Those of us interested in the extracellular matrix (ECM) are faced with significant challenges of definition. ECM proteins are large, complex and assembled into crosslinked insoluble matrices. This has meant that defining the biochemical composition of ECMs has been difficult. Nonetheless, protein chemistry and molecular biology have defined many familiar ECM proteins — collagens, proteoglycans, laminins, thrombospondins, tenascins, fibronectins, etc. With the completion of many genomes it should now be possible to develop complete “parts lists” for the ECM. Such lists are needed for analyzing data from “omic” approaches such as expression arrays, latest-generation sequencing and proteomics. These approaches generate long lists and it is typically necessary to extract from those lists the genes/proteins of interest. Anyone who attempts to do this using the commonly used gene ontology (GO) categories soon discovers that they are largely useless for defining ECM proteins. Many ECM proteins are unannotated and those which are, are sorted, with little evidence of logic or consistency, into diverse categories such “extracellular matrix,” “basement membrane,” “cell surface” and many others. The human and mouse orthologs are often found in different categories and attempts to use GO categories to extract a complete list of ECM genes or proteins from a data set are unsatisfactory at best.
Faced with this problem in the course of an ECM proteomics project, we decided we needed to develop a better list of ECM proteins (Naba et al., 2012). This turned out to be not entirely straightforward. While the familiar ECM glycoproteins, collagens and proteoglycans could be collected relatively easily, the standard procedures for collecting lists of homologs, using BLAST or domain-based searches, quickly run into problems. ECM proteins are characteristically formed from multiple domains and those domains are shared among different ECM proteins and also with non-ECM proteins (Hohenester and Engel, 2002; Adams and Engel, 2007). Two obvious examples among many are EGF and FN3 domains, both very ancient protein domains that predate the origins of extracellular matrix in metazoa. They are found in many ECM proteins but also in diverse membrane proteins and secreted factors. So a BLAST or domain search produces a confusing mix of “homologs.” It is the domain architecture (the entire domain composition, number and order) that defines families of ECM proteins.
Nonetheless, the domain composition of ECM proteins does offer the route to defining an essentially complete list of ECM proteins and sorting them from homologous but non-ECM proteins (i.e., those which share one or more domains with ECM proteins but are clearly not matrix proteins — examples would include many tyrosine kinase or phosphatase receptors or adhesion receptors). We started with a list of characteristic ECM domains and used them to pull out all genes/proteins containing those domains from the human and mouse genomes/proteomes. We then culled those lists using a list of “excluding domains” to remove “contaminants” — the excluding list contained domains such as kinase, phosphatase and protease, chosen to eliminate the obvious “contaminants.” We performed this positive/negative sweep procedure iteratively, checking to ensure that we collected all the ECM proteins we could think of (that meant adding and deleting some domains) and eliminated all the “contaminants.” The details are given in Naba et al. (2012). We ended up with a list of around 50 “including domains” and around 20 “excluding domains” that effectively collected all known (at least to us) ECM proteins and did not select most growth factor and adhesion receptors. We then screened out any proteins with transmembrane domains, with the exception of a few collagens. The eventual lists (human and mouse) each of around 300 proteins we called the “core matrisome” (Fig. 1). These lists included all the known collagens, proteoglycans and well defined ECM glycoproteins along with additional proteins that had all the structural (domain) characteristics of ECM proteins but about which essentially nothing is known — presumptive novel ECM proteins.
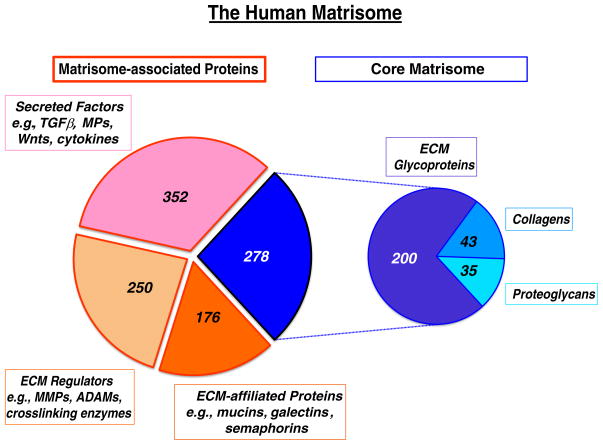
Fig. 1.
The human matrisome and its subcategories.
The core matrisome comprises three subcategories; ECM glycoproteins, collagens and proteoglycans, in each case defined by their domain structures (see text). All of these proteins are believed to assemble into extracellular matrices of one sort or another.
The three main subcategories of matrisome-associated proteins are more inclusively defined – they include proteins known to associate with assembled ECM as well as related proteins that may or may not – all were included to ensure their capture in “omic” screens of various sorts.
