To fully use human pluripotent stem cells (PSCs) in regenerative medicine, highly efficient differentiation strategies are required to drive induced PSCs into desired lineages and generate functional cell progenies. Current differentiation protocols for deriving dopaminergic neurons from PSCs involve months of stem cell culture procedures and multiple reagents. This study was motivated by a desire for a rapid and highly efficient system to generate human PSC-derived functional dopaminergic neurons.
Keywords: Induced pluripotent stem cells, Embryonic stem cells, Parkinson’s disease, Basic helix-loop-helix transcription factors, Tet-On
Abstract
Human pluripotent stem cells (PSCs) are a promising cell resource for various applications in regenerative medicine. Highly efficient approaches that differentiate human PSCs into functional lineage-specific neurons are critical for modeling neurological disorders and testing potential therapies. Proneural transcription factors are crucial drivers of neuron development and hold promise for driving highly efficient neuronal conversion in PSCs. Here, we study the functions of proneural transcription factor Atoh1 in the neuronal differentiation of PSCs. We show that Atoh1 is induced during the neuronal conversion of PSCs and that ectopic Atoh1 expression is sufficient to drive PSCs into neurons with high efficiency. Atoh1 induction, in combination with cell extrinsic factors, differentiates PSCs into functional dopaminergic (DA) neurons with >80% purity. Atoh1-induced DA neurons recapitulate key biochemical and electrophysiological features of midbrain DA neurons, the degeneration of which is responsible for clinical symptoms in Parkinson’s disease (PD). Atoh1-induced DA neurons provide a reliable disease model for studying PD pathogenesis, such as neurotoxin-induced neurodegeneration in PD. Overall, our results determine the role of Atoh1 in regulating neuronal differentiation and neuron subtype specification of human PSCs. Our Atoh1-mediated differentiation approach will enable large-scale applications of PD patient-derived midbrain DA neurons in mechanistic studies and drug screening for both familial and sporadic PD.
Introduction
Human pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), exhibit unique characteristics such as indefinite self-renewal capacity and multilineage differentiation potential. Human PSCs, especially patient-derived iPSCs, hold enormous promise for various applications in regenerative medicine, including disease modeling, drug development, and cell replacement therapy [1–3]. In order to fully use these patient-specific iPSCs in regenerative medicine, highly efficient differentiation strategies are required to drive iPSCs into desired lineages and generate functional cell progenies, such as various subtypes of neurons. Current protocols for differentiating human PSCs into lineage-specific neurons (e.g., dopaminergic [DA] neurons) are based on embryoid body formation, stromal feeder coculture, selective survival conditions, or inhibitors of SMAD signaling [4–8]. These PSC-derived neurons have allowed scientists to study molecular mechanisms underlying various neurological disorders, test potential drugs, and optimize strategies for cell replacement therapy. However, current neuron differentiation protocols for PSCs involve months of stem cell culture procedures and multiple reagents, which cause significant variation, especially for researchers who have limited practice in PSC culture and desire functional neurons at high purity for disease-in-a-dish models. A broad desire for a robust system to generate human PSC-derived neurons motivated us to develop a highly efficient strategy to generate lineage-specific functional neurons using the proneural transcription factor Atoh1.
ATOH1 (the mammalian homolog of Drosophila Atonal) belongs to the proneural transcription factors of the basic helix-loop-helix family [9, 10]. Proneural transcription factors are crucial in driving the acquisition of a generic neuronal fate and regulating neuronal subtype specification during development [9, 11]. Atoh1 proteins form heterodimers with E proteins, and these heterodimers function as transcriptional activators by binding E box motifs (CANNTG) in the regulatory regions of their target genes [10, 12]. Atoh1 is a key regulator of neurogenesis, governing the differentiation of various neuronal lineages, including cerebellar granule neurons, brainstem neurons, inner ear hair cells, and numerous components of the proprioceptive and interoceptive systems, as well as some non-neuronal cell types [13–17]. Atoh1 can activate crucial neurogenic transcription factors, such as NeuroD1, NeuroD2, NeuroD6, Nhlh1, and Nhlh2, to initiate a neuronal differentiation program that later becomes self-supporting and Atoh1-independent [10]. Certain members of the proneural transcription factor family, such as ASCL1, Ngn2, and NeuroD1, have been successfully used to generate neurons from both PSCs and somatic cells [18–22]. However, the role of Atoh1 in the neuronal differentiation and neuron subtype specification of human PSCs is largely unknown, and, as a result, Atoh1-based strategies for the neuronal conversion of human PSCs are still unavailable.
Here, we show that Atoh1 is induced during the neuronal differentiation of human PSCs. By transiently inducing ectopic Atoh1 expression, we are able to efficiently convert PSCs into neurons. Atoh1 induction, in combination with two neural patterning morphogens (Sonic Hedgehog [SHH] and fibroblast growth factor 8b [FGF8b]), leads to rapid and highly efficient conversion of PSCs into DA neurons that recapitulate key biochemical and electrophysiological features of primary midbrain DA neurons. We also demonstrate that Atoh1-induced DA neurons serve as a reliable model for analyzing 6-hydroxydopamine (6-OHDA)-induced neurotoxicity in human midbrain DA neurons. Because most symptoms of Parkinson’s disease (PD) result from the degeneration of midbrain DA neurons located in the substantia nigra [23], Atoh1-induced DA neurons provide an in vitro neuron model for mechanistic studies and drug testing for PD.
Materials and Methods
Cell Culture
The human H1 ESC line was obtained from WiCell Research Resources (WiCell Research Institute, Madison, WI, http://www.wicell.org). The human iPSC line ND27760 (passages 25–30) was derived from human skin fibroblasts from a PD patient with a SNCA triplication that were obtained from the Coriell Cell Repositories (Camden, NY, http://ccr.coriell.org). Cell reprogramming was performed using a nonintegrating 4 factor (SOX2/OCT4/KLF4/MYC) Sendai virus system (CytoTune-iPS Reprogramming Kit; Life Technologies, Rockville, MD, http://www.lifetech.com). The pluripotency of this iPSC line has been characterized by immunocytochemistry for pluripotent cell markers (NANOG, OCT4, TRA-1-60, and SSEA-3) and embryoid body differentiation. Human ESCs and iPSCs were maintained as feeder-free cultures in Essential 8 medium (Life Technologies) or mTESR1 medium (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) in 5% CO2/95% air conditions at 37°C and were passaged using dispase (Life Technologies). Karyotype analysis of G-banded metaphase chromosomes was performed to confirm the chromosomal integrity of these ESCs and iPSCs. All experiments involving human stem cells were performed with the approval of the Johns Hopkins Medicine Institutional Review Boards.
Lentiviral Transduction
Human Atoh1 cDNA was constructed using high-fidelity polymerase chain reaction (PCR) kit (Roche, Indianapolis, IN, http://www.roche.com) and cloned into pTRIPZ vector (Thermo Scientific) with AgeI and MluI. The Trans-Lentiviral Packaging System (Thermo Scientific) was used for lentivirus packaging. Cells were infected by lentivirus at an multiplicity of infection of 5 for 24 hours with the addition of TransDux virus infection solution (System Biosciences). Stable cell lines were established by puromycin selection (0.5 μg/ml). All recombinant DNA and lentivirus experiments were performed following the National Institutes of Health guidelines.
Cell Differentiation and Cryopreservation
To measure Atoh1 expression during the neuronal conversion of human PSCs, cells were differentiated following a dual-SMAD inhibition protocol [7]. Noggin in this protocol was replaced by LDN193189 (100 nM; Stemgent, Cambridge, MA, https://www.stemgent.com).
