SUMMARY
The transduction of transmembrane electric fields into protein motion plays an essential role in the generation and propagation of cellular signals. Voltage-sensing domains (VSD) carry out these functions through reorientations of S4 helix with discrete gating charges. Here, crystal structures of the VSD from Ci-VSP were determined in both, active (Up) and resting (Down) conformations. The S4 undergoes a ~5 Å displacement along its main axis accompanied by a ~60o rotation, consistent with the helix-screw gating mechanism. This movement is stabilized by a change in countercharge partners in helices S1 and S3, generating an estimated net charge transfer of ~1 eo. Gating charges move relative to a “hydrophobic gasket” that electrically divides intra and extracellular compartments. EPR spectroscopy confirms the limited nature of S4 movement in a membrane environment. These results provide an explicit mechanism for voltage sensing and set the basis for electromechanical coupling in voltage-dependent cellular activities.
INTRODUCTION
Electrical excitability derived from voltage-dependent ion channels is a fundamental biological phenomenon that underlies key physiological events, including cellular communication, information coding, motility and sensation. Few mechanisms can endow a membrane protein with the ability to respond to changes in the transmembrane electric field. As predicted early on (Hodgkin and Huxley, 1952), these are associated with the reorientation of a set of fixed charges (or dipoles) that respond to changes in transmembrane voltage.
Initial examination of the first amino acid sequences of voltage-gated channels suggested they were assembled from two separate domains, a voltage-sensing domain (VSD) comprised of four transmembrane helices (S1 to S4), and a pore domain (helices S5 and S6) responsible for gate opening and ion selectivity (Bezanilla, 2008). In most voltage-gated channels, the S4 segment is decorated with 4–8 basic amino acids (mostly arginines), a fact that made it a logical candidate to fulfill the requirements of the voltage sensor. Extensive site-directed mutagenesis experiments convincingly demonstrated the role of these S4 charges in sensing voltage. Evidence that the S4 physically moves in response to voltage changes came from a series of insightful experiments based on the reactivity of cysteine mutants to sulfhydryl-based chemical labels (Yang et al., 1996) and voltage clamp fluorometry measurements (Cha and Bezanilla, 1997; Mannuzzu et al., 1996).
The mechanism of voltage sensing in excitable cells remains one of the most challenging subjects in modern biophysics. Yet, in spite of a great deal of progress derived from multidisciplinary approaches, defining this mechanism still requires high-resolution structures of VSDs in both the activated (Up) and resting (Down) conformations. Crystal structures of the voltage-dependent K+ channels KvAP (Jiang et al., 2003a) and Kv1.2 (Long et al., 2005; Long et al., 2007), and two prokaryotic Na+ channels (Payandeh et al., 2011; Zhang et al., 2012) have clarified many of the basic concepts regarding VSD the architecture of the VSD and its associated pore domain. Based on their voltage dependences, not surprisingly, these VSDs all appear to populate the same Up state conformation at 0 mV in the absence of an electric field at the crystal conditions. The Down conformation has been vigorously pursued but has remained structurally unattainable.
The recent discoveries of voltage-sensing phosphatases (VSP) (Murata et al., 2005) and voltage-gated proton channels (Hv1) (Ramsey et al., 2006; Sasaki et al., 2006) have revealed that VSDs are not exclusive to ion channels with canonical pore domains, but can operate as independent functional domains. The voltage sensitive phosphatase from C. intestinalis (Ci-VSP) has a PTEN-related phosphoinositide phosphatase that is controlled by a standard VSD with high sequence similarity to the S1–S4 segments found in Na+ and K+ channels (Figure 1A). To address the fundamental questions regarding how membrane proteins sense transmembrane voltages, we set out to determine the structure of a voltage-sensing domain under conditions that stabilize both the Up and the Down conformations in the same experimental system. To this end, we have focused our attention on Ci-VSD, the voltage sensing domain of Ci-VSP in the absence of its phosphatase.
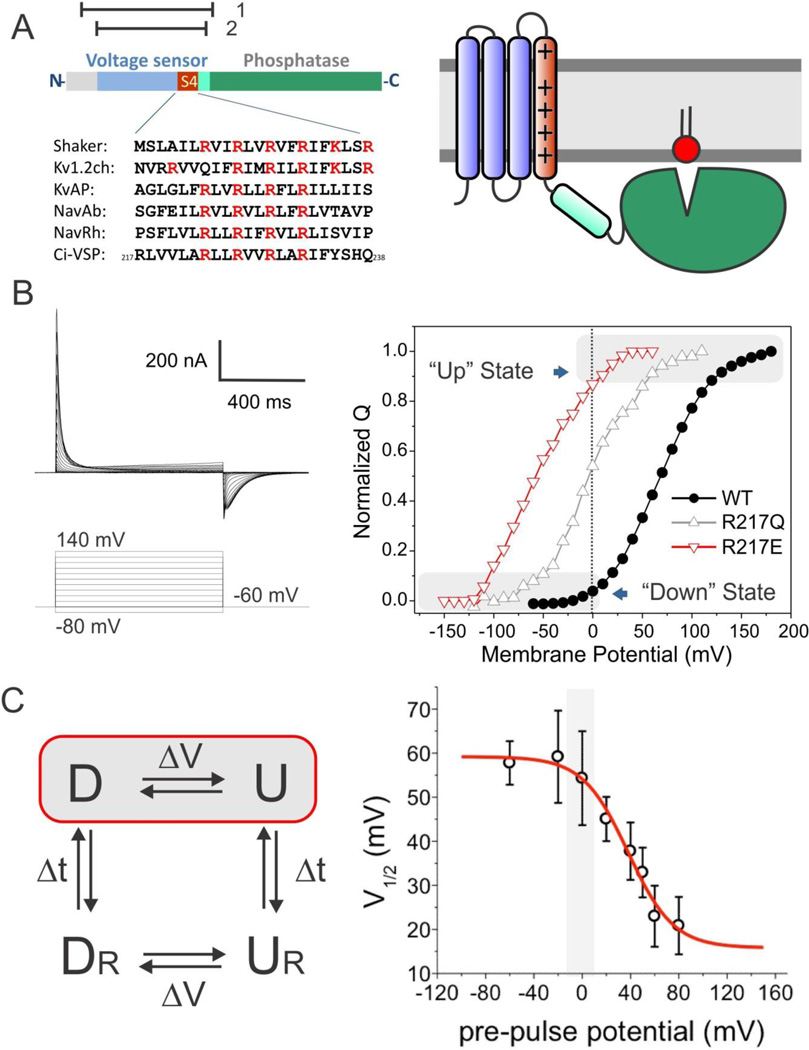
Figure 1.
Stabilizing two conformations of Ci-VSD by local field engineering. A. Ci-VSP is composed of a voltage sensing domain and a phosphatase domain. Left, linear schematics of Ci-VSP sequence regions. Highlighted in red is the conserved S4 helix containing positive charges every three residues. The relative location the protein constructs for crystallization is shown on top: residues 89–260 (1) for R217E mutant and 106–244 (2) for WT. Right, general cartoon of Ci-VSD embedded in a lipid membrane. B. Representative traces of WT Ci-VSP gating (sensing) currents. The voltage dependence of Ci-VSP can be dramatically left shifted by mutations on the extracellular end of S4 at position 217: V½ = +58 mV for WT (black circle); − 15 mV for R217Q (white triangle); −62 mV for R217E (red triangle). Under 0 mV conditions, Ci-VSD populates the activated (“Up”) state for R217E mutant and the resting (“Down”) state for WT. C. Positive holding potential can promote transition of VSDs into “relaxed” state that generates a leftward shift of its voltage dependence. Left, basic kinetic cycle of canonical gating transitions. Right, voltage dependence of the transition into the relaxed state. Noticeable differences only take place for holding potentials higher than 20 mV. Therefore, at 0 mV, Ci-VSD WT and R217E mutants likely populate the canonical Down and Up states respectively.
