Abstract
The amnion membrane is developed from embryo-derived cells, and amniotic cells have been shown to exhibit multidifferentiation potential. These cells represent a desirable source for stem cells for a variety of reasons. However, to date very few molecular analyses of amnion-derived cells have been reported, and efficient markers for isolating the stem cells remain unclear. This paper assesses the characterization of amnion-derived cells as stem cells by examining stemness marker expressions for amnion-derived epithelial cells and mesenchymal cells by flow cytometry, immunocytochemistry, and quantitative PCR. Flow cytometry revealed that amnion epithelial cells expressed CD133, CD 271, and TRA-1-60, whereas mecenchymal cells expressed CD44, CD73, CD90, and CD105. Immunohistochemistry showed that both cells expressed the stemness markers Oct3/4, Sox2, Klf4, and SSEA4. Stemness genes' expression in amnion epithelial cells, mesenchymal cells, fibroblast, bone marrow–derived mesenchymal stem cells (MSCs), and induced pluripotent stem cells (iPSCs) was compared by quantitative reverse-transcription polymerase chain reaction (RT-PCR). Amnion-derived epithelial cells and mesenchymal cells expressed Oct3/4, Nanog, and Klf4 more than bone marrow–derived MSCs. The sorted TRA1-60–positive cells expressed Oct3/4, Nanog, and Klf4 more than unsorted cells or TRA1-60–negative cells. TRA1-60 can be a marker for isolating amnion epithelial stem cells.
Introduction
The amnion is a fetal origin tissue and is composed of a single layer of epithelial cells on a thicker basement membrane and spongy collagen layer containing mesenchymal cells that are derived from the inner cell mass (ICM) in the blastocyst. It has been reported that embryonic stem cells (ESCs) derived from blastocysts have normal karyotypes, express high levels of telomerase activity, express all embryonic stem cell markers, and can develop to all three germ layers (Thomson et al., 1998). Amnion membrane-derived cells are also reported to be multipotent cells that can replicate as undifferentiated cells as they express stem cell genes, such as Oct3/4, Sox2, and c-Myc and that have the potential to differentiate into various tissue (Bilic et al., 2008; Diaz-Prado et al., 2010; Izumi-Yoneda et al., 2009; Murphy et al., 2010; Nagura et al., 2013; Nogami et al., 2012; Otaka et al., 2013; Takashima et al., 2004; Toda et al., 2007; Tsuno et al., 2012; Wei et al., 2003, 2009; Zhao 2005). In addition, they do not express human leukocyte antigen (HLA) class II and secrete HLA-G and CD59, which are immunologic suppression factors (Adinolfi et al., 1982; Akle et al., 1981; Kamiya et al., 2005; Wolbank et al., 2007). It has also been shown that the conditioned medium of amnion-derived cells have immunosuppressive activity (Cargnoni et al., 2014). Moreover, they do not attract ethical concern because they are usually discarded after parturition. Thus, amnion-derived cells are anticipated to be a valuable cell source for cell therapy (Corgnoni et al., 2009; De Coppi et al., 2007; Hu et al., 2009; Murphy et al. 2010; Parolini et al., 2009, 2010).
However, few molecular biological analyses have been performed to characterize amnion-derived cells. Here we report a comparison analysis of human amnion-derived epithelial (HAE) cells and human amnion-derived mesenchymal (HAM) cells. Although amnion-derived cells have stem cell characteristics and differentiation potency for several cell types, they are a heterogeneous cell population that includes stem cells, progenitors of certain cells, and differentiated cells. It has been shown that they have multidifferentiation potential, but their differentiation efficiency is low. If the stem cells are isolated from the heterogeneous population, the differentiation efficiency may increase and those cells could represent a better cell source for cell therapy. TRA1-60 is known to be one of the markers of ESCs (Thomson et al., 1998). Also, it is known that some amnion cells express TRA1-60. Thus, the isolation of stem cells from the heterogeneous population using TRA1-60 as a marker was attempted. The analysis of the isolated cells showed a higher expression of stemness genes relative to unsorted cells.
Materials and methods
Cell isolation
The amniotic membrane was mechanically peeled from the chorion of a placenta obtained, with informed consent, after an uncomplicated cesarean section. The study and the use of the amnion membrane were approved by the Research Ethics Committee of the University of Toyama as described previously (Wei et al., 2003). The tissue was minced and treated with trypsin (2 mg/mL) at 37°C for 20 min to isolate HAE cells. After repeating this treatment several times, the epithelial cells were completely removed. The tissue pieces were placed in Dulbecco's Modified Eagle Medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) containing collagenase (0.75 mg/mL) and DNase (0.075 mg/mL) and were incubated at 37°C for 60 min to isolate HAM cells. The dispersed HAE or HAM cells were collected by filtration of the mixture through gauze and centrifugation.