For the Atoh1-induced neuron differentiation protocol, cells were plated (8 × 104 cells per cm2) on Matrigel (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com) in Essential 8 medium (Life Technologies) with the ROCK inhibitor (Y-27632, 10 µM; Stemgent). Atoh1 was induced by doxycycline (0.5 μg/ml; Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) in culture medium from day 1 to day 5. From day 1 to day 3, cell culture medium was changed every day and gradually shifted from Essential 6 medium (Life Technologies) to N2 medium (Dulbecco’s modified Eagle’s medium/F-12 medium with N2 supplement; Life Technologies). Cells were cultured in N2 medium until day 7, dissociated using Accutase (Sigma-Aldrich), and replated (3 × 105 cells per cm2) on dishes precoated with poly-d-lysine (1 μg/ml) and laminin (1 μg/ml) using neuron culture medium (Neurobasal medium with B27 supplement, brain-derived neurotrophic factor [20 ng/ml; PeproTech, Rocky Hill, NJ, http://www.peprotech.com], glial cell line-derived neurotrophic factor [20 ng/ml; PeproTech], transforming growth factor type β3 [1 ng/ml; R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com], ascorbic acid [0.2 mM; Sigma-Aldrich], dibutyryl cAMP [0.5 mM; Sigma-Aldrich], and γ-secretase inhibitor DAPT [10 µM; Stemgent]). From day 8 to day 36, half of the cell culture medium was replenished every 3–4 days. For Atoh1-induced DA neuron differentiation protocol, the protocol above was modified by adding SHH (SHH C25II, 100 ng/ml; R&D Systems) and FGF8b (100 ng/ml; PeproTech) from day 1 to day 5.
Atoh1-induced DA neuron precursors at differentiation day 7 were dissociated using Accutase. Then, 1 × 106 cells were cryopreserved in 1 ml of freezing medium (40% Neurobasal medium with B27 supplement, 50% fetal bovine serum, and 10% DMSO) using a freezing container (Nalgene) in −80°C for 24 hours and stored in liquid nitrogen.
Quantitative Real-Time PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen). Reverse transcription was performed using murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) and oligo(dT) primers. Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the IQ5 RT-PCR detection system (Bio-Rad, Hercules, CA, http://www.bio-rad.com). All primer sequences are listed in supplemental online Table 1. Relative expression of each gene was normalized to the 18S rRNA.
Western Blot
Total cellular proteins were extracted with RIPA buffer (Sigma-Aldrich) containing a protease and phosphatase inhibitor cocktail (Calbiochem, San Diego, CA, http://www.emdbiosciences.com). SDS-polyacrylamide gel electrophoresis was performed with 50 µg of total cellular proteins per lane using 4%–12% gradient Tris-glycine gels (Lonza, Walkersville, MD, http://www.lonza.com). Western blot was performed using a Quantitative Western Blot System (LI-COR Biosciences, Lincoln, NE, http://www.licor.com/) following the manufacturer’s instructions. The primary antibodies were: mouse anti-FALG M2 (Sigma-Aldrich), rabbit anti-Atoh1 (Millipore, Billerica, MA, http://www.millipore.com), and mouse anti-β-actin (Sigma-Aldrich). Secondary antibodies were labeled with IRDye IR dyes, and protein levels were quantified with Odyssey IR Imaging System (LI-COR Biosciences).
Immunofluorescence and Cell Counting
Differentiated cells were fixed in 4% paraformaldehyde/1% sucrose in phosphate-buffered saline (pH 7.4) at room temperature and blocked with 5% normal goat serum and 0.2% Triton X-100. Primary antibodies (listed in supplemental online Table 2) were diluted in 5% normal goat serum and incubated with samples overnight at 4°C. Cy3- and Alexa Fluor 488-labeled secondary antibodies were applied for 2 hours. Samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted on glass slides using ProLong antifade kit (Life Technologies).
The percentage of marker-positive cells was determined in samples derived from at least three independent experiments. In Adobe Photoshop software, images from 10 randomly selected fields were used for counting the number of DAPI-positive cells expressing a specific marker.
Electrophysiological Recordings
Voltage-clamp or current-clamp recordings were performed at 35°C in a chamber perfused with regular artificial cerebrospinal fluid (124 mM NaCl, 2.5 mM KCl, 1.3 mM MgCl2, 2.5 mM CaCl2, 1 mM NaH2PO4, 26.2 mM NaHCO3, 20 mM glucose, pH 7.4, equilibrated with 95% O2 and 5% CO2, ∼310 mosm), which flowed at 4 ml/minute. Patch electrodes were pulled from borosilicate glass and had resistances of 2–4.0 MΩ when filled with an intracellular solution (135 mM KMeSO4, 5 mM KCl, 5 mM HEPES, 0.25 mM EGTA-free acid, 2 mM Mg-ATP, 0.5 mM GTP, 10 mM phosphocreatine-tris, pH 7.3, ∼290 mosm).
Neurons were identified using a ×10 objective mounted on an upright microscope with transmitted light, and their neuronal somata were then visualized through a ×40 water immersion objective using IR differential interference contrast optics. The cell somatic recordings were made using an Axopatch 200B amplifier in combination with pClamp 9.0 software (Molecular Devices). Neurons were voltage-clamped at −80 mV. Rseries and Rinput were monitored using a 2.5-mV 100-ms depolarizing voltage step in each recording sweep. Current traces were filtered at 5 kHz, digitized at 10 kHz using a Digidata 1322A interface, and stored for off-line analysis. Leak and capacitative currents were corrected by subtracting a scaled current elicited by a +2.5-mV step from the holding potential.
For current clamp recording, the same Axopatch 200B amplifier was used; whole-cell mode was achieved initially in the voltage clamp configuration. Then the recording was switched into current clamp mode. The resting membrane potential was monitored for more than 5 minutes. The experiment was discontinued if the resting membrane potential became more positive than −40 mV. The action potential was continually monitored for 5 minutes, and if there was no threshold change, the reagent perfusion commenced. All reagents were bought from Sigma-Aldrich except tetrodotoxin (TTX; Abcam, Cambridge, U.K., http://www.abcam.com) and ML252 (Vanderbilt Center for Neuroscience Drug Discovery, Nashville, TN, http://www.vcndd.com/index.php).
High-Performance Liquid Chromatography Analysis
On day 36 of differentiation, medium was replaced by Hanks’ balanced saline solution buffer with the addition of 56 mM KCl (200 µl per well in 24-well plates) and incubated for 15 minutes at 37°C. Medium was collected and centrifuged (15,000g for 15 minutes at 4°C) to clear cell debris. Samples were immediately frozen in liquid nitrogen and stored at −80°C. For high-performance liquid chromatography (HPLC) analysis, samples were thawed and concentrated using a vacuum (Savant SDP 121P; Thermo Fisher Scientific, Waltham, MA, http://www.thermofisher.com) connected with refrigerated vapor trap (Savant RVT 5105; Thermo Fisher Scientific), and the freeze-dried samples were resuspended in 10 mM perchloric acid. Monoamines were analyzed by HPLC electrochemical detection by dual channel Coulochem III electrochemical detector (model 5300; ESA Inc., Chelmsford, MA, http://www.esainc.com), and monoamines were separated by using a reverse phase C18 column (3-mm × 150-mm C-18 RP-column; Acclaim Polar Advantage II; Thermo Fisher Scientific) with a flow rate of 0.600 ml/minute. Monoamine concentrations were quantified by comparison of the area under the curve to known standard dilutions.