RESULTS
Manipulating the conformational landscape of Voltage Sensors at 0 mV
When expressed in Xenopus oocytes, Ci-VSP produces nonlinear capacitive currents (Figure 1B, left) similar to those observed in voltage-gated channels (Armstrong and Bezanilla, 1973; Murata et al., 2005) and this behavior is fully recapitulated by the sensor alone (Kohout et al., 2008; Villalba-Galea et al., 2008). Yet, in comparison to VSDs from ion channels, the Ci-VSP’s Q–V curve is greatly shifted to more positive potentials, with a V½ near +60 mV (Murata et al., 2005) (Figure 1B, right). A fortunate consequence of a shifted Q-V curve is that, in the absence of a membrane potential, Ci-VSD should stably populate the Down conformation of the voltage sensor at zero mV. Furthermore, the Q–V curve of Ci-VSP can be easily shifted left by neutralizing a basic residue at position 217 (Kohout et al., 2008) (Figure 1B). Adding an acidic residue (R217E) generates a negative shift of ~120 mV in the hyperpolarizing direction (V½ near −60 mV), and provides a straightforward strategy to stabilizing the Ci-VSD “Up” conformation, also at zero mV (Figure 1B).
However, when exposed to long-term depolarization, VSDs transition to new “relaxed” conformations of the Up and Down states (UR, DR) (Villalba-Galea et al., 2008). In these states, the voltage dependence is sharply shifted towards negative potentials as a result of changes in the kinetics of charge return. Therefore, transitions to relaxed states occur in parallel to the standard voltage-driven D ↔ U equilibrium, according to a simple kinetic cycle (Figure 1C, left) (Villalba-Galea et al., 2008). We wondered whether the long-term influence of zero mV conditions would stabilize the relaxed states in Ci-VSD in our biochemical sample. Figure 1C (right) shows the dependence of a steady-state pre-pulse on the V½ of the Q-V curve. The results indicate that the transition to the relaxed states appears to only take place at large positive potentials.
These results underlies the fact that the engineering of local field through one particular residue in Ci-VSD allows the study of its canonical (D ↔ U) structural rearrangements at zero mV. On the basis of this strategy, we present the crystal structures for Ci-VSD in the Up conformation (R217E mutant) at 2.5 Å resolution and Down conformation (WT) at 3.6 Å resolution. In spite of the different resolutions, with additional spectroscopic and computational arguments, these structures provide rich insights into the mechanisms underlying voltage sensing and electromechanical coupling in voltage sensing domains.
Structure of Ci-VSD in its activated conformation
Crystallization of the E. coli-expressed (Li et al., 2012) Ci-VSD was accomplished in complex with synthetic Fab as crystallization chaperon (Fellouse et al., 2007). Ci-VSD R217E in complex with Fab 33F12_4 from two distinct crystallization conditions generated crystals that diffracted to resolutions of 2.5 and 2.8 Å respectively (Table 1, Figure S1A). The essentially identical structures with different water molecules inside voltage sensor were solved by molecular replacement using the Fab molecule and refined to Rwork/Rfree of 20.0/24.0 (2.5 Å data) and 21.2/26.4 (2.8 Å data). The Fab binds to an epitope formed by the S3–S4 loop and the top of S4 (Figure S1B). The head groups of three detergent molecules and a solvent molecule were observed in the vicinity of the S3–S4 loop (Figure S1D), consistent with the hydrophilic environment anticipated around the extracellular surface.
Table 1.
Crystallographic data collection and refinement statistics.
| R217E + 33F12_4 | R217E + 33F12_4 | WT + 39D10_18 | |
|---|---|---|---|
| PDB ID | 4G7V | 4G7Y | 4G80 |
| Space group | P6522 | P6522 | P1 |
| Cell dimensions | |||
| a, b, c (Å) | 121, 121, 230 | 121, 121, 230 | 77.3, 94.2, 193.9 |
| α7 β, γ (◦) | 90, 90, 120 | 90, 90, 120 | 102.6, 93.5, 105.3 |
| Resolution (Å) | 50-2.5 (2.66-2.52)a | 50-2.8 (2.85-2.80) | 50-3.6 (3.66-3.60) |
| Rsym | 16.8 (134.7) | 16.8 (95.8) | 14.4 (87.1) |
| I/σ/ | 8.2 (2.0) | 9.2 (2.2) | 12.2 (1.0) |
| Completeness (%) | 99.8 (100) | 100 (100) | 95.1 (83.2) |
| Redundancy | 5.1 (5.3) | 8.2 (7.2) | 2.9 (2.2) |
| Refinement | |||
| Resolution (Å) | 38.6-2.5 | 37.4-2.8 | 50-3.6 |
| No. reflections | 33756 | 24429 | 55790 |
| Rwork/Rfree (%) | 17.3/23.6b | 20.6/26.1 | 24.8/29.2 |
| No. atoms | |||
| Protein | 4303 | 4262 | 17446 |
| Ligand/ion | 153/1 | 16/2 | |
| Water | 197 | 35 | |
| B-factors (Å2) | |||
| Protein | 48 | 54 | 150 |
| Ligand/ion | 39/32 | 48/44 | |
| Water | 41 | 39 | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.011 | 0.011 | 0.005 |
| Bond angles (◦) | 1.316 | 1.338 | 0.38 |
| Ramachandranc | |||
| Favored | 97.0% | 92.1% | 93.8% |
| Allowed | 2.5% | 7.3% | 5.7% |
| Outlier | 0.5% | 0.5% | 0.5% |
Values in parenthesis are for the highest resolution shell.
5 % of randomly chosen data was used in the Rfree calculation.
Values were calculated by MolProbity (Chen et al., 2010).
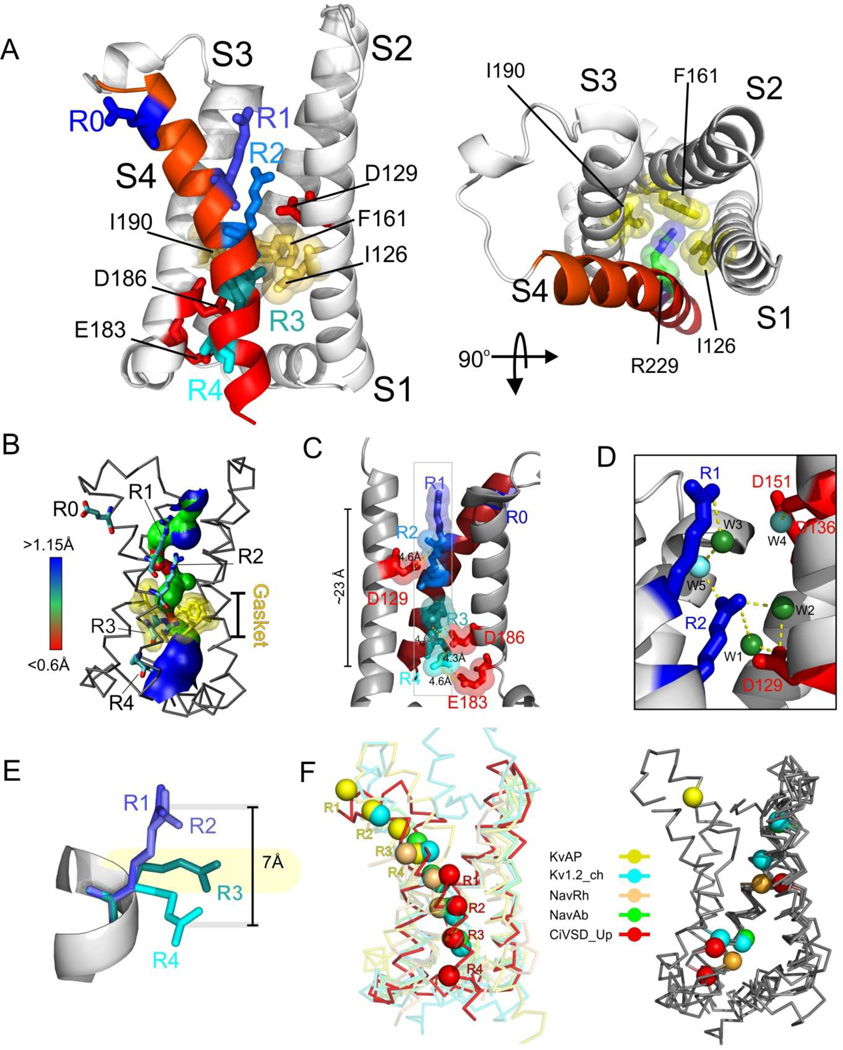
The Ci-VSD R217E is arranged as an antiparallel four helix bundle (Figure 2A), in agreement with the basic three-dimensional architecture of all known VSDs (Jiang et al., 2003a; Long et al., 2007; Payandeh et al., 2011; Zhang et al., 2012). S1 is preceded by a short, 3½-turn amphipatic helix S0, and is likely oriented parallel to the plane of the membrane. The relative alignment of helices S1–S4 sets up a fairly large water-filled extracellular cavity and a smaller opening on the intracellular side (Figure 2B). There are a total of five charges in Ci-VSD’s S4 helix. The topmost, R217 (mutated to E in this structure), points straight away from the outer vestibule and presumably engages in direct interactions with the lipid head groups. Electrophysiological evidence suggests that R217 does not contribute to charge transfer (Villalba-Galea et al., Submitted), so we refer to it as R0. The putative gating charges are thus, R223 (R1), R226 (R2), R229 (R3) and R232 (R4). The four gating charges appear to be stabilized by a combination of hydrogen bonding and direct electrostatic ion pairing. R1 and R2 are in a fully extended rotameric conformation, pointing straight towards the extracellular vestibule (Figure 2A). From the two R217E structures, we identified five crystallographic waters in the external cavity, with two of them establishing H-bonding interactions with R1 (Figure 2D). There are only three obvious countercharges: D129 in S1 (D1) and D186 and E183 in S3 (D2 and E3, respectively). R2 is within 4.6 Å of D1 and is simultaneously in range for H-bonding with two waters (Figure 2C 2D). R3 is located well within range (4.6 Å) of ion pair interactions with D2, while R4 appears neatly intertwined between D2 and E3 (4.3 Å and 4.6 Å, respectively). Surprisingly, and albeit the strong sequence and structural conservation with VSDs in K+ and Na+ channels, we see no viable countercharges in S2 of Ci-VSD, a fact that points to an interesting heterogeneity in the way gating charges might traverse the transmembrane electric field in other VSDs.