Flow cytometric analysis and cell sorting
Cells were blocked with 5% bovine serum albumin (BSA; Sigma-Aldrich) in phosphate-buffered saline (PBS) for 30 min at room temperature and stained with antibodies at a concentration of 20 μL/1×106 cells at room temperature for 1 h. Antibodies against CD14, CD29, CD34, CD45, CD49f, CD105, HL-DR (Beckman Coulter, Brea, CA, USA), CD24, CD44, CD73, TRA1-60, TRA1-81, SSEA3, SSEA4 (BD Pharmingen, Franklin Lakes, NJ, USA), CD90 (Immune tech, Cedex, France), CD133, or CD271 (Miltenyi Biotech, Bergisch Gladbach, Germany) were used. Flow cytometry was performed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) using Cell Quest software, and data were analyzed using WinMDI ver 2.9.
MACS separation (Miltenyi Biotec) was used for cell sorting according to the manufacturer's protocols. Anti-TRA1-60-FITC at a concentration of 20 μL/1×106 cells was used for selection.
Immunocytochemistry
Cells were seeded in a 24-well plate at a density of 1×105 cells/well and cultured for 7 days. Cells were fixed with 4% paraformaldehyde and treated with 0.3% Triton-100. After washing with 0.05 M PBS, cells were stained with primary antibodies against vimentin (1:200), Oct 3/4 (1:200), c-Myc (1:200), Sox-2 (1:50), Klf4 (1:200), Nanog (1:200), SSEA-3 (1:200), SSEA-4 (1:200) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), TRA1-60 (1:200), TRA1-81 (1:200), and pan-CK (1:200) (Millipore, CA, USA) overnight at 4°C. Cells were incubated with Alexa Fluor 488 anti-mouse immunoglobulin M (IgM), IgG, rabbit IgG, or rat IgM (Life Technologies, CA, USA) (1:1000 in PBS) for 30 min at room temperature. Then, cells were counterstained with Hoechst 33342 solution (Wako, Okasa, Japan) (1:1000 in PBS) and examined with a fluorescence microscope (Zeiss, Oberkochen, Germany) with Axiovision LE.
Reverse-transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from the cells using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. Aliquots of 1 μg of total RNA were treated with 0.1 U/μL deoxyribonuclease I (DNase I, Sigma-Aldrich, Inc.) at room temperature for 15 min. cDNAs were synthesized using 0.5 μg of DNase I–treated RNA by using a Rever Tra Ace qPCR RT Kit (Toyobo Co., Ltd., Osaka, Japan). The resulting cDNA was subjected to quantitative PCR using Brilliant II Fast QPCR Master Mix (Agilent Technology, Santa Clara, CA, USA) on MxP3000 (Agilent) with primers for Oct3/4, c-Myc, Sox2, Klf4, Nanog, Lin28 (Nagura et al., 2013), and β2-microglobin (B2M) (Takara Bio, Tokyo, Japan) or primers for matrix metallopeptidase 1 (MMP1), leukemia inhigibitory factor (LIF), MGP, or apolipoprotein D (APOD) (Applied Biosystems, Carlsbad, CA, USA) (Igarashi et al. 2007; Ishii et al., 2005). Primers of B2M were as follows: forward, 5′-cgggcattcctgaagctga-3′; reverse, 5′-ggatggtgaaacccagacacatag-3′. The levels of a given mRNA were normalized to the internal control gene B2M mRNA level.
Statistical analysis
All values are expressed as mean±standard error. Comparison parameters for more than three groups were made by Student's t-test using statistical software (SPSS Statistics ver. 20 for Mac; IBM, Tokyo, Japan).
Results
The characteristics of HAE and HAE cells were analyzed. Morphologically, cultured HAE cells are small and round in shape, and cultured HAM cells are spindle-shaped (Fig. 1). To confirm that we had obtained epithelial cells and mesenchymal cells, HAE and HAM cells were stained with antibodies against the epithelial marker pan-CK or mesenchymal marker vimentin, respectively. More than 90% of HAE cells expressed the epithelial marker pan-CK, whereas less than 5% of HAM cells did. On the other hand, less than 5% of HAE cells expressed vimentin, the mesenchymal marker, whereas over 90% of HAM cells expressed it (Fig. 1). These results confirmed that we had obtained epithelial cells and mesenchymal cells, respectively.
FIG. 1.
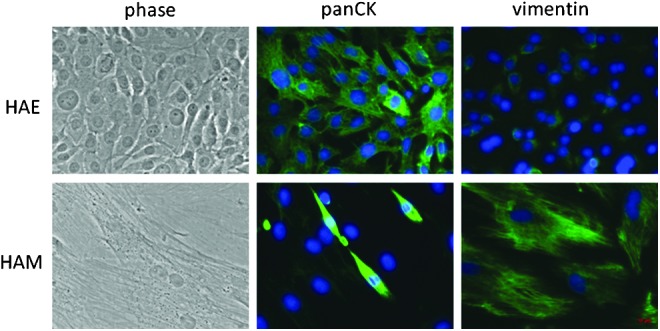
Morphology of HAE and HAM cells. HAE and HAM cells are stained with anti-pan-CK or vimentin antibodies, respectively (green). Nuclear staining was performed with Hoechst (blue).