6-OHDA Treatment in DA Neurons and Lactate Dehydrogenase Analysis
Neuron culture medium was changed to Neurobasal medium before treatment. 6-OHDA was freshly prepared in vehicle solution (0.15% ascorbic acid in H2O) and quickly added to the neuron culture. Control cells were treated with vehicle solution alone. After 15 minutes at 37°C, the medium was removed, and the neurons were gently washed twice with Neurobasal medium. Two hundred microliters of neuron culture medium (Neurobasal medium with B27 supplement) was added to each well and further incubated for 24 hours. Cytotoxicity induced by 6-OHDA was measured using a lactate dehydrogenase (LDH) cytotoxicity detection kit (Roche) following the manufacturer’s instructions. The percentage of cytotoxicity was calculated using the following equation: Cytotoxicity (%) = (Experiment value − Low control)/(High control − Low control) × 100, where low control indicates culture medium, and high control indicates total cell lysate.
Data Analysis and Statistics
All results reported here represent at least three independent replications. Statistical analysis was performed using Prizm software (GraphPad, San Diego, CA, http://www.graphpad.com). Post hoc tests included the Student’s t test and the Tukey multiple comparison tests as appropriate. All of the data are represented as mean values ± SEM.
For neurophysiological recordings, the recorded data were first visualized with Clampfit 9.2 and exported to MATLAB (Mathworks, Natick, MA, http://www.mathworks.com) for further analysis and plotting. The recording traces are visualized with Igor 6.0 (WaveMetrics, Portland, OR, http://www.wavemetrics.com). All group data are reported as means ± SD except otherwise stated.
Results
Atoh1 Is Induced During the Neuronal Differentiation of Human PSCs
We followed a dual-SMAD inhibition protocol [7] for differentiating human iPSCs into neurons. Differentiated cells first expressed neurectodermal marker (PAX6) and neural rosette marker (NESTIN) at day 10 of differentiation (Fig. 1A, left). Mature neurons at day 40 of differentiation expressed neuronal marker β-Tubulin III (TUJ1) and MAP2 (Fig. 1A, right). We further examined the expression of various markers at differentiation days 0, 10, and 20 by qRT-PCR, which confirmed the inhibition of pluripotency marker (NANOG) and the induction of neural markers (PAX6, Ngn2, and NEUROD1) (Fig. 1B). Next, we examined Atoh1 expression by qRT-PCR and found that ATOH1 was induced at differentiation days 10 and 20 when compared with undifferentiated cells (Fig. 1B). Western blotting also confirmed the induction of Atoh1 protein at differentiation day 20 (Fig. 1C). These data suggested that Atoh1 is involved in the neuronal conversion of human PSCs, which warranted further study.
Figure 1.
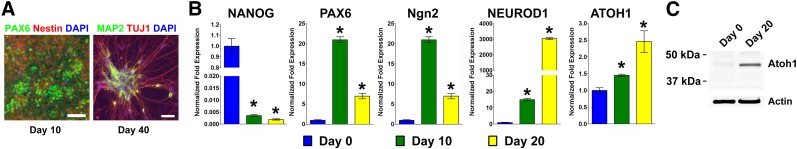
Atoh1 is induced during the differentiation of human pluripotent stem cells into neurons. (A): Human induced pluripotent stem cells (iPSCs) were differentiated into neurons following the dual-SMAD inhibition protocol. By day 10 of differentiation, cells expressed neural lineage markers (PAX6 and NESTIN). By day 40 of differentiation, iPSC-derived neurons expressed neuronal markers (TUJ1 and MAP2). Cell nuclei were counterstained with DAPI. Scale bars = 50 μm. (B, C): Markers for pluripotent cells (NANOG), neural (PAX6), and neuronal (Ngn2 and NEUROD1) lineages were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) during iPSC differentiation at days 0, 10, and 20. Atoh1 expression was analyzed by qRT-PCR (B) and Western blotting (C). The data represent means ± SEM. ∗, p < .01 compared with day 0. Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole.
Ectopic Atoh1 Expression Induces Highly Efficient Neuronal Conversion of Human PSCs
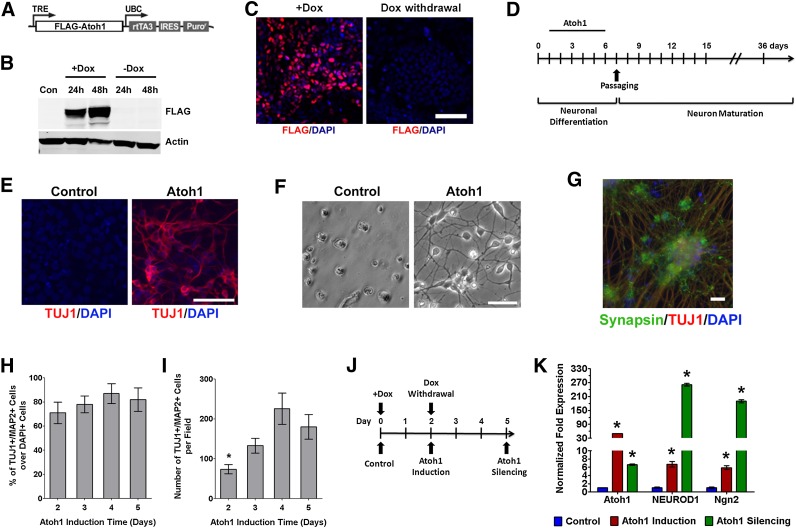
To address whether Atoh1 induction is sufficient for the neuronal differentiation of human PSCs, we established a lentivirus-mediated gene delivery system to achieve doxycycline (Dox)-inducible Atoh1 expression in human PSCs. We constructed a Tet-On lentiviral vector that harbors human Atoh1 transgene with N-terminal FLAG tag (Fig. 2A). Human iPSCs and ESCs were infected with Atoh1 lentivirus to establish stable cell lines (Atoh1-iPSC and Atoh1-ESC) after puromycin selection. Dox treatment for 48 hours in Atoh1-iPSCs induced Atoh1 expression as determined by immunoblotting against FLAG tag, and transgenic Atoh1 expression was turned off after Dox withdrawal (Fig. 2B). Immunostaining against FLAG tag also confirmed Atoh1 induction after 3-day Dox treatment and the silencing of Atoh1 transgene after Dox withdrawal for 3 days (Fig. 2C).
Figure 2.