Figure 2.
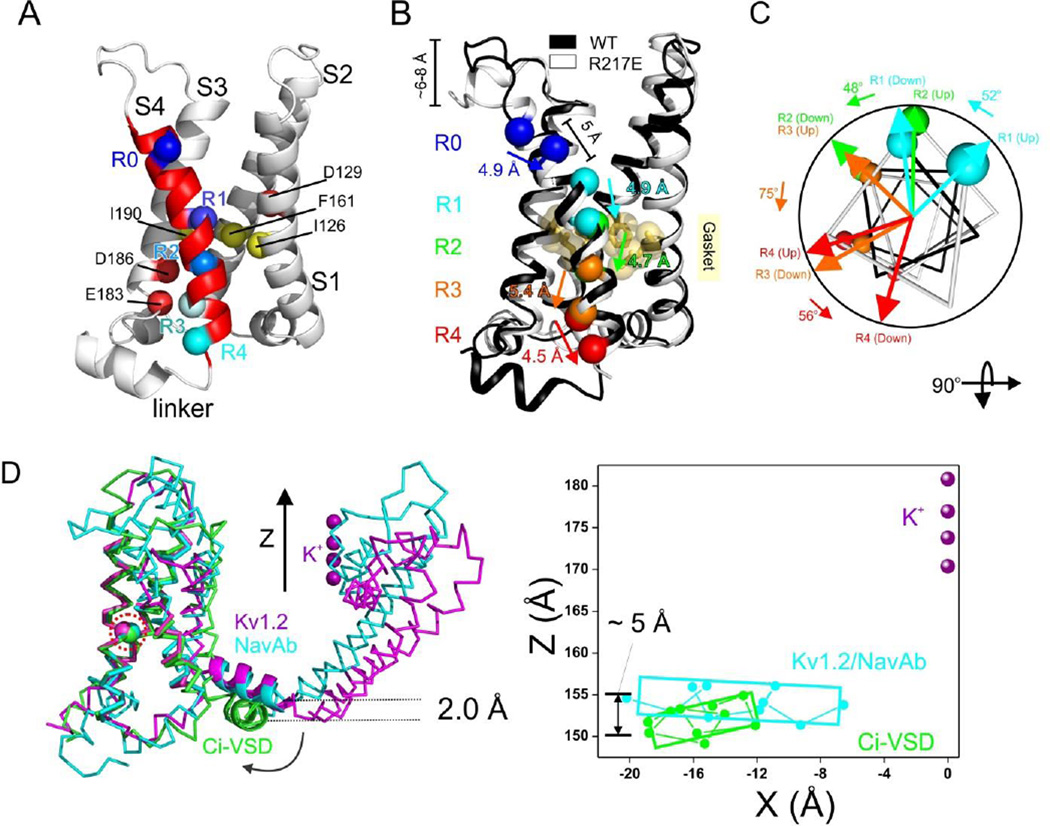
Structure of Ci-VSD R217E in the activated (“Up”) conformation. A. Side (left) and top (right) view of the Up conformation of Ci-VSD R217E in cartoon representation with S4 colored red. The four positive charges (R223, R226, R229 and R232) are depicted as sticks and colored in a series of blue shades. A well-defined hydrophobic gasket was formed by I126, F161, I190 with a gating charge R229 (R3). B. Architecture of the internal and external vestibules as calculated with the program HOLE61. The narrowest region is surrounded by the hydrophobic gasket at R3 (in yellow). C. With the subtle tilt and bend of S4, all four gating charges form a neat line perpendicular the hydrophobic gasket. The negative counter charges (D129:D1, D186:D2 and E183:E3) are located in S1 and S3. D. The top two arginines, R223 (R1) and R226 (R2) above the hydrophobic gasket are stabilized by hydrogen bonds with crystallographic waters between the negative charges D129 and D151. E. Left, heterogeneity of arginine positions within S4 was highlighted with VSD alignment in reference to Kv1.2 chimera (emerald): KvAP (yellow, RMSD=1.4 Å), NavRh (orange, RMSD=3.0 Å), NavAb (green, RMSD=1.1 Å) and Ci-VSD R217E (red, RMSD=1.1 Å). RMSD is calculated with Cα of aligned region (S1+S2+S3a) indicating the general agreement of VSD scaffold. The gating charges, shown in sphere, follow an arched path along S4 and but are distinctly different among the homologs even though all of them were expected to populate at Up state. Right, heterogeneity of putative counter charge positions within the VSD scaffold. Based on the crystal structures, only countercharges potentially involved in direct ion pairing with gating charges are shown.
In spite of being located on a standard α-helix, the slight bend and tilt of S4 place all four gating arginines along a remarkably straight line (Figure 2C). We argue that this has important implications to the potential mechanism of charge transfer, as the aligned R1–R4 charges appear to fit through an in-plane semi-circular arrangement of hydrophobic side chains formed by residues I126, F161 and I190 (Figure 2A, right). This is the obvious equivalent of the so-called hydrophobic plug (Campos et al., 2007) or charge transfer center (Tao et al., 2010) in K+ channels as an effective dielectric barrier to the permeation of water and ions and defining the extra and intracellular sides of the transmembrane electric field (Figure 2B). R3 is in the unique position of being in plane with the hydrophobic gasket, presumably at the center of the focused electric field, whereas both R1 and R2 are located well above and R4 is below the gasket.
A structural comparison of the Up conformation of Ci-VSD with all existing voltage sensor structures provides key insights in relation to the general organization of the gating charges and countercharges. Currently available sensors were aligned through backbone of the S1+S2+S3a helices, these include VSDs from two distinct archetypes of K channels (KvAP and Kv1.2-chimera) and two prokaryotic Na channels (NavAb and NavRh) (Figure 2E). Beyond decent agreement on VSD scaffold, three main observations can be derived from this comparison. First, in spite of the fact that all structures present (in principle) the activated conformation of each sensor, there is a remarkable difference in the positions of the gating charges. Second, gating charges appear to follow a neat arched path, where KvAP represents the topmost range of positions and Ci-VSD the bottom. Indeed, the distance between the R1 backbone of KvAP and Ci-VSD at activated states is around 21 Å, which is larger than the proposed conformation change for any VSD. Third, positional heterogeneity also extends to the number and placement of the putative countercharges that serve as electrostatic partners to the individual gating charges (Figure 2E, right). These countercharges are located throughout S1, S2 and/or S3 among different sensors, according to their propensity to establish stable ion pairings with selected gating charges. Since the heterogeneity between VSD systems could be larger than the extent of individual conformation changes associated with voltage gating, the present alignment strongly argues against the direct comparison of structural information across VSD systems.