Cell-surface marker expression
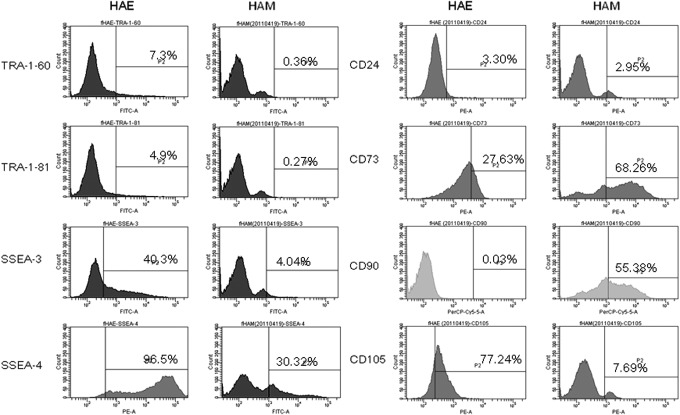
We analyzed by flow cytometry the HAE and HAM cells with mesenchymal stem cell (MSC) markers CD105, CD90, CD73, and CD44; hepatopoietic markers CD45, HLA-DR, CD14, and CD34; mesenchymal cell–related markers, CD24, CD133, and CD271; and stemness markers, TRA1-60, TRA1-81, SSEA3, and SSEA4 (Fig. 2).
FIG. 2.
Histograms of flow cytometric analysis against TRA-1-60, TRA-1-81, SSEA3, SSEA4, CD24, CD73, CD90, and CD105 using HAE and HAM cells.
Almost none of the HAE cells expressed hematopoietic markers CD34, CD45, HLA-DR, or CD14, but surprisingly 3% of HAM cells expressed CD34, 17% of HAM cells expressed CD45, 14% of HAM cells were positive for HLA-DR, and 10% were positive for CD14. Less than 1% of HAE cells expressed CD105, CD90, or CD44, less than 5% of HAE cells expressed CD24 or CD133, and 69% of HAE cells expressed CD73, 89% were positive for CD29, 91% for CD49f, 38% for CD271, 7.3% for TRA1-60, 2.8% TRA1-81, 0.6% for SSEA3, and 61% for SSEA4, respectively. Less than 10% of HAM cells expressed CD105, CD44, CD24, or CD133, whereas 20% of HAM cells were positive for CD90, 43% for CD73, 99% for CD29, 69% for CD49f, 50% for CD271, 0.5% for TRA1-60, 0.5% for TRA1-81, 3.6% SSEA3, and 43% for SSEA4, respectively (Table 1). Both HAE and HAM cells expressed CD73, CD29, CD49f, CD271, SSEA3, and SSEA4. Only HAE cells expressed TRA1-60 and TRA1-81. Only HAM cells expressed CD105, CD90, and CD44.
Table 1.
Marker Expression by HAE and HAM Cells
| HAE cells | HAM cells | |
|---|---|---|
| CD 105 | 0% | 8%±2.2% |
| CD 90 | 0% | 20%±9.5% |
| CD 73 | 69%±2.2% | 43%±16% |
| CD 44 | 0% | 3%±0.2% |
| CD 34 | 0% | 3%±0.9% |
| CD 45 | 0% | 17%±3.9% |
| HLA-DR | 0% | 14%±2.7% |
| CD 14 | 0% | 10%±1.9% |
| CD 24 | 2%±2% | 8%±2.1% |
| CD 29 | 89%±12% | 99%±1% |
| CD 49f | 91%±14% | 65%±14% |
| CD 133 | 3.2%±5% | 2%±0.2% |
| CD 271 | 38%±20% | 50%±9.1% |
| TRAI-60 | 7.3%±3.3% | 0.5%±0.2% |
| TRAI-81 | 2.8%±1.4% | 0.5%±0.2% |
| SSEA3 | 0.6%±0.5% | 3.6%±0.5% |
| SSEA4 | 61%±19% | 43%±7.2% |
HAE cells, human amnion-derived epithelial cells; HAM cells, human amnion-derived mesenchymal cells.
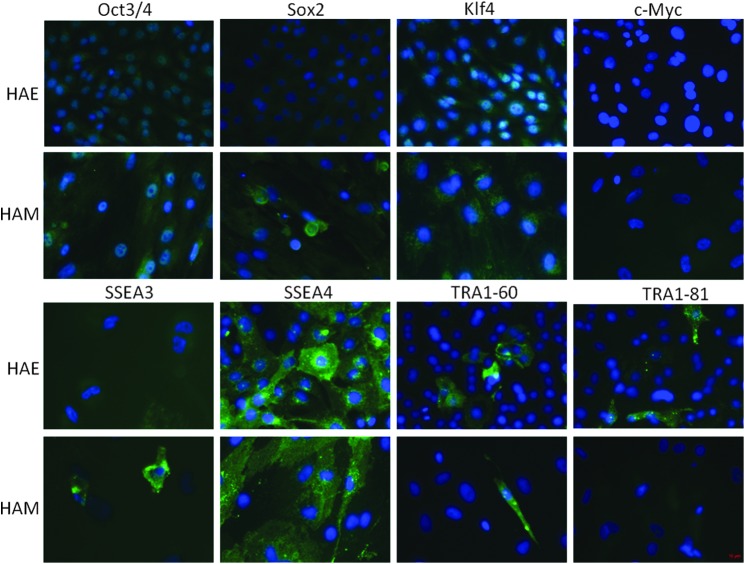
HAE and HAM cells were stained with antibodies against the stemness markers Oct3/4, Sox2, Klf4, c-Myc, Nanog, TRA1-60, TRA1-81, SSEA3, and SSEA4, respectively. Both HAE and HAM cells expressed all stemness markers, Oct3/4, Sox2, Klf4, c-Myc, TRA1-60, TRA1-81, SSEA3, and SSEA4, but not all of the cells were stained with antibodies against stemness markers. Overall, more HAE than HAM cells expressed stemness markers, as more than 80% of HAE cells expressed Oct3/4, Sox2, Klf4, and SSEA4. Nearly 70% of HAE cells expressed c-Myc. More than 90% of HAM cells expressed Oct3/4. About 75% of HAM cells expressed Sox2 and Nanog. Klf4 positive HAMS were about 60% and c-Myc positive and SSEA4 positive HAMS were 30%, respectively (Fig. 3). From the fact that not all the cells expressed markers, it is suggested that they are heterogeneous, including stem cells that express stemness markers.