Ectopic Atoh1 expression drives neuronal conversion in induced pluripotent stem cells (iPSCs). (A): Diagram of the lentiviral vector for Dox-inducible Atoh1 expression. (B): Dox controls the on/off switch of Atoh1 expression. Human iPSCs were infected with lentivirus harboring Dox-inducible Atoh1. Stable Atoh1-iPSCs after puromycin selection were treated with or without Dox for 48 hours and transferred to Dox-free medium. Whole cell lysates collected on each indicated time point were subjected to immunoblot using anti-FLAG antibody. (C): Atoh1-iPSCs were treated with Dox (+Dox) for 3 days and changed to Dox-free medium (Dox withdrawal) for 3 days. Cells were immunostained with FLAG antibody. (D): Diagram of Atoh1-induced neuron differentiation protocol. Atoh1 is induced by Dox from days 1 to 5. (E): Immunostaining from cell cultures at differentiation day 6 shows TUJ1 expression in Atoh1-induced cells but not in control cells. (F): Bright-field microscope images show cell adhesion and neuronal process formation in Atoh1-induced cells on differentiation day 10. (G): Immunostaining shows the coexpression of TUJ1 and Synapsin in Atoh1-induced neurons on differentiation day 36. (H, I): During a 5-day time period, an equal number of Atoh1-iPSCs received different lengths of Dox treatment (from 1 to 5 days). After being matured for 30 days, cells were immunostained against neuronal marker TUJ1 and MAP2. The percentage of TUJ1+/MAP2+ cells over DAPI+ cells and the total number of TUJ1+/MAP2+ cells were quantified in 10 random-selected microscopic fields (∗, p < .01 compared with cells that had 4- and 5-day Atoh1 induction). (J, K): Atoh1-iPSCs were treated with Dox for 2 days and returned to Dox-free medium for 3 days (J). The expression of Atoh1, NEUROD1, and Ngn2 was measured by quantitative real-time polymerase chain reaction in control, Atoh1 induction, and Atoh1 silencing samples (∗, p < .01 compared with control). In (C) and (E–G), cell nuclei were counterstained with DAPI. Scale bars = 20 μm. The data represent means ± SEM. Abbreviations: Con, control; DAPI, 4′,6-diamidino-2-phenylindole; Dox, doxycycline; FLAG-Atoh1, FLAG-tagged Atoh1; IRES, internal ribosome entry site; Puror, puromycin selection marker; rtTA3, reverse tet-transactivator; TRE, tet-inducible promoter; UBC, human ubiquitin C promoter.
Next, we induced ectopic Atoh1 expression in PSCs for neuronal differentiation following a protocol outlined in Figure 2D (also see details in Materials and Methods). Atoh1-iPSCs were maintained in a feeder-free culture system, and Atoh1 was induced by Dox for 5 days to drive neuronal conversion. After Dox withdrawal, neuronal precursors were passaged and allowed to further mature in vitro. On differentiation day 6, Atoh1 induced robust expression of the neuronal differentiation marker TUJ1, which was not detected in Dox-untreated cells (Fig. 2E). On day 7, cells were dissociated and replated on surfaces precoated for neuron culture. Two days after cell passaging, Atoh1-induced cells adhered and formed neuronal processes. In contrast, control cells failed to attach or grow in neuron culture medium (Fig. 2F). After further maturation in vitro for 30 days, Atoh1-induced neurons coexpressed the synaptic vesicle protein Synapsin and neuronal marker (TUJ1), demonstrating the establishment of synaptic terminals and neuronal maturation (Fig. 2G). We also replicated these results in Atoh1-ESC, in which Atoh1 also initiated the neuronal differentiation process and generated mature neurons (supplemental online Fig. 1).
To further optimize the Atoh1-mediated differentiation strategy, we asked what is the minimum time of Atoh1 induction for successful neuronal conversion. Equal numbers of Atoh1-iPSCs received different durations of Dox treatment (1–5 days), after which cells were replated and allowed to mature in vitro for additional 30 days. Cells with Dox treatment for 1 day failed to attach after replating. In contrast, Dox treatment for 2–5 days successfully differentiated iPSCs into neurons expressing TUJ1 and MAP2 (supplemental online Fig. 2). By quantifying the number of TUJ1+/MAP2+ neurons, we found that longer Atoh1 induction time did not increase the purity of Atoh1-induced neurons (Fig. 2H) but did significantly increase the yield of neurons, especially when comparing Atoh1 induction for 4–5 days to 2-day induction (Fig. 2I). To compare the level of neurogenic signaling before and after silencing ectopic Atoh1 expression, we treated Atoh1-iPSCs with Dox for 2 days and then withdrew Dox for 3 days (Fig. 2J, 2K). As determined by qRT-PCR, Atoh1 showed 49-fold up-regulation after Dox treatment and decreased after Dox withdrawal. Two neurogenic transcription factors (NEUROD1 and Ngn2) showed six- and fivefold induction, respectively, in response to Atoh1 induction. After Dox withdrawal, their expression did not decrease but increased further to 261- and 189-fold higher than control cells, respectively. These results suggest that ectopic Atoh1 expression in PSCs initiates a neurogenic program that becomes self-sustaining after the withdrawal of ectopic Atoh1.
To determine the subtype specification of Atoh1-induced neurons, we characterized neurons induced from Atoh1-iPSCs with various neuron subtype markers (Fig. 3). By day 36 of differentiation, ∼35% of Atoh1-induced neurons expressed tyrosine hydroxylase (TH), the rate-limiting enzyme in DA synthesis and a widely used DA neuron marker. Less than 10% of total cells expressed glutamate decarboxylase (GAD67) and serotonin, markers for GABAergic and serotonergic neurons, respectively. Glutamatergic neurons expressing vesicular glutamate transporter 1 were not detected in Atoh1-induced neurons.
Figure 3.
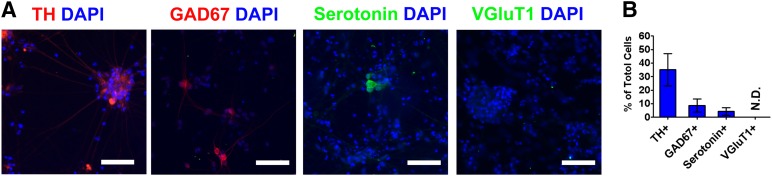
Neuron subtype specification in Atoh1-induced neurons. (A): Atoh1-induced neurons derived from Atoh1-induced pluripotent stem cells were allowed to mature in vitro, and cells at differentiation day 36 were immunostained with antibodies detecting dopaminergic (TH), GABAergic (GAD67), serotonergic (serotonin), and glutamatergic (VGluT1) neuron subtypes. Cell nuclei were counterstained with DAPI. Scale bars = 50 μm. (B): Immunostained neurons from 10 random-selected microscopic fields were counted to calculate the percentage of TH+, GAD67+, serotonin+, and VGluT1+ cells over DAPI+ cells. The data represent means ± SEM. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; N.D., not detected; TH, tyrosine hydroxylase; VGluT, vesicular glutamate transporter 1.