Conformational changes in a membrane environment
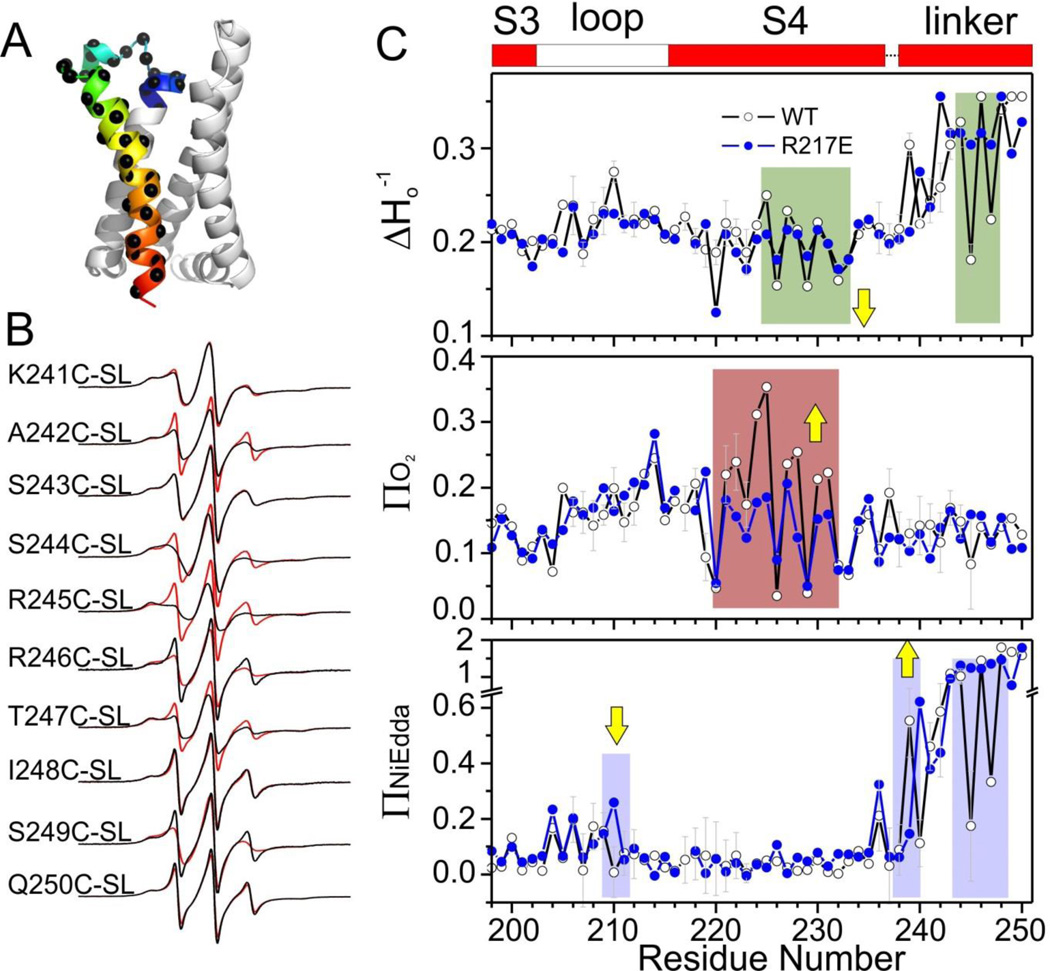
There are extensive evidences that the lipidic environment plays a critical role on the conformation, stability and functional behavior of VSDs (Ramu et al., 2006; Schmidt et al., 2006; Zheng et al., 2011). So it is worth asking whether the obtained crystal structures would be a reflection of the voltage-driven events in a biological membrane and what kind of structural rearrangement we expect between the Up and Down conformations of Ci-VSD. We addressed these issues by evaluating the conformation of the S3–S4 hairpin through site-directed spin labeling EPR spectroscopy of membrane-reconstituted Ci-VSD WT and R217E. For this data set, all measurements (Figure S2) were made in the absence of a membrane potential, so the spin-labeled sensors are expected to populate the Up state in R217E and the down state for WT Ci-VSD.
41 cysteine mutants covering the S3–S4 loop, S4 and S4-phosphatase linker (Figure 3A) in both WT and R217E background were purified, spin-labeled, and reconstituted in POPC:POPG liposomes (3:1, mol:mol (Li et al., 2012)). We focused on two types of dynamic EPR structural information: First, the spin-probe motional freedom was estimated from the inverse of the central line width of the first derivative absorption spectra (Δ Ho−1). Second, the spin-probe solvent accessibility was evaluated by collisional relaxation methods. Here, non-polar oxygen (ΠO2) serves as a contrast agent to evaluate membrane exposure, while polar Ni(II) ethylenediaminediacetic acid (Π NiEdda) reports on the extent of aqueous exposure. The periodicity of ΠO2 oscillations indicates the secondary structure of S4 as α-helix and low ΠNiEdda levels clearly defines the boundaries of S4, both of which agree very well with Ci-VSD R217E structure, strongly suggesting that the obtained Ci-VSD R217E crystal structure samples its native conformation in a biological membrane.
Figure 3.
The nature of S4 rearrangement in a lipid environment. A. The region studied by site-directed spin labeling EPR spectroscopy covering the S3–S4 loop and entire S4 and the S4-phosphatase linker. B. Representative EPR spectra of the S4-phosphatase linker (residues 241–250) on both Ci-VSD WT (black) and R217E (red) background. C. Mobility, oxygen accessibility and NiEDDA accessibility of WT (black) and R217E (blue). The characteristic large ΠO2 oscillation and low ΠNi of transmembrane region are consistent with the boundary of S4 (shown on the top as reference) defined by crystal structure. Comparing the ΠNi, there is a decrease at the top of S4 with a simultaneous increase at the bottom of S4 from R217E to WT. This is consistent with a modest down movement of S4 from the Up state (R217E) to the Down state (WT). Residues 245 and 247 have significantly lower mobility and ΠNi in WT indicating stronger tertiary contact.
As a first approximation, the spectral data set of Ci-VSD WT (Figure S2) confirms its similarly dynamic nature (Figure 3C top), with the exception of gating charges R2, R3 and R4, which become further motionally-restricted. This is accompanied by an increase in lipid exposure in the center of S4 (Figure 3C, center). Most importantly, aqueous accessibilities (ΠNiEdda) show simultaneous reduction in at the extracellular end of S4 (residue 210) together with an increase at its intracellular end (residue 239) (yellow arrows, Fig 3C bottom). This ΠNiEdda boundary shifts explicitly demonstrates the downward S4 helix movement between Up (R217E) and Down (WT) conformations. In addition, these accessibility changes suggest a modest vertical displacement (about one residue corresponding to ~2 Å).
Larger spectroscopic changes can be seen at the inter-domain linker (after residue 236, not present in the R217E crystal structure). In particular, the lower ΠNiEdda accessibility and lower mobility show that residues 245 and 247 in the S4-phosphatase linker of Ci-VSD WT are likely involved in additional tertiary contacts (Figure 3C). The nature and extend of these changes are a clear demonstration of propagation of the S4 helix movement into the linker region.