The secreted protein category includes a list of growth factors, cytokines and other secreted proteins — some are known to bind to ECM at least part of the time; others are included in the expectation that many of them will also be discovered to bind to ECM.
The ECM regulator category includes proteases, protease inhibitors and ECM crosslinking enzymes. Again, many are known to bind to and modify ECM proteins and structures in important ways — their homologs are likely also to do so and have been included for completeness.
The final category, designated “ECM-affiliated” includes protein families that some scientists (but not others) may consider as ECM proteins (e.g., mucins, C-type lectins, syndecans, glypicans), some that could be viewed as secreted factors but which also associate with solid-phase complexes (e.g., semaphorins and their homologous receptors, plexins, collagen-related proteins such as C1q and homologs) and a few families that appear repeatedly in ECM-enriched preparations for currently unknown reasons (e.g., annexins, galectins). Complete lists of the proteins in each subcategory for both human and mouse, together with gene and protein identifiers as well as protein sequence files are given in Naba et al. 2012 and at http://web.mit.edu/hyneslab/matrisome/ and summary tables are given in Hynes and Naba (2012).
However, another question of definition arose immediately. How do we define extracellular matrix? Does it include bound growth factors or bound ECM-modifying enzymes? And what about protein families such as mucins, galectins and semaphorins or proteins containing short stretches of collagen triple helix (C1q, collectins, ficolins, acetylcholine esterase)? Operationally, many of these proteins fractionate/co-purify with extracellular matrix and certainly contribute to its biological functions. Scientists may differ on whether or not to include such protein families (and others) in the definition of ECM proteins. So we decided to develop some additional “matrisome-associated” categories, using similar strategies of “including” and “excluding” domains. Specifically we defined a list of secreted factors — known growth factors and their homologs (TGF-β, BMPs, PDGFs, FGFs, Wnts, Hedgehogs, S100 proteins, chemokines etc.). We were deliberately inclusive and did not restrict these lists to growth factors, cytokines and chemokines known to bind to ECM proteins but also all their homologs as well as other families of secreted factors that might bind to ECM. Similarly we developed a list of ECM regulators (MMPs and other proteases, including membrane-bound proteases such as ADAMs and ADAM-TS proteins as well as other protease families and protease inhibitors) and ECM crosslinking enzymes (transglutaminases, lysyl oxidases and prolyl hydroxylases and regulators of these modifiers). Finally, we developed a list of “ECM-affiliated” proteins to include proteins that some might consider ECM-associated, whereas others would not. This list includes mucins, C-type lectins, semaphorins, syndecans, glypicans, as well as some protein families that we included because some members of the family co-enriched with genuine ECM proteins in our experiments (annexins, galectins).
We believe that the “core matrisome” categories (collagens, proteoglycans and ECM glycoproteins) are robust and not likely to change much with further analyses, at least for mammals and probably other vertebrates (other taxonomic groups clearly do contain additional ECM proteins). However, the “matrisome-associated” categories (secreted factors, regulators and affiliated proteins) are, by their nature, less firmly established and we suspect that they may well evolve in light of subsequent analyses. These latter categories were deliberately “inclusive” — although many proteins within those categories undoubtedly do bind reproducibly to ECM, others may not (see Fig. 1). Our aim was to define categories that would capture all candidate components of the ECM.
Having generated these categorical lists (see Hynes and Naba, 2012; Naba et al., 2012 and http://web.mit.edu/hyneslab/matrisome/) we would like to suggest that these six subcategories offer a much more workable way to select out sub-lists of ECM proteins and ECM-associated proteins from data sets, be they derived from proteomics, genomics, expression profiling or any other genome-scale analyses. We believe these lists will be straightforward to use. We expect them to evolve and welcome suggestions for additions or other modifications. It is our intention to maintain the “matrisome” web site and incorporate additional information in due course.
References
- Adams JC, Engel J. Bioinformatic analysis of adhesion proteins. Methods Mol Biol. 2007;370:147–172. doi: 10.1007/978-1-59745-353-0_12. [DOI] [PubMed] [Google Scholar]
- Hohenester E, Engel J. Domain structure and organisation in extracellular matrix proteins. Matrix Biol. 2002;21:115–128. doi: 10.1016/s0945-053x(01)00191-3. [DOI] [PubMed] [Google Scholar]
- Hynes RO, Naba A. Hynes RO, Yamada KM, editors. Overview of the matrisome — an inventory of extracellular matrix constituents and functions. Extracellular Matrix Biology Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a004903. http://dx.doi.org/10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed]
- Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.014647. (M111.014647) [DOI] [PMC free article] [PubMed] [Google Scholar]