FIG. 3.
HAEs and HAMs are stained with anti-Oct3/4, Sox2, Klf4, c-Myc, SSEA3, or SSEA4, antibodies, respectively (green). Nuclear staining was performed with Hoechst (blue).
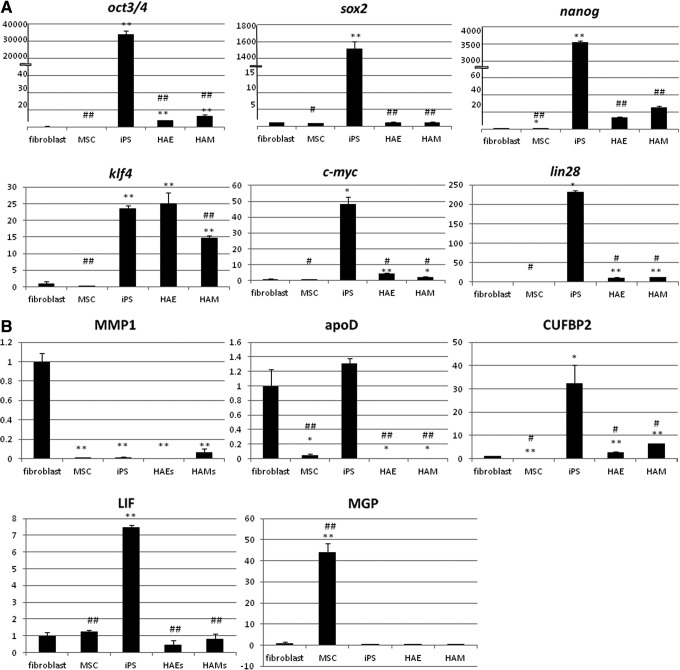
MSC marker expression was measured by quantitative RT-PCR. MMP1 and ApoD are negative MSC markers and are supposed to be expressed more in fibroblasts than in MSC (Ishii et al., 2005). The expression of MMP1 was lower in all bone marrow (BM)-MSCs, induced pluripotent stem cells (iPSCs), HAE cells, and HAM cells than in fibroblasts. Interestingly, iPSCs expressed ApoD as well as fibroblasts, but HAE and HAM cells expressed less than fibroblasts, about 100-fold lower than BM-MSCs. LIF and MGP are positive MSC markers and are supposed to be expressed higher in BM-MSC than in fibroblasts. The expression of LIF in BM-MSCs was only 1.2-fold higher than that in fibroblasts, although iPSCs expressed it 7.5-fold more. HAE cells expressed LIF 0.4-fold that of fibroblasts and HAMs 0.8-fold. BM-MSCs expressed MGP 44-fold compared with fibroblasts, whereas iPSCs expressed MGP 0.4-fold, HAEs 0.004-fold, and HAMs 0.02-fold (Fig. 4A). These results suggest that HAE cells, HAM cells, and BM-MSCs have different characteristics.
FIG. 4.
Quantitative RT-PCR was performed using cDNAs from fibroblasts, MSCs, iPSCs, HAE cells, or HAM cells. Relative expression patterns compared with the expression level of fibroblasts are shown. Bars, mean±standard error (SE). Statistical significance: (*) p<0.05, (**) p<0.01 compared to fibroblasts, (#) p<0.05, (##) p<0.01 compared to iPSCs are shown. (A) MSC markers, MMP1, ApoD, LIF, or MGP. (B) Stemness markers Oct3/4, Sox2, Klf4, c-Myc, Nanog, or Lin28.