Structure of Ci-VSD at rest
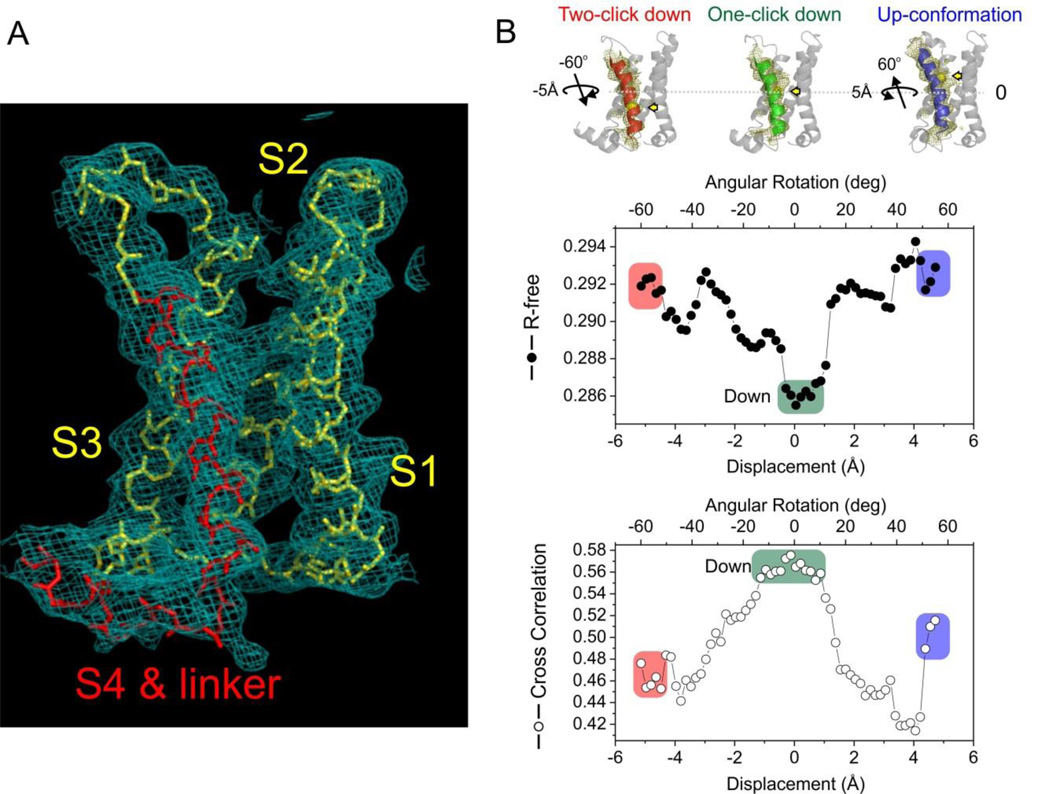
Although we followed a similar strategy to determine the structure of Ci-VSD WT in its Down conformation, the crystallization proved to be a keen challenge (supplementary information). The working data set for Ci-VSD WT in complex with Fab 39D10_18 was ultimately merged from 8 fractional data sets. The structure was solved by molecular replacement and refined to a final Rwork/Rfree of 26.9/30.3, including data to 3.6 Å (I/σ = 1.0 with 83% completeness at the highest shell) for the nominal 3.8 Å data (Figure 4A, S3A, S3B, S3C). In the Down conformation complex, the Fab binds to a different epitope formed by the external surface between S2 and S3.
Figure 4.
Structure of WT Ci-VSD in the resting (“Down”) conformation. A. Electron density map (2Fo-Fc at σt1.0) of WT Ci-VSD at 3.6 Å resolution. The backbone of S4 and inter-domain linker are colored red. B. Top: Three potential models within the electron density map of Ci-VSD WT: Two-click down (S4 in red), One-click down (S4 in green) and Up-conformation (S4 in blue). The rotation angle and vertical displacement of S4 were measured in reference to the One-click down model. The first Arginines R223 of three models were shown with yellow sphere and arrow to illustrate the vertical displacement on S4 helix. The independent R-free values and correlation coefficients were averaged among frames of every 2° rotation for simplicity.
Ci-VSD WT structure was built at backbone level within the continuous electron density map (Figure 4A) assisted with R217E structure. Initial analysis of the Ci-VSD WT structure revealed a similar scaffold as that of R217E mutant, but with substantial changes associated with the conformation of the S3–S4 loop (Figure S5A) and the addition of distinct electron density towards the intracellular face of the bundle (Figure 4A, Fig S5A). We interpreted the additional density as a short (2 ½ turn) helix (Hobiger et al., 2012) corresponding to the inter-domain linker that connects the VSD to the phosphatase. At the present resolution, however, crystallographic uncertainty associated with S4 registry needs to be addressed to fully interpret the substantial deviations from R217E structure. Repeated attempts on generating and identifying electron density markers along S4 were unsuccessful due to large and systematic reductions in the quality and resolution of the Ci-VSD WT crystals (supplementary materials). Therefore, we developed alternative lines of evidence in support of the present interpretation of the Ci-VSD WT electron density map.
First, EPR data demonstrated that gating charges R2, R3 and R4 in WT are shielded from membrane exposure (low O2 accessibility) by strong tertiary contacts (low to−1). This limits the position of the S4 helix to only three possibilities inside the WT electron density map (Figure 4B, top): an Up conformation (same S4 position as R217E structure), a 3-residue downward movement of S4 (one “click” down), or a 6-residue downward movement of S4 (two “clicks” down).
Second, taking into account that in the Ci-VSD WT data the additional electron density from the S4-phosphatase linker includes all residues in the expressed Ci-VSD construct (up position 244) and reaches the reported N-terminal end of the phosphatase domain (Kohout et al., 2010), it represents a useful anchor not only for determining the registry of the S4 helix but also for any future effort to model full-length Ci-VSP. In the reference to the one-click down model, the hypothetical Up-conformation model shifts the S4 helix 3-residue upward and leaves unaccounted electron density after the last residue 244 around S4-phosphatase linker (Figure S5C, S5D, S5E). Fitting the two-click down model shortens the S3–S4 loop, which obviously deviates from the continuous electron density map of the loop (Figure S4B). Furthermore, the two-click down model positions the residue R232 in a water-exposed environment (Figure 4B, top left), which is inconsistent with the strong tertiary contact derived from EPR spectroscopy. By excluding the two of above options, we argue that the one-click down model is the only compatible S4 arrangement to fit both S3–S4 loop and S4-phosphatase linker in the electron density map. Furthermore, the one click down model is also in agreement with the direction and modest extent of S4 displacement indicated from the shift in water accessibility (Πin wat).
Third, to quantitatively evaluate explicit S4 positions within current experimental electron density maps, the crystallographic uncertainties were addressed directly by an independent Molecular Dynamics Flexible Fitting (MDFF) based (Trabuco et al., 2008; Trabuco et al., 2009) structure refinement procedure xMDFF (Figure S4, supplementary information). In brief, the xMDFF refinement started with a tentative phase model constructed from medium-confidence homology information. During refinement, the initial model underwent a large-scale deformation of RMSD 5 Å yielding structures with S4 helices in excellent agreement to those from the WT Ci-VSD. To further confirm explicit S4 positions, potential structural models were generated by gradual shift and rotation of S4 helix from the Up-conformation model to the two-click down model in 2000 even steps (in all, a ~10 Å vertical displacement and ~110° rotation). Each of the 2000 structures as initial phase model was independently refined and two independent parameters were calculated for model evaluation: 1. crystallographic R-free value; 2. Correlation Coefficient (CC) between the experimental density map of the S4 region and the calculated electron density from the refined model. Figure 4B shows that the global R-free minimum and CC maximum coincide in a region that unambiguously determines the most probable position of the S4 helix. xMDFF clearly differentiates the pattern of side chain associated with specific S4 helix position while the individual side chain is not visible at 3.6 Å resolution. The resulting S4 position is 3-residue lower than that of the R217E structure, positioning R2, R3 and R4 in a protein environment and away from lipids, and in excellent agreement with the one-click down model.
