | ↓ 40% Glycylglycinec | ↓ 9% Phenylalanineb | ↓ 5% γ-Aminobutyric acidc |
| ↓ 24% L-Aspartic acidb | ↓ 9% Ethanolaminec | ↓ 5% Serineb | |
| ↑ 19% O-phosphoethanolaminec | ↑ 8% Indole-3-acetatea | ↓ 5% Ornithinec | |
| ↑ 16% Methioninea | ↑ 7% Serotoninc | ↓ 4% L-Lysinec | |
| ↑ 12% Phenylacetic acida | ↓ 6% Taurinec | ↓ 2% L-Histidinec | |
| ↑ 11% Hydrocinnamic acida | ↓ 6% Leucineb | — | |
| ↓ 10% L-Glutamic acidb | ↓ 5% L-Valineb | — | |
| Carbohydrates and carbohydrate conjugates | ↑ 5% Glucosea | ↑ 7% Threitola | ↓ 10% Xylulosea |
| ↓ 13% 3-Hydroxybutanoic acida | — | — | |
| Other | ↑ 7% Conduritol-β-epoxidea | ↑ 9% Maleimidea | — |
↑ = Metabolite increased after aspirin exposure; ↓ = metabolite decreased after aspirin exposure. Median percentage change in metabolite levels is provided. For metabolites measured by both GC-MS and LC-MS, percentage change from LC-MS is provided.
GC-MS, gas chromatography-mass spectrometry; LC-MS, liquid chromatography-mass spectrometry.
Measured by GC-MS.
Measured by GC-MS and LC-MS.
Measured by LC-MS
Although establishing signatures of drug exposure can reveal important biological insights, comparison of metabolomic profiles between good and poor responders to pharmacotherapy can provide critical information regarding response variability as well as potentially identify novel pathways/biomarkers involved in drug function. We performed untargeted gas chromatography– mass spectrometry–based metabolomic profiling of serum samples from 76 healthy individuals from the Heredity and Phenotype Intervention study who were either good (n = 40) or poor (n = 36) responders (see Supplementary Figure S1 online).4 Good and poor responders were defined as participants who were from the first (good responders) and fourth (poor responders) quartiles of postaspirin collagen-stimulated platelet aggregation adjusted for participant age and pretreatment collagen- stimulated platelet aggregation level (see Supplementary Figure S2 online). Consistent with our data regarding signature of aspirin exposure, we observed significant differences in purine metabolite levels between good and poor responders. Specifically, aspirin intervention resulted in greater changes in inosine and adenosine levels in poor responders as compared with good responders, resulting in significantly higher inosine and adenosine concentrations in poor responders after aspirin intervention (P = 0.003 and 0.02, respectively). Furthermore, in an independent cohort consisting of 19 good and 18 poor responders, we validated our findings regarding postaspirin inosine levels being significantly higher in poor as compared with good responders (P = 0.05); however, adenosine was unable to be measured in the replication sample.4
PHARMACOMETABOLOMICS-INFORMED PHARMACOGENOMICS APPROACH
Despite being highly heritable, few genetic variants are robustly associated with aspirin response. With this in mind, we extended our investigation by implementing our “pharmacometabolomics- informed pharmacogenomics” paradigm2 in order to identify novel genetic determinants of aspirin response in candidate genes that regulate biosynthesis, transport, and degradation of purine metabolites. Detailed methods of this approach have been previously reported.4 We tested for association between ex vivo collagen-stimulated platelet aggregation and nine purine candidate genes in 718 Heredity and Phenotype Intervention Study participants who underwent aspirin intervention. Using this approach, we identified 51 single-nucleotide polymorphisms in the adenosine kinase (ADK) gene region that had associations with platelet aggregation in response to aspirin therapy (P < 5 × 10−4), the strongest of which was the intronic variant rs16931294 (P = 3.4 × 10−4; β = 0.8).4 The association between rs16931294 and platelet aggregation was replicated in an independent sample of 341 participants from the Pharmacogenomics of Antiplatelet Intervention Study (P = 0.002; β = 1.9).