The expression of Oct3/4, Sox2, Klf4, c-Myc, Nanog, and Lin28 genes in HAE and HAM cells was analyzed by quantitative RT-PCR. Compared to human fibroblasts, HAE and HAM cells expressed eight-fold and 13-fold more Oct3/4, whereas BM-MSCs expressed 0.7-fold, and iPSCs expressed 34,535-fold. Similar results were seen with Nanog, as HAE cells expressed 13-fold, HAM cells expressed 26-fold, BM-MSCs 0.9-fold, and iPSCs 3601-fold. They also showed similar expression patterns with the Lin28 gene. HAE and HAM cells expressed 12-fold, MSCs 0.9-fold, and iPSCs expressed 232-fold compared to fibroblasts. Both HAE and HAM cells expressed Sox2 about the same as fibroblasts, whereas BM-MSCs expressed 0.7-fold and iPSCs 1445-fold. As for Klf4, HAE cells expressed 25-fold and HAM cells 15- fold, whereas BM-MSCs expressed 0.3-fold and iPSCs expressed 23-fold (Fig. 4B). HAE and HAM cells expressed Oct3/4, Sox2, Klf4, c-Myc, Nanog, and Lin28 more than BM-MSCs. Especially, they expressed Klf4 as much as iPSCs.
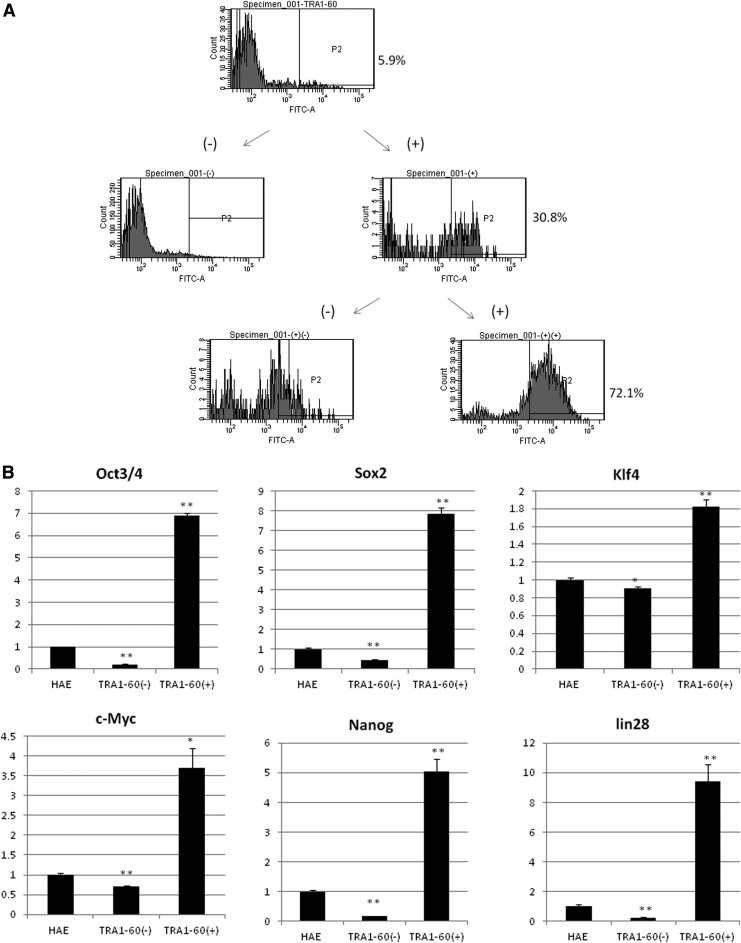
These results imply that HAE and HAM cells are heterogeneous cell populations containing stem cells, progenitors, and differentiated cells. Isolation of stem cells was attempted. HAE cells were sorted with anti-TRA1-60 antibody using magnetic-activated cell sorting (MACS) beads. TRA1-60–positive cells were concentrated up to 72% by sorting twice (Fig. 5A). Quantitative PCR of stemness genes was performed to compare their expression levels between the TRA1-60–positive cell population, TRA1-60–negative population, and unsorted population (Fig. 5B). TRA1-60–positive cells expressed the Oct3/4 gene 6.9-fold higher than unsorted cells and 33-fold higher than TRA1-60–negative cells. TRA1-60–positive cells expressed the Sox2 gene 7.8-fold higher than unsorted cells and 18-fold higher than TRA1-60–negative cells, the Klf4 gene 1.8-fold that of unsorted cells and two-fold of TRA1-60–negative cells, the c-Myc gene 3.7-fold that of unsorted cells, 5.6-fold of TRA1-60–negative cells, the Nanog gene five-fold of unsorted cells, 29.4-fold of TRA1-60–negative cells, and Lin28 gene 9.4-fold that of unsorted cells and 37.9-fold of TRA1-60–negative cells, respectively. TRA1-60–negative cells expressed all stemness genes less than unsorted cells.
FIG. 5.
(A) HAEs were sorted by MACS beads twice using anti-TRA-1-60 antibody. (B) Quantitative RT-PCR was performed using cDNAs from HAEs, TRA-1-60–negative cells, and TRA-1-60–positive cells. Relative expression patterns compared with the expression level of HAEs are shown. (*) p<0.05, (**) p<0.01 compared to unsorted cells are shown.
All stemness gene expression was higher in TRA1-60–positive cells compared to unsorted HAE cells or TRA1-60–negative cells. The small difference in expression level of Klf4 between TRA1-60–positive cells, TRA1-60–negative cells, and unsorted cells would be because the expression level of Klf4 was originally quite high. This result suggests sorting HAE cells using anti TRA1-60 antibody concentrated the epithelial stem cells.