Molecular basis of gating charge transfer in voltage sensing domains
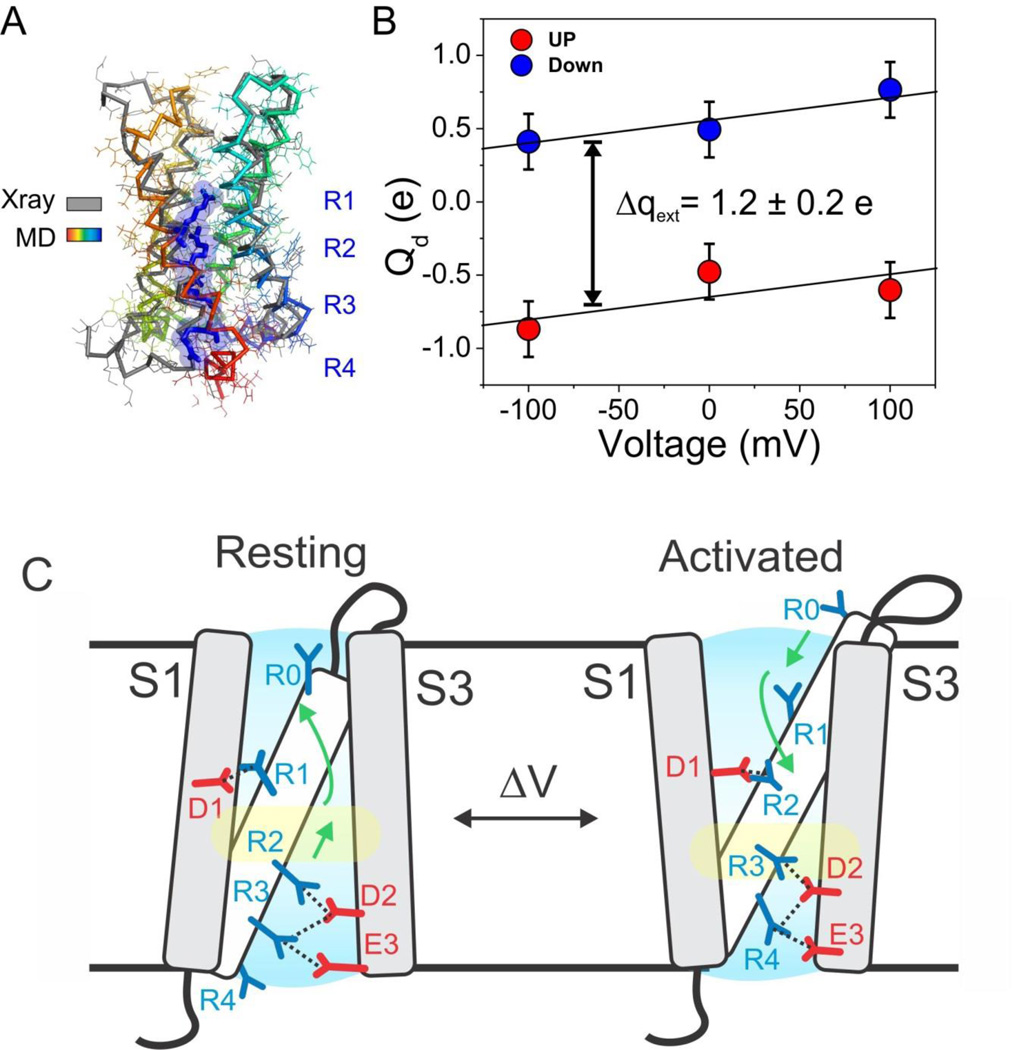
The structure of Ci-VSD in its Down state offers an opportunity to address long sought-out questions regarding the mechanism by which voltage influences the conformation of S4. This relative displacement of S4 helix becomes apparent when aligning WT (black ribbon) structure with R217E structure (white ribbon) (Figure 5A): ~5 Å downward in every gating charge (including R0), from the Up to the Down conformation. There are no apparent changes in S4 α-helical secondary structure and minimal angular reorientation in relation to S1–S3 during this conformation change. A helical projection of each gating charge Conin the WT and R217E structures reveals that this downward movement is accompanied by, a counterclockwise rotation that varied between 48°–75°, viewed from extracellular side (Figure 5B). During deactivation, the movement of S4 is enabled by a simultaneous reorientation in the S3–S4 loop (Figure S5A). This involves the limited unwinding of a short helical turn (residues 207–210), together with a sharp upwards reorientation (6–8 Å) of most of the loop residues.
Figure 5.
Structural change between Ci-VSD WT and R217E. A. Overlap of Ci-VSD R217E (white) and WT (black) backbones reveals the conformational changes S3–S4 linker, relative position change in S4 and the existence of inter-domain linker in WT. The axial translations of Cα of the arginines are about ~5 Å. The four arginines rotates 48~75° relative to the helical axis. C. Ci-VSD WT has nearly identical scaffold (ribbon) and the conserved phenylalanine (sphere, highlighted in red circle), but a distinctly lower inter-domain linker than Kv1.2 chimera and NavAb. Based on the Z-axe established by potassium ions in Kv1.2 chimera filter, the Ci-VSD WT linker is ~ 2 Å lower on average and ~ 5 Å lower at the end of S4.
The nature of the S4 movement described above places the Cα of each individual Up conformation arginine, essentially in the same position as the following arginine of the Down conformation (three residues away, sequence-wise). This way, a ~5 Å oneclick charge displacement is achieved in relation to the hydrophobic gasket. In this gating “click”, S4 charges transition from a position with electrostatic partners R1-D1, R2-D2 and R3-E3 in the down state (with R4 fully solvated by water), to on where the pairings are R2-D1, R3-D2 and R4-E3 in the Up conformation. R1 thus pushes into the external cavity and becomes fully hydrated. Neither the hydrophobic gasket nor the countercharges in S1 and S3 display sizable changes during S4 movement.
The S3 segment itself does not appear to move substantially nor does its overall conformation appear to be affected by the movements in S4. From a mechanistic standpoint, the present structures and associated conformational rearrangements provide no evidence for a “paddle”-like correlated movement of S3–S4 hairpin (Jiang et al., 2003b; Ruta et al., 2005). In fact, the movements described above are a straightforward representation of the classic “helix screw” or “sliding helix” concepts independently proposed (Catterall, 1986; Guy and Seetharamulu, 1986) more than 25 years ago.
The presence of inter-domain linker at the intracellular base of the Down conformation of Ci-VSD can serve as a reference point to evaluate the process by which S4 movement triggers downstream rearrangements in the phosphatase domain. Yet, in spite of its higher resolution and same interaction in crystal lattice, the Up conformation Ci-VSD structure offers no clues as to the structure and overall conformation of the linker (residues 237–244). Nevertheless, we can partially address this question by comparing the Ci-VSD linker conformation with those found on full-length ion channels. Figure 5C shows a superposition of Ci-VSD in the Down conformation with VSDs from Kv1.2 and NavAb in their Up conformation, with RMSD = 1.1 Å on the backbone of S1+S2+S3a helices. While the orientation of the S4–S5 linker relative to the aligned sensors is the same for NavAb and Kv1.2, the Ci-VSD WT linker has swung some 30° (clockwise) and shows an average ~2 Å downward displacement relative to the S4–S5 linker. It is interesting to point out the vertical difference at the end of S4 between activated sensors (NavAb and Kv1.2) and resting sensor (Ci-VSD WT) is ~5 Å (Figure 5C, right) which coincides with the extent of conformation change between Up and Down state of Ci-VSD (Figure 5A). We suggest that this downward reorientation could represent a physiologically relevant linker movement and provide a useful window into the putative structural rearrangements that take place during electromechanical coupling.
Electrostatic consequences of S4 movement
Beyond the backbone motion, side chain orientation also contains interesting details toward the mechanism of voltage sensing. Superposition of all gating charge residues from 2.5 Å R217E crystal structure at the backbone level reveals a remarkable rotameric behavior: as we move down the S4 helix, from R1 to R4, arginine side chains undergo a downwards reorientation (up to ~7 Å) as they first interact and then cross the hydrophobic gasket (Figure 6A). A survey of the existing high-resolution VSD structures suggests that this appears to be a common characteristic of sensor arginines (Figure S6). Hence, we propose that this rotameric reorientation mechanism could be an additional contributor to the total gating charge translocation in VSDs.
Figure 6.