Atoh1-Mediated Differentiation of Human PSCs Into DA Neurons
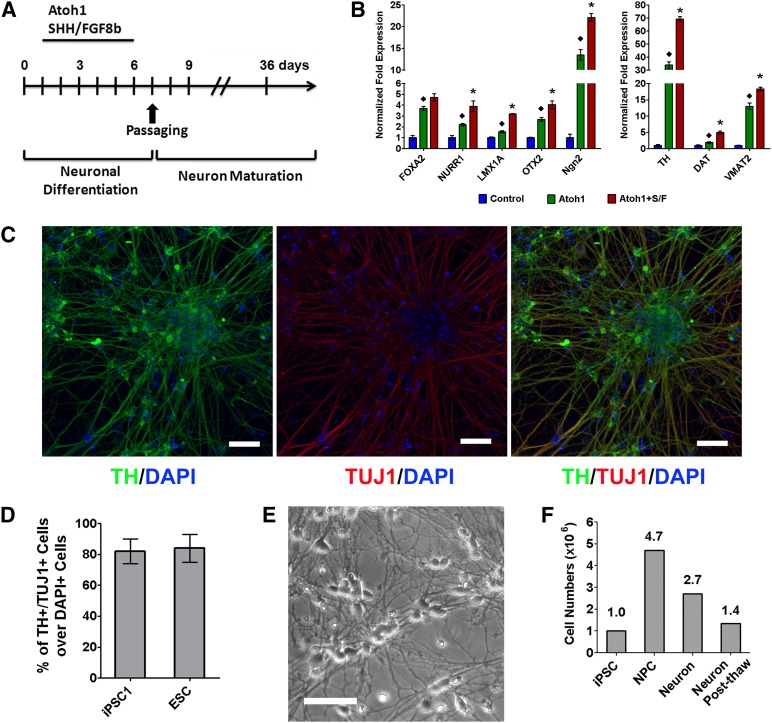
We found that ectopic Atoh1 expression preferentially drives the differentiation of human PSCs to TH-expressing neurons, suggesting a DA lineage specification. Two morphogens (SHH and FGF-8b) for neural patterning have been widely used to drive DA lineage specification during the neuronal conversion of human PSCs [5, 7, 24, 25]. We combined Atoh1 induction with these two morphogens to differentiate PSCs into DA neurons, following a protocol outlined in Figure 4A. Ectopic Atoh1 expression alone induced multiple DA neuron markers, such as FOXA2, NURR1, LMX1A, OTX2, Ngn2, TH, dopamine transporter (DAT), and VMAT2, most of which were further upregulated significantly by combining Atoh1 induction with SHH and FGF-8b (Fig. 4B). At day 36 of differentiation, Atoh1-induced neurons derived from both iPSCs and ESCs coexpressed the neuronal marker (TUJ1) and the DA neuron marker (TH) (Fig. 4C; supplemental online Fig. 3). The Atoh1-mediated protocol yielded DA neurons from human iPSCs and ESCs with 82% ± 8% and 84% ± 9% purity, respectively, as determined by the percentage of TH+/TUJ1+ cells over DAPI+ cells (Fig. 4D).
Figure 4.
Ectopic Atoh1 expression and cell extrinsic factors induces dopaminergic (DA) neurons from PSCs. (A): Diagram of DA neuron differentiation induced by Atoh1, SHH, and FGF8b. (B): Atoh1-iPSCs were differentiated by Atoh1 induction alone or in combination with SHH and FGF8b. The expression of DA lineage markers was analyzed by quantitative real-time polymerase chain reaction using cells at differentiation day 6. Control cells followed the same differentiation protocol but did not receive Dox treatment. Atoh1 induction in combination with SHH and FGF8b more robustly induced DA lineage markers than Atoh1 alone or untreated cells. The data represent means ± SEM. ∗, p < .01 compared with Atoh1 alone; ♦, p < .01 compared with control. (C, D): Atoh1-iPSCs were differentiated following the protocol shown in (A). Atoh1-induced neurons at differentiation day 36 were immunostained for neuronal marker (TUJ1) and DA neuron marker (TH). Cell nuclei were counterstained with DAPI. Scale bars = 20 μm. TH+/TUJ1+ DA neurons derived from Atoh1-iPSCs and Atoh1-ESCs from 10 random-selected microscopic fields were counted to calculate the percentage of TH+/TUJ1+ cells over DAPI+ cells. The data represent means ± SEM. (E): Bright-field microscope image of Atoh1-induced iPSC-derived DA neurons 7 days after being recovered from cryopreservation. Scale bars = 20 μm. (F): Atoh1-iPSCs (1 × 106) were differentiated following the protocol shown in (A). Cells were counted to calculate the number of NPCs (differentiation day 7), DA neurons (differentiation day 14), and post-thaw DA neurons (frozen at differentiation day 7 and cultured 7 days after cryopreservation). Abbreviations: Atoh1+S/F, Atoh1 induction in combination with SHH and FGF8b; DAPI, 4′,6-diamidino-2-phenylindole; DAT, dopamine transporter; ESC, embryonic stem cell; FGF, fibroblast growth factor; iPSC, induced pluripotent stem cell; NPC, neuron precursor cell; SHH, Sonic Hedgehog; TH, tyrosine hydroxylase.
In order to store Atoh1-induced DA neurons, Atoh1-induced DA neuron precursor cells (NPCs) at differentiation day 7 were cryopreserved, and these cells showed high viability and neuronal morphology upon being recovered from cryopreservation and cultured for 7 days (Fig. 4E). From 1 × 106 iPSCs, the Atoh1-mediated protocol generated 4.7 × 106 DA NPCs and yielded 2.7 × 106 or 1.4 × 106 DA neurons after direct cell passaging or cryopreservation, respectively (Fig. 4F).
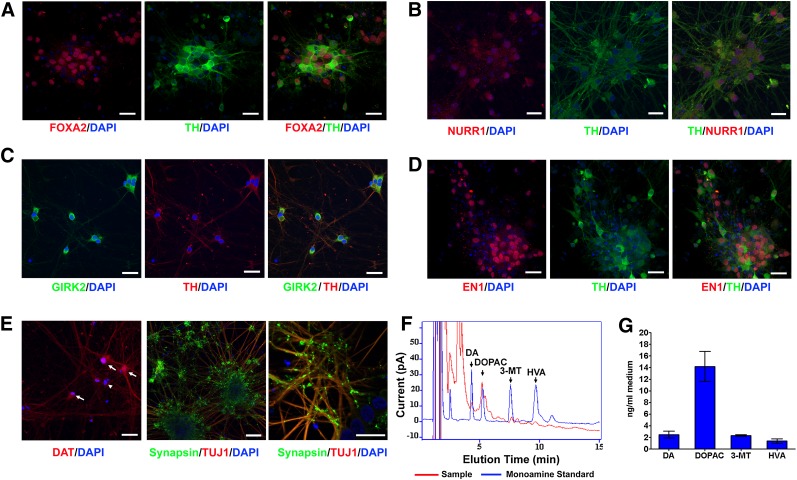
Next, we analyzed the expression of midbrain DA neuron markers in Atoh1-induced neurons. By differentiation day 36, these neurons expressed the midbrain DA neuron markers FOXA2, NURR1, Engrailed 1 (EN1), TH, G protein-regulated inward-rectifier potassium channel 2 (GIRK2), and DAT (Fig. 5A–5E), which are also expressed in midbrain DA neurons located in substantia nigra pars compacta. These Atoh1-induced DA neurons show extensive TUJ1+ nerve fiber growth and robust expression of synaptic vesicle protein Synapsin (Fig. 5E). GABAergic (GAD67+) or serotonergic (serotonin+) neurons were not detected in Atoh1-induced neurons derived from iPSCs, and undifferentiated iPSCs (SOX2+ or OCT4+) were also not detected (supplemental online Fig. 4).
Figure 5.
The expression of midbrain DA neuron markers and dopamine release in Atoh1-induced neurons. (A–E): Atoh1-induced pluripotent stem cells were differentiated following the protocol shown in Fig. 4A. DA neurons at differentiation day 36 were immunostained for midbrain DA neuron markers (FOXA2, NURR1, EN1, TH, GIRK2, and DAT) and mature neuron marker (Synapsin). Cell nuclei were counterstained with DAPI. The arrows and arrowhead in (E) indicate DAT+ and DAT− neurons, respectively. Scale bars = 20 μm. (F, G): Representative HPLC chromatogram (F) and quantification (G) of DA and its metabolites (DOPAC, 3-MT, and HVA) released from Atoh1-induced DA neurons at differentiation day 36 in response to KCl-evoked depolarization for 15 minutes. The data represent means ± SEM (n = 2). Abbreviations: DA, dopaminergic; DAPI, 4′,6-diamidino-2-phenylindole; DAT, dopamine transporter; DOPAC, 3,4-dihydroxy-phenylacetic acid; GIRK2, G protein-regulated inward-rectifier potassium channel 2; HVA, homovanillic acid; 3-MT, 3-methoxytyramine; TH, tyrosine hydroxylase.