Using the results of our pharmacogenomic analysis, we went back to our pharmacometabolomic data set to probe the relationship between rs16931294 and purine metabolite levels. We observed that rs16931294 was significantly associated with adenosine monophosphate (P = 0.04), xanthine (P = 0.01), and hypoxanthine (P = 0.02) levels before aspirin intervention. After aspirin intervention, this single-nucleotide polymorphism was also associated with levels of inosine (P = 0.007) and guanosine (P = 0.02). These secondary investigations that make use of both pharmacometabolomic and pharmacogenomic data not only further our understanding of aspirin’s mechanism of action but also allow continual refinement of our hypotheses.
PERSPECTIVES AND CONCLUSION
Despite the wide use of aspirin in a growing number of clinical settings, the mechanisms by which it exerts its therapeutic effects are, in some cases, poorly understood. In this article, we highlight the potential of combining pharmacometabolomic protocols with integrative systems biology approaches such as pharmacogenomics in order to identify novel pathways, metabolites, genes, and genetic variants implicated in response to antiplatelet therapy. Although these results represent only a first step and require independent validation, our findings suggest, for the first time, not only that metabolites of the purine pathway are important in establishing metabolomic signatures of aspirin exposure but also that the circulating posttreatment levels of purines may aid in differentiating good from poor aspirin responders. Furthermore, by integrating pharmacometabolomic and pharmacogenomic approaches, we identified genetic variants in ADK that alter platelet aggregation in response to aspirin and subsequently showed that these variants are significantly associated with purine levels. Of note, although our initial studies primarily focused on purine metabolism, our results indicate that several other pathways, including fatty acids, glycerol metabolites, amino acids, and carbohydrate-related metabolites, also play a role in defining the metabolic signature of aspirin exposure. Additional studies evaluating the effect on aspirin exposure on these pathways are also warranted.
Although our results have unveiled new biology regarding aspirin exposure and response variability, we acknowledge a couple of important limitations. First, aspirin is rapidly deacetylated in the body into salicylic acid, and, given the known anti-inflammatory properties of salicylic acid,5 it is possible that the changes in metabolite levels we observed in this study are mediated by salicylic acid and not by aspirin directly. To evaluate this, further investigations comparing the metabolomic effects of aspirin and salicylic acid seem warranted. Another limitation of this investigation is that only healthy individuals were evaluated in this study. We believe that comprehensive pharmacometabolomic investigation of patients with coronary artery disease on aspirin therapy is critical to validate the generalizability of our results and to define the role of these pathways/metabolites/genes in on-treatment cardiovascular event risk. Indeed, clinical studies of cardiovascular patients that utilize these types of integrative pharmacometabolomic paradigms are currently under way by our group.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health grants RC2GM092729, U01-HL72515, K23-GM102678, and U01 HL105198; the Netherlands Metabolomics Centre, as part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research; the University of Maryland General Clinical Research Center (GCRC, M01-RR-16500), the Johns Hopkins University GCRC (M01-RR-000052), the Clinical Nutrition Research Unit of Maryland (P30-DK072488), the Pharmacometabolomics Research Network, and the National Center for Research Resources.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
CONFLICT OF INTEREST
R.K.-D. has patents in the field of metabolomics. The other authors declared no conflict of interest.
References
- 1.Abo R, et al. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes’ single-nucleotide polymorphism imputation: selective serotonin reuptake inhibitor response pharmacogenomics. Pharmacogenet Genomics. 2012;22:247–253. doi: 10.1097/FPC.0b013e32835001c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji Y, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell BD, et al. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am Heart J. 2008;155:823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yerges-Armstrong LM, et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2013;94:525–532. doi: 10.1038/clpt.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu XM, Sansores-Garcia L, Chen XM, Matijevic-Aleksic N, Du M, Wu KK. Suppression of inducible cyclooxygenase 2 gene transcription by aspirin and sodium salicylate. Proc Natl Acad Sci USA. 1999;96:5292–5297. doi: 10.1073/pnas.96.9.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.