A molecular mechanism of charge translocation in voltage sensor. A. Comparison of the rotamer distribution of gating arginines in Up conformation resolved in the 2.5 Å crystal structure. There is a large downward reorientation going down the S4 and translocating through the hydrophobic gasket. B. Crystal structure of Ci-VSD WT (grey) is stable after 105 ns of all-atom MD simulation (spectrum). C. Calculation of the displacement charge between the Up and Down conformation of Ci-VSD at +100, 0 and −100 mV yielding a gating charge of 1.2 ± 0.2 eo, which is in agreement of the apparent valence z=1~1.5 estimated by the activation slope of gating current. D. Four existing metal bridges constrains for the down state in Shaker: bridge 1, 2 (Campos et al., 2007), bridge 3 (Lin et al., 2010) and bridge 4 (Tao et al., 2010). The three statedependent bridges 1, 2 and 4 are only compatible at Down state, but not the Up state of Ci-VSD. E. A mechanistic model of voltage sensing in Ci-VSD. A ~5 Å S4 down movement is accompanied by a counterclockwise ~60° rotation of the entire helix. arginines in S4 are stabilized by successive negative counter charges on S1 and S3. A potential rotameric reorientation of the arginines might additionally contribute to the overall gating charge transfer.
To better evaluate the structural rearrangements informed by the Down and Up Ci-VSD conformations, we modeled the full complement of side chains in the Down structure to generategenerated all-atom models for both Ci-VSD WT and R217E, which are stable after a ~105 ns MD simulation in explicit membrane and solvent (Figure 6B). Based on the all-atom models, we set out to determine the amount of charge transferred by the one-click gating rearrangement from the average displacement charge difference ΔQd between two conformations (the Q-route formalism, (Roux, 2008)). The time-averaged displacement charge during 20ns of simulation at each of three transmembrane potentials (−100, 0 and +100 mV) is shown on Figure 6C. From these calculations, the magnitude of the calculated gating charge was estimated at 1.2 ± 0.2 eo, remarkably close to the expected value from gating current measurements (~1 eo) (Murata et al., 2005; Villalba-Galea et al., 2008). The present calculation is an indication that the one-click charge translocation is able to account for the transfer of at least one electronic gating charge, in agreement with the characteristic shallow voltage dependence of Ci-VSD (Figure 1B) (Murata et al., 2005).
DISCUSSION
Implications for the gating mechanism in other voltage sensors
It seems to be generally agreed that in spite of long evolutionally distances and the obvious differences in structure and functional set points, voltage sensing in VSDs is based on a common structural blueprint. The determination of the crystal structures of Ci-VSD in the Up and Down conformations reveals an explicit mechanism of voltage activation that recapitulates the basic tenets of the helical screw or sliding helix models (Catterall, 1986; Guy and Seetharamulu, 1986) in voltage-sensing phosphatases. But is this mechanism an effective model to understand ion channel-based electrical excitability?
A quick survey of the voltage dependence of ion channels, proton channels and voltage-dependent enzymes illustrates the large heterogeneity that exists in regards to the estimated charge transferred (zo from ~1 to ~3.6 eo per VSD) and the amount of energy required to activate the system (V½ from −120 to +80 mV). We point out that the key mechanistic differences in regards to voltage sensing are likely derived from heterogeneity in gating charge positions in the VSD scaffold and their interactions with the co-evolved countercharges. These interactions define not only the overall charge transferred required for function, but also the extent of S4 movement needed to fulfill a given charge translocation.
Several lines of evidence suggest that relatively modest S4 movements like those observed in Ci-VSD conform to the extensive experimental data that exists for voltage-gated channels. Most of the available cysteine reactivity data (see (Tombola et al., 2006) for a survey) can be readily accommodated with S4 movements in the vicinity of 5 Å. In addition, distance estimations on Shaker R1 from tethered charges of different length suggest that S4 moves ~4 Å (Ahern and Horn, 2005). The present gating model is also compatible with recent LRET and fluorescence data (Posson and Selvin, 2008) showing 5–7 Å S4 vertical displacements in Shaker channels. Explicit distance constraints for the Down conformation of the Shaker VSD have been recently obtained from metal bridging experiments that link S4 with S1 (Cd2+ Bridge 1, (Campos et al., 2007)), S4 with S2 (Cd2+ Bridge 2, (Campos et al., 2007)) and S2 with S3 (Mg2+ Bridge 3, (Lin et al., 2010)). In addition, direct electrostatic interactions were identified between R1 and F194 in S3 (Bridge 4, (Tao et al., 2010)). Taking as a reference Ci-VSD residue R223 in S4 as the functional equivalent of Shaker R1, we tested the compatibility of these distance constraints with the Up and Down Ci-VSD structures (Figure 6D). When evaluated on the R217E structure, the observed distances exceeded 10 Å, well beyond the range for cysteine-driven Cd2+ bridges. But when the same exercise was carried out on the WT (Down) structure, all distances were within the experimental range of Cd2+ bridges. The compatibility of Bridge 3 with both Up and Down structures is expected, since the relative positions of S2 and S3 do not change during gating.
In order to account for the 3–4 electronic charges needed to gate Shaker (and other K+ channels), we can interpret our structures in two different contexts. On one hand, it is possible that Ci-VSD undergoes a larger motion than what is reflected in the present crystal structures. In this case WT Ci-VSD would represent an intermediate state, on its way to the Down conformation. The limited changes in accessibility from EPR experiments of membrane-embedded Ci-VSD tend to argue against this option. An alternative possibility is that while sharing a similar helical screw mechanism, the S4 in voltage dependent channels would move further to accommodate the additional charge transfer. For instance, a “two click” gating motion would extend the movement of S4 to about 10 Å and its rotation to 110–180°, an option supported by recent metal-ion bridge experiments (Henrion et al., 2012). Indeed, a comparison of the WT Ci-VSD structure to that of computationally-derived Down conformations (Jensen et al., 2012; Vargas et al., 2011), requires an additional 8–10 Å displacement to match the gating charges. This suggests a larger sliding helix movement for voltage-gated channels. However, very recent experiments with Ci-VSD/Vch chimeras (KvSynth) beautifully show that voltage-dependent rearrangements of Ci-VSD, putatively a “one click” sensor, are sufficient and necessary to gate a K-selective pore (Arrigoni et al., 2013).
Our structures are incompatible with data supporting simple “field rearrangement” models, where S4 movement is minimal (Cha et al., 1999). Similarly, fluorescence data predicting ~180° S4 rotations (Glauner et al., 1999) would require 2–3 additional gating-translocating “clicks” to comply with this rotational flip. The overall exposure of S4 to the lipid environment is incompatible with the original description of the S4 “gating canal” (Yang et al., 1996) or canaliculus (Goldstein, 1996). Finally, observations in support of very large S4 movements from biotin/streptavidin trapping experiments (Jiang et al., 2003b; Ruta et al., 2005) or mass tagging approaches (Koag and Papazian, 2009; Zheng et al., 2011) require separate consideration, given the present lack of evidence for the functional existence of an S3–S4 “paddle”.
In summary, we suggest that voltage-dependence activation in Ci-VSP is best illustrated through the basic diagram in Fig 6C. Upon depolarization, the S4 segment undergoes a ~5 Å outward translation along its main axis in concert with a ~60° clockwise rotation. As a consequence, linearly aligned gating charges R1-R4 undergo a “one click” displacement that translocates R2 through a dielectrically tight hydrophobic gasket, while exchanging electrostatic partners with countercharges D1, D2 and E3 in helices S1 and S3. As S4 translocates, the flexible arginine side chains in R2 and R3 putatively undergo a rotameric upward flip that potentially contributes to the charge transfer process. We propose that relatively subtle variations of this general mechanism should help explain voltage-dependent activation in many excitable systems.
EXPERIMENTAL PROCEDURES
Electrophysiology
WT and mutant constructs of Ci-VSP (R217E and R217Q) were made on a phosphatase-inactive mutant C363S background and expressed in Xenopus laevis oocytes. Transient sensing currents were recorded using a cut-open oocyte voltage-clamp following published methods (Bezanilla and Stefani, 1998). The net charge was calculated by integrating the “off” sensing currents at each potential within the whole 500 ms pulse. Data acquisition and analysis were carried out with in-house programs.
Protein expression and purification
The voltage sensor domain of Ci-VSP (Ci-VSD) was expressed and purified following standard procedures as described (Li et al., 2012). The constructs for the reported crystal structures correspond to residues 106–244 for WT and 89–260 for the R217E mutant. Expression and purification protocols for these truncations are the same as for Ci-VSD-1-260.