Discussion
We have shown here the stemness marker expression in HAE and HAM cells. Our immunohistochemistry results showed that both HAE and HAM cells express all stemness markers, Oct3/4, Sox2, Klf4, c-Myc, Nanog, and SSEA-4. Some cells were stained strongly with antibodies against marker proteins, some weakly, and some were not stained among both HAE and HAM cells. This that suggests HAE and HAM cells are heterogeneous populations consisting of stem cells, progenitor cells, and differentiated cells. Flow cytometric analysis showed that HAE and HAM cells expressed CD73, CD29, CD49f, and SSEA-4. The expression patterns of CD 24, CD133, and SSEA4 are consistent with the results of Miki et al. (2005, 2006, and 2007). Because only HAM cells expressed CD105, CD90, and CD44, MSCs would be enriched in this population.
HAE and HAM cells expressed stemness marker genes Oct3/4, Klf4, c-Myc, and Nanog more than BM-MSCs. This suggests that they are closer to iPSCs than BM-MSCs as stem cells. HAE and HAM cells expressed Klf4 as much as iPSCs. This suggests that their stem cell characters are closer to that of iPSCs than BM-MSC, thus supporting the idea that amnion-derived cells being more efficiently reprogrammed than fibroblasts (Easley et al., 2012). One means of efficiently reprogramming amnion-derived cells is through introduction of Oct4, Sox2, and Nanog (Zao et al., 2010).
Izumi et al. showed that 17- to 19-week amnia expressed Nanog and Sox2 more than term amnia (Izumi et al., 2009). Because our HAE and HAM cells were isolated from a 35- to 36-week amnion membrane, it is possible that cells would express stemness markers more than our HAE and HAM cells if isolated from the amnion membrane at an earlier stage. Also amnia at an earlier stage might contain more stem cells than amnia at later stages or at term. However, ethical problems would arise from using earlier-stage amnia because they are a consequence of abortion. Considering the ethical issues, HAE and HAM cells appear to be the best amniotic cell source at this point.
It was interesting that iPSCs highly expressed MSC-negative markers, ApoD, or expressed less of the MSC-positive marker MGP. We assumed that somatic stem cells such as MSCs and ESCs or iPSCs have different characteristics that do not have one direction of stemness. This idea is supported by previous studies demonstrating that iPSCs and ESCs have different signatures of gene expression (Chin et al., 2009; Kim et al., 2008; Maguire et al., 2013; Marchetto et al., 2009).
To isolate stem cells from the heterogeneous population of amnion epithelial cells, anti-TRA1-60 antibody was used. TRA1-60–positive cells expressed stemness markers more than unsorted cells. TRA1-60–negative cells expressed all stemness marker genes less than unsorted cells. The expression of Klf4 was only 1.5-fold that of unsorted cells, because Klf4 expression is as high as in iPSCs in unsorted HAE cells. This suggests that TRA1-60–positive HAE cells have more stem cell-like characteristics. Thus, TRA1-60–positive amnion-derived epithelial cells are the concentrated stem cell population. We concluded that TRA1-60 can be a marker for isolating stem cells from amnion epithelial cells. More studies will be needed to identify the markers for amniotic MSCs.
Conclusion
We analyzed the characteristics of HAE and HAM cells. Both HAE and HAM cells expressed the stemness markers Oct3/4, Sox2, Klf4, c-Myc, Nanog, TRA1-60, TA1-81, SSEA3, and SSEA4. Especially, the relative expression of the Klf4 gene in HAE cells was as high as in iPSCs. TRA-1-60–positive HAE cells expressed those stemness markers more than unsorted cells and TRA-1-60–negative cells. Our results suggested that HAE and HAM cells have stem cell characteristics and TRA-1-60 can be a marker for isolating HAE stem cells.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20390430, 22790277, and 25282143).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Adinolfi M., Akle C.A., McColl I., Fensom A.H., Tansley L., Connolly P., His B.L., Faulk W.P., Travers P., and Bodmer F.W. (1982). Expression of HLA antigens, beta 2-microglobulin and enzymes by human amniotic epithelial cells. Nature, 295, 325–327 [DOI] [PubMed] [Google Scholar]
- Akle C.A., Adinolfi M., Welsh K.L., Leibowitz S., and McColl I. (1981). Immunogenicity of human amniotic epithelial cells after transplantation into volunteers. Lancet, 2, 1003–1005 [DOI] [PubMed] [Google Scholar]