Functional Characterization of Atoh1-Induced DA Neurons
We asked whether Atoh1-induced DA neurons exhibit key physiological properties of mature midbrain DA neurons. Dopamine release was quantified in Atoh1-induced DA neurons at differentiation day 36. HPLC analysis demonstrated the release of dopamine and its metabolites evoked by KCl depolarization (Fig. 5F, 5G).
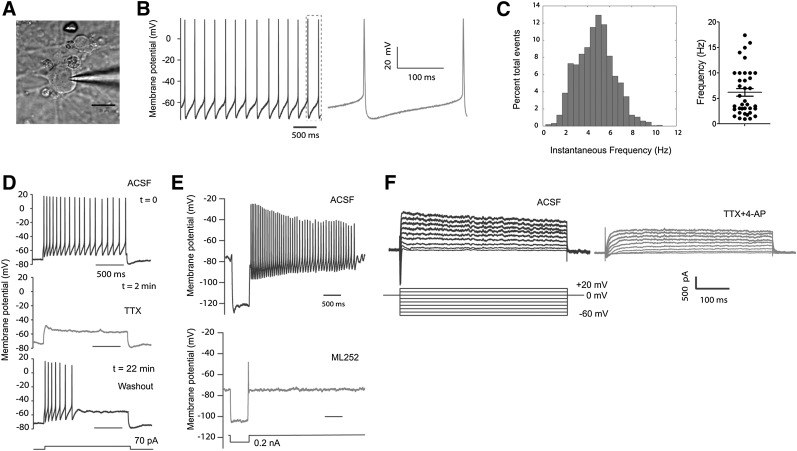
It has been well established that midbrain DA neurons are pacemaker neurons that discharge spontaneously at a rate between 1 and 10 Hz with an average rate of 4.5 Hz [26–30]. To test whether Atoh1-induced DA neurons display similar electrophysiological properties to primary midbrain DA neurons, we performed patch-clamp recording in Atoh1-induced DA neurons (n = 57) derived from human iPSCs at differentiation days 36–49 (Fig. 6A). In voltage-clamp experiments, the series input resistance of these Atoh1-induced DA neurons was 7.7 ± 3 MΩ; the input resistance was 295.8 ± 174.5 MΩ, and the average resting membrane potential was 75.3 ± 9.9 mV. Of these neurons, 64.9% showed spontaneous spiking activity (Fig. 6B; n = 37) with an amplitude of 66.1 ± 18.3 mV and a mean frequency of 6.2 ± 4.7 Hz (Fig. 6C; n = 37). Of these neurons, 26.3% discharged action potentials during current injection either by depolarization or hyperpolarization (n = 15), and only 8.7% (n = 5) of these neurons did not have typical action potential either by positive or negative current injection.
Figure 6.
Electrophysiological properties of Atoh1-induced dopaminergic (DA) neurons. (A): Differential interference contrast image of a patched Atoh1-induced DA neuron. Scale bar = 20 μm. (B, C): Atoh1-induced DA neurons derived from Atoh1-iPSCs showed spontaneous spiking activity. This cell has a resting membrane potential of −65 mV (B) (a zoomed view is shown in the right panel) and an average spiking frequency of 4.8 Hz (C) (left panel). The spontaneous spiking frequencies from 37 neurons were plotted in the right panel of (C) with the means ± SEM marked inside. (D): Whole cell current-clamp recording of action potentials evoked by 70 pA current injection (top panel). Action potentials were suppressed by sodium channel blocker (TTX) (middle panel). Action potentials recovered after TTX withdrawal (bottom panel). (E): An hyperpolarized injection of current (0.2 nA) evoked hyperpolarization and rebound tonic spiking. A typical hyperpolarization sag was observed in the upper panel, which was dampened by ML252 (5 µM, a KCNQ2 inhibitor, lower panel). (F): Voltage-clamp recording of Atoh1-induced neurons. Depolarized sodium and potassium currents were evoked by elevation the membrane potential to different levels (left panel). Both sodium and potassium currents were attenuated by sodium and potassium channel inhibitors (TTX [0.5 µM] and 4-AP [25 µM], respectively). Abbreviations: ACSF, artificial cerebrospinal fluid; TTX, tetrodotoxin.
We further investigated the maturation of intrinsic ion channels in Atoh1-induced DA neurons. As shown in Figure 6D, we first injected current (70 pA) into a neuron to depolarize the membrane potential. This induced a train of action potentials (Fig. 6D, top), which were completely blocked by the sodium channel blocker TTX (0.5 µM, 5 minutes of administration; Fig. 6D, middle). This effect was reversed by TTX washout, after which the action potentials recovered in 17 minutes (Fig. 6D, bottom).
Midbrain DA neurons have been found to also have KCNQ potassium channels that contribute to their tonic spontaneous activity, and during hyperpolarization these neurons display a typical sag in voltage [31]. Here, we injected negative current to hyperpolarize the membrane of Atoh1-induced neurons (Fig. 6E). This negative current injection produced hyperpolarization sag and rebound action potentials that resemble the tonic spontaneous spiking activity. Both events were blocked by ML252 (5 µM, a KCNQ2 inhibitor) [32]. We further investigated the voltage-sensitive sodium and potassium currents in voltage-clamp mode by elevating membrane potentials to different levels (Fig. 6F, left). After the treatment of sodium and potassium channel inhibitors (TTX [0.5 µM] and 4-AP [25 µM], respectively), both sodium and potassium currents are significantly attenuated (Fig. 6F, right).
Atoh1-Induced DA Neurons Are Sensitive to 6-OHDA Treatment
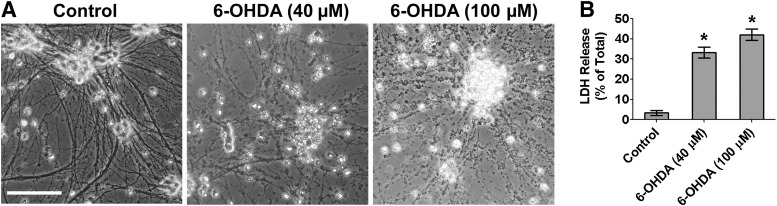
6-OHDA is a neurotoxin widely used to induce neurotoxicity both in vivo and in vitro to model DA neuron loss in PD pathogenesis [33–35]. In neuron cultures from the substantia nigra of neonatal rat brains, 6-OHDA treatment at 40 µM causes selective DA neuron loss without affecting GABA neurons [33]. Here, we tested the response of Atoh1-induced DA neurons to 6-OHDA treatment. 6-OHDA treatment (40 and 100 µM) for 15 minutes caused morphological signs of neuron death, including cell condensation and neurite fragmentation (Fig. 7A), and neuron death was also confirmed by LDH cytotoxicity assay (Fig. 7B). Thus, Atoh1-induced DA neurons derived from human iPSCs are sensitive to 6-OHDA treatment at a concentration that selectively damages primary DA neurons isolated from substantia nigra.
Figure 7.
Neurotoxicity induced by 6-OHDA in Atoh1-induced dopaminergic (DA) neurons. (A): Atoh1-induced DA neurons derived from Atoh1-iPSCs at differentiation day 36 were treated with 6-OHDA for 15 minutes. Bright-field microscope images show the morphological signs of neuron death at 24 hours after treatment. Scale bar = 100 μm. (B): Cell death was quantified by LDH cytotoxicity assay. The data represent means ± SEM (n = 3). ∗, p < .01 compared with control. Abbreviations: LDH, lactate dehydrogenase; 6-OHDA, 6-hydroxydopamine.