Antibody screening, expression and purification
WT and R217E mutant of Ci-VSD-1-260 constructs were biotinylated through amine coupling with NHS-SS-PEG4-Biotin (Thermo Scientific). Phage display selections were performed using a synthetic antibody library built on the 4D5 Fab scaffold as described previously (Rizk et al., 2011). Target protein concentrations were serially adjusted using 100 nM in the first round and 10 nM and 5 nM in subsequent rounds to ensure proper stringency. The conformation specific binders were selected using a competitive selection strategy where 1 μM nonbiotinylated competitor (such as Ci-VSD WT) was added to the phage library prior to the incubation with actual selection target (such as Ci-VSD R217E). A single point competitive ELISA step used to identify positive clones. Fabs were expressed in E. coli strain 55244 in phosphate depleted media as described previously (Rizk et al., 2011). Fabs were purified using automated two step protocol employing a custom build AKTA purification system with 5ml rProteinA Sepharose 4 Fast Flow and 1ml Resource S column (GE Healthcare) columns. Fractions were verified by SDS-PAGE, pooled and dialyzed against PBS.
Protein crystallization
The Ci-VSD-R217E-89-260 with Fab 33F12_4 complex was purified in 20 mM Tris pH 8.0, 150 mM NaCl and 4 mM LDAO. Crystals were grown by hanging-drop vapor diffusion at 20°C by mixing 0.1 μL of protein and 0.1 μL reservoir solutions with the Mosquito robotic system (TTP LabTech). Crystals were grown with 100 μL reservoir solution of 0.1 M SPG pH 8.0, 20% PEG 1500, 10% glycerol or 0.1 M Bicine pH 9.0, 0.3 M MgCl2, 24% PEG 2000, 10% glycerol. The Ci-VSD-WT-106-244 with Fab 39D10_18 complex was purified in 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM LDAO and 2 mM DM. Crystals were grown with a reservoir solution of 0.1 M Na Citrate pH 5.5, 0.1 M Li2SO4, 20% PEG 1000, 10% glycerol. Cryoprotection was achieved by increasing glycerol concentration to 5% for both cases.
Data collection and structure determination
All data were collected at the Advanced Photon Source synchrotron (beamlines 24-ID-E, 24-ID-C, 23-ID-B or 23-ID-D) under a nitrogen stream at 100 K, and processed with HKL2000. The phase was determined by molecular replacement with constant domain and variable domain of Fab (PDB ID 3PGF). Coot (Emsley and Cowtan, 2004), Refmac5 (Murshudov et al., 1997) and Phenix (Adams et al., 2010) were used for model building and refinement. The atomic coordinates and structural factors have been deposited in the RCSB Protein Data Bank with the accession number 4G7V (R217E, 2.5 Å), 4G7Y (R217E, 2.8 Å) and 4G80 (WT, 3.8 Å).
EPR spectroscopy
41 single cysteine mutants comprising residues 200–250 on both Ci-VSD-1-260 WT and R217E background were purified, spin-labeled with (1-oxyl-2,2,5,5,-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate and reconstituted into POPC:POPG = 3:1 liposomes for continuous-wave EPR measurements. Details of single Ci-VSD cysteine mutants purification, reconstitution and EPR spectroscopy analyses on Ci-VSD has been described previously (Li et al., 2012).
Molecular Dynamic simulation
The procedures on all-atom simulation for both Ci-VSD WT and R217E, as well as the development of XMDFF methods, were described in details in supplementary information.
Supplementary Material
HIGHLIGHTS.
Two conformations were captured in crystal structures in the Ci-VSD system.
Structural details highlight the heterogeneity of voltage sensing domains.
Conformational difference reveals an explicit mechanism of voltage sensing.
ACKNOWLEDGEMENTS
We are thankful to K Rajashankar and the staff at the NE-CAT 24-ID beamline, as well as to R. Sanishvili and the staff at the GM/CA 23-ID beamline in the Advanced Photon Source, Argonne National Laboratory. We thank Perozo, Bezanilla and Roux labs for illuminating discussions and invaluable experimental advice. A. Koide and S. Koide for the antibody library. We are grateful to E.J. Adams, R. Keenan, P. Rice and X. Yang for helpful crystallographic advice. Anton computer time was provided by the National Resource for Biomedical Supercomputing and the Pittsburgh Supercomputing Center through Grant RC2GM093307 from the National Institutes of Health. This work was supported in part by NIH grants R01-GM57846 (to E.P.), U54-GM74946 (to E.P.) and R01-GM062342 (to B.R.).
Footnotes
The authors declare no competing financial interests.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- Arrigoni C, Schroeder I, Romani G, Van Etten JL, Thiel G, Moroni A. The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J Gen Physiol. 2013;141:389–395. doi: 10.1085/jgp.201210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. How membrane proteins sense voltage. Nat Rev Mol Cell Biol. 2008;9:323–332. doi: 10.1038/nrm2376. [DOI] [PubMed] [Google Scholar]
- Bezanilla F, Stefani E. Gating currents. Methods Enzymol. 1998;293:331–352. doi: 10.1016/s0076-6879(98)93022-1. [DOI] [PubMed] [Google Scholar]
- Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci U S A. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- Goldstein SA. A structural vignette common to voltage sensors and conduction pores: canaliculi. Neuron. 1996;16:717–722. doi: 10.1016/s0896-6273(00)80092-6. [DOI] [PubMed] [Google Scholar]
- Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci U S A. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrion U, Renhorn J, Borjesson SI, Nelson EM, Schwaiger CS, Bjelkmar P, Wallner B, Lindahl E, Elinder F. Tracking a complete voltage-sensor cycle with metal-ion bridges. Proc Natl Acad Sci U S A. 2012;109:8552–8557. doi: 10.1073/pnas.1116938109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobiger K, Utesch T, Mroginski MA, Friedrich T. Coupling of Ci-VSP modules requires a combination of structure and electrostatics within the linker. Biophys J. 2012;102:1313–1322. doi: 10.1016/j.bpj.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003a;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003b;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- Koag MC, Papazian DM. Voltage-dependent conformational changes of KVAP S4 segment in bacterial membrane environment. Channels (Austin) 2009;3:356–365. doi: 10.4161/chan.3.5.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout SC, Bell SC, Liu L, Xu Q, Minor DL, Jr, Isacoff EY. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat Chem Biol. 2010;6:369–375. doi: 10.1038/nchembio.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- Li Q, Jogini V, Wanderling S, Cortes DM, Perozo E. Expression, purification, and reconstitution of the voltage-sensing domain from Ci-VSP. Biochemistry. 2012;51:8132–8142. doi: 10.1021/bi300980q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MC, Abramson J, Papazian DM. Transfer of ion binding site from ether-ago- go to Shaker: Mg2+ binds to resting state to modulate channel opening. J Gen Physiol. 2010;135:415–431. doi: 10.1085/jgp.200910320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posson DJ, Selvin PR. Extent of voltage sensor movement during gating of shaker K+ channels. Neuron. 2008;59:98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- Rizk SS, Paduch M, Heithaus JH, Duguid EM, Sandstrom A, Kossiakoff AA. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat Struct Mol Biol. 2011;18:437–442. doi: 10.1038/nsmb.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. The membrane potential and its representation by a constant electric field in computer simulations. Biophys J. 2008;95:4205–4216. doi: 10.1529/biophysj.108.136499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas E, Bezanilla F, Roux B. In search of a consensus model of the resting state of a voltage-sensing domain. Neuron. 2011;72:713–720. doi: 10.1016/j.neuron.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Galea CA, Frezza L, Sandtner W, Ramsey IS, Bezanilla F. Sensing charges of the Ciona Intestinalis Voltage Sensing Phosphatase. doi: 10.1085/jgp.201310993. (Submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Galea CA, Sandtner W, Starace DM, Bezanilla F. S4-based voltage sensors have three major conformations. Proc Natl Acad Sci U S A. 2008;105:17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Liu W, Anderson LY, Jiang QX. Lipid-dependent gating of a voltage-gated potassium channel. Nat Commun. 2011;2:250. doi: 10.1038/ncomms1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.