- Bilic G., Zeisberger S.M., Mallik A.S, Zimmermann R., and Zisch A.H. (2008). Comparative caracherization of cultured human term amnion epithelial and mesenchymal stromal cells for application in cell therapy. Cell Transplant. 17, 955–968 [DOI] [PubMed] [Google Scholar]
- Cargnoni A., Gibelli L., Tosini A., Signoroni P.B., Nassuato C., Arienti D., Lombardi G, Alberini A., Wengler G.S., and Perolini O. (2009). Transplantation of allogenic and xenogenic placenta-derived cells reduces bleomycine-induced lung fibrosis. Cell Transplant. 18, 405–422 [DOI] [PubMed] [Google Scholar]
- Cargnoni A., Piccinelli E.C., Ressel L., Rossi D., Magatti M., Tosci I., Cesari V., Albertini M., Mazzola S., and Parolini O. (2014). Conditioned medium from amniotic membrane-derived cells prevents lung fibrosis and preserves blood gas exchanges in bleomycin-injured mice—specificity of the effects and insights into possible mechanisms. Cytotherapy 16, 17–32 [DOI] [PubMed] [Google Scholar]
- Chin M.H., Mason M.J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J., Khvorsotov I., Ott V., Grunstein M., Lavon N., Benvenisty N., Croce C.M., Clark A.T., Baxter T., Pyle A.D., Teitell M.A., Pelegrini M., Plath K., and Lowry W.E. (2009). Induced pluripotent cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coppi P., Bartsch G., Jr, Siddiqui M.M., Xu T., Santos C.C., Perin L., Motstoslavsky G., Serre A.C, Snyder E.Y, Yoo J.J., Furth M.E., Soker S., and Atala A. (2007). Isolation of amniotic stem cell lines with potential for therapy. Nat. Biotechnol. 25, 100–106 [DOI] [PubMed] [Google Scholar]
- Diaz-Prado S., Mulinos-Lopez E., Hermida-Gomez T., Rendal-Vazquez M.E., Fuentes-Boquete I., de Toro F.J., and Blanco F.J. (2010). Multilineage differentiation potential of cells isolated from the human amniotic membrane, J. Cell. Biochem. 111, 846–857 [DOI] [PubMed] [Google Scholar]
- Easley C.A., Miki T., Castro C.A., Ozolek J.A., Minervini C.F., Ben-Yehudah A., and Schatten G.P. (2012). Human amniotic epithelial cells are reprogrammed more efficiently by induced pluripotency than adult fibroblasts. Cell. Reprogram. 14, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Cai Z., and Zhou Z. (2009). Progress in studies on the characteristics of human amnion mesenchymal cells. Progr. Natural Sci. 19, 1047–1052 [Google Scholar]
- Igarashi A., Segoshi K., Sakai Y., Pan H., Kanawa M., Higashi Y., Sugiyama M., Nakamura K., Kurihara H., Yamaguchi S., Tsuji K., Kawamoto T., and Kato Y. (2007). Selection of common markers for bone marrow stromal cells from various bones using real-time RT-PCR: Effects of passage number and donor age. Tiss. Eng. 13, 2405–2417 [DOI] [PubMed] [Google Scholar]
- Ishii M., Koike C., Igarashi A., Yamanaka K., Pan H., Higashi Y., Kawaguchi H., Sugiyama M., Kamata N., Iwata T., Matsubara T., Nakamura K., Kurihara H., Tsuji K., and Kato Y. (2005). Molecular marker distinguish bone marrow mesenchymal stem cells from fibroblast. Biochem. Biophys. Res. Commun. 332, 297–303 [DOI] [PubMed] [Google Scholar]
- Izumi M., Pazin B.J., Minervini C.F., Gerlach J., Ross M.A., Stolz D.B., Turner M.E., Thompson R.L, and Miki T. (2009). Quantitative comparison of stem cell markers positive cells in fetal and term human amnion. J. Reprod. Immunol. 81, 39–43 [DOI] [PubMed] [Google Scholar]
- Izumi-Yoneda N., Toda A., Okabe M., Koike C., Takashima S., Yoshida T., Konishi I., Saito S., and Nikaido T. (2009). Alpha 1 antitrypsin activity is decreased in human amnion in premature repture of the fetal membranes. Mol. Hum. Reprod. 15, 49–57 [DOI] [PubMed] [Google Scholar]
- Kamiya K., Wang M., Uchida A., Amano S., Oshika T., Sakuragawa N., and Hori J. (2005). Topical application of culture supplement from human amniotic epithelial cells suppresses inflammatory reactions in cornea. Exp. Eye Res. 80 671–679 [DOI] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., and Orkin S.H. (2008). An extended transcriptional network for pluripotency of embrionic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire C.T., Demarest B.L., Hill J.T., Palmer J.D., Brothman A.R., Yost H.J., and Condic M.L. (2013). Genome-wide analysis reveals the unique stem cell identity of human amniocytes. PLoS One 8, e53372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M.C., Yeo G.W., Kainohama O., Marchetto M.C., N, Yeo G.W., Kainohara O., Marsala M., Gage F.H, and Muotri A.R. (2009). Transcriptional signature and memory retention of human-induced pluripotent stem cells. PloS One 4, E7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Lehmann T., Cai H., Stolz D.B., and Storm S.C. (2005) Stem cell characteristics of amniotic epithelial cells. Stem Cell 23, 1549–1559 [DOI] [PubMed] [Google Scholar]