Discussion
Human iPSCs provide a unique cell resource for establishing patient-specific disease models and for testing potential therapies. Because of the limited resource of human neurons, lineage-specific neurons derived from human iPSCs are the most desirable cells for modeling various neurological disorders, such as midbrain DA neurons for PD, striatal GABAergic neurons for Huntington’s disease, and cholinergic motor neurons for amyotrophic lateral sclerosis [1]. It is critical to develop highly efficient protocols for neuronal conversion in PSCs, to translate current iPSC-derived neuron models from small-scale laboratory applications to large-scale personalized drug testing platforms. Proneural transcription factors are core drivers of neurogenesis, and multiple members in this family (e.g., ASCL1, Ngn2, and NeuroD1) have been used to differentiate PSCs into neurons and more recently transdifferentiate somatic cells into neurons [1, 19, 20, 22, 36, 37]. We now show that Atoh1 is a highly efficient driver for neuronal conversion in PSCs, and Atoh1 induction in combination with cell extrinsic factors rapidly differentiates human PSC to functional DA neurons at high purity.
Multiple proneural transcription factors (e.g., ASCL1, Ngn2, and NeuroD1) are activated during the neuronal conversion of human PSCs, which initiate and sustain a neurogenic transcriptional network [24]. We identified Atoh1 as a proneural transcription factor that is also upregulated during this neuronal conversion process. By using a Tet-On gene expression system to transiently induce ectopic Atoh1 expression in PSCs, we found that Atoh1 induction alone is sufficient for highly efficient neuronal conversion in PSCs. We further determined that 2 days is the minimal amount of time and that 4–5 days is ideal for transient Atoh1 induction to achieve successful neuronal conversion in PSCs. Several studies have suggested that Atoh1 and other proneural transcription factors are able to activate a neurogenic transcription factor network that over time becomes self-supporting. We also found that the neurogenic transcription factor NeuroD1 and Ngn2 were induced by ectopic Atoh1, and their expression was sustained after the silencing of exogenous Atoh1. This demonstrates that Atoh1-induced neuronal differentiation program in PSCs can become self-supporting and independent of exogenous Atoh1. It is noteworthy to mention that although exogenous Atoh1 was not detectable by Western blotting after Dox withdrawal, Atoh1 expression did not return to baseline. This result is consistent with our previous result in Figure 1B showing that endogenous Atoh1 is upregulated during the neuronal conversion of PSCs. The persistence of endogenous Atoh1 expression can be explained by the evidence that Atoh1 protein binds to its own enhancer to establish an autoregulation loop for maintaining its expression [38]. Overall, our results support the mechanism that transient Atoh1 expression in PSCs can activate a cell intrinsic program for neuronal commitment (a neuroprogramming process). This process might share similar features to somatic cell reprogramming, in which transient expression of reprogramming transcription factors induces the remodeling of epigenetic markers and drives cells into a self-sustaining pluripotent status [39]. The epigenetic dynamics during this neuroprogramming process warrants further studies, and a deep understanding of this process might lead to more potent approaches for converting both PSCs and somatic cells into neurons.
Proneural transcription factors have been shown to coordinately control the acquisition of a generic neuronal fate and the neuron subtype specification [9]. The functions of proneural transcription factors during neural development are strongly influenced by the spatial and temporal context including multiple modifiers such as transcriptional cofactors and cell extrinsic factors [11]. Atoh1 has been found to drive the differentiation of numerous neuronal populations (e.g., cerebellar granule neurons, spinal cord neurons, and inner ear hair cells), as well as diverse non-neuronal cell types (e.g., Merkel cells and intestinal secretory lineages) [13–17], suggesting that the functions of Atoh1 depend on specific developmental contexts. When ectopic Atoh1 was expressed in the context of human PSCs, we detected a high percentage of neurons expressing the DA marker TH, and multiple DA neuron markers were induced in response to ectopic Atoh1 expression. Moreover, SHH and FGF8b, two neural patterning morphogens for DA specification further promoted the expression of DA neuron markers and increased the efficiency of Atoh1-induced DA neuron conversion. These results suggest that Atoh1-induced neurons respond to extrinsic factors for generating lineage-specific neurons. A recent report shows that in embryonic bodies derived from mouse ESCs, Atoh1 induction in combination with extrinsic factors promotes the generation of cerebellar granule neurons [40]. Atoh1 induction has also been found to induce inner ear hair cell-like cells from mouse ESC-derived embryonic bodies [41]. Overall, it is possible to derive different neuron subtypes from human or mouse PSCs by controlling the temporal induction of Atoh1 in various differentiation stages of PSCs and combining Atoh1 induction with different cell intrinsic and extrinsic factors (e.g., neuron subtype-specific transcription factors or morphogens). Other proneural transcription factors also show this plasticity in specifying various neuron subtypes. For example, ASCL1 has been used to generate glutamatergic/GABAergic, DA, and cholinergic neurons [18–21].
We established a highly efficient Atoh1-mediated approach for generating lineage-specific functional neurons from human PSCs. Inducible Atoh1 transgene was delivered using a single-vector Tet-On lentivirus and is stable in PSCs after >15 passages with puromycin selection (data not shown). Atoh1-induced DA neuron cultures derived from human iPSCs and ESCs showed >80% pan-neuronal purity and >80% DA subtype purity. This protocol yields DA neurons from PSCs with a rate of return of >250% or >100% after the cryopreservation of Atoh1-induced NPCs. The cryopreservation of Atoh1-induced DA NPCs will enable us to establish patient-specific DA neuron banks. After in vitro maturation, Atoh1-induced DA neurons expressed midbrain DA neuron markers (such as GIRK2, NURR1, FOXA2, and DAT) and exhibited robust synapse formation. The functional maturation of these DA neurons was further confirmed by DA release and spontaneous spiking activity. Overall, Atoh1-induced DA neurons derived from human iPSCs recapitulate key features of primary midbrain DA neurons, making this Atoh1-mediated approach particularly applicable for PD modeling using patient-derived iPSCs. It has been reported that primary DA neurons but not GABAergic neurons, from the substantia nigra of neonatal rat brains are sensitive to 6-OHDA treatment at low concentration (40 µM) [33]. We also demonstrated that Atoh1-induced DA neurons showed similar 6-OHDA sensitivity to primary midbrain DA neurons, supporting that Atoh1-induced DA neurons can serve as a reliable neurotoxicity model for PD.
Because of the use of genome-integrating lentivirus for ectopic Atoh1 expression, our Atoh1-induced DA neurons will not be optimal for cell replacement therapy. However, we found that transient Atoh1 expression for 3–5 days is sufficient for highly efficient neuronal conversion in PSCs. Thus, nonintegrating viruses (e.g., adenovirus and Sendai virus) should be suitable to overcome the limitation because of using lentivirus. More recently, multiple virus-free systems based on mRNA or protein delivery have been established to generate transgene-free human iPSCs [42–44], and these approaches can be applied to generate transplant-ready Atoh1-induced DA neurons. It is also noteworthy that chemical compounds that induce Atoh1 expression have been identified and patented (patent publication number US20090232780A1), thus providing another potential method for Atoh1 induction in PSCs that warrants further testing.
Conclusion
Atoh1 is a potent driver for highly efficient neuronal conversion in human PSCs. Atoh1 induction in combination with cell extrinsic factors differentiates PSCs into functional DA neurons in high purity. Atoh1-induced DA neurons derived from human iPSCs recapitulate key features of primary midbrain DA neurons and provide a useful cell model for studying the pathogenesis of both familial PD and, more importantly, sporadic PD and testing potential PD therapies.
Supplementary Material
Acknowledgments
We thank the Stem Cell Core Facility in the Johns Hopkins Institute for Cell Engineering for help in pluripotent stem cell culture. We thank Dr. Craig W. Lindsley for kindly providing ML252 and Dr. Werner Graf for generous support. This work was supported by Maryland Stem Cell Research Fund (M.Y., J.L., T.M.D., and V.L.D.), American Brain Tumor Association Discovery Grant (M.Y.), NIH Grant R01NS076759 R01NS073611 (J.L.), and NIH/National Institute of Neurological Disorders and Stroke Grant P50 NS038377 (T.M.D.). We acknowledge the Adrienne Helis Malvin Medical Research Foundation and its direct engagement in the continuous active conduct of medical research in conjunction with the Johns Hopkins Hospital, the Johns Hopkins University School of Medicine, and the Foundation’s Parkinson’s Disease Programs. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases at Johns Hopkins University School of Medicine.
Author Contributions
J.S. and J.X.: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; X.Z. and S.S.K.: collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.T.: collection and/or assembly of data, final approval of manuscript; L.C.: provision of study material or patients, final approval of manuscript; V.L.D. and T.M.D.: provision of study material or patients, manuscript writing, final approval of manuscript; J.L.: conception and design, financial support, manuscript writing, final approval of manuscript; M.Y.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.Y. and J.L. were named on an uncompensated provisional patent for Atoh1-driven neuronal conversion methods submitted by the Johns Hopkins University.
References
- 1.Qiang L, Fujita R, Abeliovich A. Remodeling neurodegeneration: Somatic cell reprogramming-based models of adult neurological disorders. Neuron. 2013;78:957–969. doi: 10.1016/j.neuron.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Ito D, Okano H, Suzuki N. Accelerating progress in induced pluripotent stem cell research for neurological diseases. Ann Neurol. 2012;72:167–174. doi: 10.1002/ana.23596. [DOI] [PubMed] [Google Scholar]
- 3.Jung YW, Hysolli E, Kim KY, et al. Human induced pluripotent stem cells and neurodegenerative disease: Prospects for novel therapies. Curr Opin Neurol. 2012;25:125–130. doi: 10.1097/WCO.0b013e3283518226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawasaki H, Mizuseki K, Nishikawa S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 5.Swistowski A, Peng J, Liu Q, et al. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrier AL, Tabar V, Barberi T, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundberg M, Bogetofte H, Lawson T, et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells. 2013;31:1548–1562. doi: 10.1002/stem.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 10.Klisch TJ, Xi Y, Flora A, et al. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci USA. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akazawa C, Ishibashi M, Shimizu C, et al. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 13.Bermingham NA, Hassan BA, Price SD, et al. Math1: An essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Arie N, Bellen HJ, Armstrong DL, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 15.Miesegaes GR, Klisch TJ, Thaller C, et al. Identification and subclassification of new Atoh1 derived cell populations during mouse spinal cord development. Dev Biol. 2009;327:339–351. doi: 10.1016/j.ydbio.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Bermingham NA, Finegold MJ, et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Arie N, Hassan BA, Bermingham NA, et al. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- 18.Qiang L, Fujita R, Yamashita T, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Caiazzo M, Dell’Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 20.Pfisterer U, Kirkeby A, Torper O, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers SM, Qi Y, Mica Y, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore DJ, West AB, Dawson VL, et al. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 24.Kriks S, Shim JW, Piao J, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hargus G, Cooper O, Deleidi M, et al. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc Natl Acad Sci USA. 2010;107:15921–15926. doi: 10.1073/pnas.1010209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grace AA, Bunney BS. Nigral dopamine neurons: Intracellular recording and identification with L-dopa injection and histofluorescence. Science. 1980;210:654–656. doi: 10.1126/science.7433992. [DOI] [PubMed] [Google Scholar]
- 27.Grace AA, Bunney BS. The control of firing pattern in nigral dopamine neurons: Burst firing. J Neurosci. 1984;4:2877–2890. doi: 10.1523/JNEUROSCI.04-11-02877.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grace AA, Bunney BS. The control of firing pattern in nigral dopamine neurons: Single spike firing. J Neurosci. 1984;4:2866–2876. doi: 10.1523/JNEUROSCI.04-11-02866.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman JN, Sánchez-Padilla J, Chan CS, et al. Robust pacemaking in substantia nigra dopaminergic neurons. J Neurosci. 2009;29:11011–11019. doi: 10.1523/JNEUROSCI.2519-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roeper J. Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci. 2013;36:336–342. doi: 10.1016/j.tins.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Shi L, Bian X, Qu Z, et al. Peptide hormone ghrelin enhances neuronal excitability by inhibition of Kv7/KCNQ channels. Nat Commun. 2013;4:1435. doi: 10.1038/ncomms2439. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Xu K, Zou B, et al. Identification of a novel, small molecule inhibitor of KCNQ2 channels. In: Probe Reports From the NIH Molecular Libraries Program. Bethesda, MD: National Center for Biotechnology Information, 2010. [Google Scholar]
- 33.Ding YM, Jaumotte JD, Signore AP, et al. Effects of 6-hydroxydopamine on primary cultures of substantia nigra: Specific damage to dopamine neurons and the impact of glial cell line-derived neurotrophic factor. J Neurochem. 2004;89:776–787. doi: 10.1111/j.1471-4159.2004.02415.x. [DOI] [PubMed] [Google Scholar]
- 34.Deumens R, Blokland A, Prickaerts J. Modeling Parkinson’s disease in rats: An evaluation of 6-OHDA lesions of the nigrostriatal pathway. Exp Neurol. 2002;175:303–317. doi: 10.1006/exnr.2002.7891. [DOI] [PubMed] [Google Scholar]
- 35.Bové J, Prou D, Perier C, et al. Toxin-induced models of Parkinson’s disease. NeuroRx. 2005;2:484–494. doi: 10.1602/neurorx.2.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu ML, Zang T, Zou Y, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thoma EC, Wischmeyer E, Offen N, et al. Ectopic expression of neurogenin 2 alone is sufficient to induce differentiation of embryonic stem cells into mature neurons. PLoS One. 2012;7:e38651. doi: 10.1371/journal.pone.0038651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helms AW, Abney AL, Ben-Arie N, et al. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- 39.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava R, Kumar M, Peineau S, et al. Conditional induction of Math1 specifies embryonic stem cells to cerebellar granule neuron lineage and promotes differentiation into mature granule neurons. Stem Cells. 2013;31:652–665. doi: 10.1002/stem.1295. [DOI] [PubMed] [Google Scholar]
- 41.Ouji Y, Ishizaka S, Nakamura-Uchiyama F, et al. Induction of inner ear hair cell-like cells from Math1-transfected mouse ES cells. Cell Death Dis. 2013;4:e700. doi: 10.1038/cddis.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee YH, Ko JY, Chang MY, et al. Protein-based human iPS cells efficiently generate functional dopamine neurons and can treat a rat model of Parkinson disease. J Clin Invest. 2011;121:2326–2335. doi: 10.1172/JCI45794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.