- Miki T., and Strom S.C. (2006). Amnion-derived pluripotent multipotent stem cells. Stem Cell Rev. 2, 133–141 [DOI] [PubMed] [Google Scholar]
- Miki T., Mitamura K., Ross M.A., Stolz D.B., and Storm S.C. (2007). Identification of stem cell marker positive cells by immunofluorescence in term human amnion. J. Reprod. Immunol. 75, 91–96 [DOI] [PubMed] [Google Scholar]
- Murphy S., Rosli S., Acharya R., Mathias L., Lim R., Wallance E., and Jenkin G. (2010). Amnion epithelial cell isolation and characterization for clinical use. Curr. Protoc. Stem Cell Biol. 13, 1–25 [DOI] [PubMed] [Google Scholar]
- Nagura S., Ohtaka S., Koike C., Okabe M., Yoshida T., Fathy M.Fukahara K., Yoshimura N., Misaki T., and Nikaido T. (2013). Effect of exogenous Oct4 overexpression on cardiomyocte differentiation of human amniotic mesenchymal cells. Cell. Reprogram. 15, 471–480 [DOI] [PubMed] [Google Scholar]
- Nogami M., Tsuno H., Koike C., Okabe M., Yoshida T., Seki S., Matsui Y., Kimura T., and Nikaido T. (2012). Isolation and characterization of human amniotic mesenchymal stem cells and their chontrogenic differentiation. Transplantation 93, 1221–1228 [DOI] [PubMed] [Google Scholar]
- Otaka S., Nagura S., Koike C., Okabe M., Yoshida T., Fathy M., Yanagi K., Misaki T., and Nikaido T. (2013). Selective isolation of Nanog positive human amniotic mesenchymal cells and differentiation into cardiomyocytes. Cell. Reprogram. 15, 80–91 [DOI] [PubMed] [Google Scholar]
- Parolini O., Soncini M., Evangelista M., and Schmidt D. (2009). Amniotic membrane and amniotic fluid-derived cells: Potential tools for regenerative medicine? Regen. Med. 4, 275–291 [DOI] [PubMed] [Google Scholar]
- Parolini O., Alvaino F., Bergwerf I., Boraschi D., De Bari C., De Waele P., Dominici M., Evangelista M., Falk W., Hennerbichler S., Hess D. C., Lanzoni G., Liu B., Marongiu F., McGuckin C., Mohr S., Nolli M., L., Ofir R., Ponsaerts P., Romagnoli L., Solomon A., Soncin M., Strom S., Surbek D., Venkatachalam S., Wolbank S., Zeisberger S., Zeitlin A., Zisch A., and Borlongan C.V. (2010). Toward cell therapy using placenta-derived cells: Disease mechanisms, cell biology, preclinical studies, and regulatory aspects at the round table. Stem Cells Dev. 19, 143–154 [DOI] [PubMed] [Google Scholar]
- Takashima S., Ise H., Zhao P., Akaike T., and Nikaido T. (2004). Human amniotic epithelial cells possess hepatocyte-like characteristics and functions. Cell Struct. Funct. 29, 73–84 [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., and Jones J.M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- Toda A., Okabe M., Yoshida T., and Nikaido T. (2007). The poteintial of amniotic membrane/amnion-derived cells for regeneration of various tissues. J. Pharmacol Sci. 105, 215–228 [DOI] [PubMed] [Google Scholar]
- Tsuno H., Yoshida T., Nogami M., Koike C., Okabe M., Noto Z., Arai N., Noguch M., and Nikaido T. (2012). Application of human amniotic mesenchymal cells as an allogenic transplantation cell source in bone regenerative therapy. Mater. Sci.Eng. C 32, 2452–2458 [Google Scholar]
- Wei J.P., Zhang T.S., Kawa S., Aizawa T., Ota M., Akaike T., Kato K., Konishi I., and Nikaido T. (2003). Human amnion-isolated cells normalize blood glucose in streptozotocin-induced diabetic mice. Cell Transplant. 12, 545–552 [DOI] [PubMed] [Google Scholar]
- Wei J.P., Nawata M., Wakitani S., Kametani K., Ota M., Toda A., Konishi I., Ebara S., and Nikaido T. (2009). Human amniotic mesenchymal cells differentiate into chondrocytes. Cloning Stem Cells 11, 19–26 [DOI] [PubMed] [Google Scholar]
- Wolbank S., Peterbauer A., Fahener M., Hennerbichier S., van Griensven M., Stadler G., Redl H., and Gabriel C. (2007). Dose-dependent immunomodulatory effect of human stem cells from amniotic membrane: A comparison with human mesenchymal stem cells from adipose tissue. Tiss. Eng. 13, 1173–1183 [DOI] [PubMed] [Google Scholar]
- Zao H.X., Li Y., Jin H.F., Liu C., Jiang F., Luo Y.N, Yin G.W., Li Y., Wang J., Li L.S., Yao Y.Q., and Wang X.H. (2010). Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation 80, 123–129 [DOI] [PubMed] [Google Scholar]
- Zhao P., Ise H., Hongo M., Ota M., Konishi I., and Nikaido T. (2005). Human amniotic mesenchymal cells have some characteristics of cardiomyocytes. Transplantation, 79, 528–535 [DOI] [PubMed] [Google Scholar]