Signaling networks mediated by microRNAs and epithelial-mesenchymal transition (EMT)-inducing transcription factors connect the EMT program with the core stem cell regulatory machineries. These signaling networks are also regulated by extrinsic niche signals that induce and maintain cancer stem cells (CSCs), contributing to metastatic colonization and promoting the reactivation of dormant tumor cells. Targeting these CSC pathways is likely to improve the efficacy of conventional chemo- and radiotherapies.
Keywords: Breast cancer stem cells, Epithelial-mesenchymal transition, Metastasis, Tumor dormancy
Abstract
Accumulating evidence has shown that cancer stem cells (CSCs), the cancer cells that have long-term proliferative potential and the ability to regenerate tumors with phenotypically heterogeneous cell types, are important mediators of tumor metastasis and cancer relapse. In breast cancer, these cells often possess attributes of cells that have undergone an epithelial-mesenchymal transition (EMT). Signaling networks mediated by microRNAs and EMT-inducing transcription factors connect the EMT program with the core stem cell regulatory machineries. These signaling networks are also regulated by extrinsic niche signals that induce and maintain CSCs, contributing to metastatic colonization and promoting the reactivation of dormant tumor cells. Targeting these CSC pathways is likely to improve the efficacy of conventional chemo- and radiotherapies.
Introduction
It has long been recognized that tumors form organ-like structures composed of malignant cancer cells and stromal cells [1]. Various stromal cell types are recruited to tumors to establish complex tumor microenvironments that promote tumor growth and spread [2]. In addition, tumor cells themselves are also highly heterogeneous, in that not all tumor cells have identical abilities to propagate tumor growth, seed metastases, and resist cancer therapies [3–6]. This intratumor heterogeneity has been explained by two nonmutually exclusive models. The first is the clonal evolution model, which postulates that during the course of tumor evolution, subclones of cancer cells randomly acquire different somatic mutations or epigenetic changes that confer distinct molecular characteristics and biological properties to the cells. The second is the hierarchical model (or cancer stem cell model), which proposes that cancer cells are organized in a cellular hierarchy similar to that in normal tissues. Residing at the top of the hierarchy are highly tumorigenic tumor cells called cancer stem cells (CSCs). These CSCs have both the ability to self-renew to generate additional CSCs and the ability to produce differentiated progeny cells that are poorly tumorigenic or nontumorigenic, that is, differentiated cancer cells. In many tumors, differentiated cancer cells comprise the bulk of tumor mass, whereas CSCs are a minor cell population [3–6].
Although the concept of the CSC model is based on parallels between the hierarchical organizations of normal tissues and tumors, this model does not stipulate that tumors have to originate from normal tissue-specific stem cells. In fact, evidence has shown that progenitors, or even mature cell types, can give rise to malignancies [7–10]. In addition, CSCs may not have the same characteristics as normal stem cells, such as multilineage differentiation potentials. The term “cancer stem cell” is used to operationally define two central characteristics of these cells: (a) the ability to self-renew and (b) the ability to regenerate phenotypically heterogeneous tumors. Here, I will discuss recent findings regarding the characteristics of breast cancer stem cells (BCSCs), the intrinsic and extrinsic signals regulating BCSCs, and the role of BCSCs in tumor metastasis and therapeutic resistance.
Mammary Epithelial Lineages and Origin of CSCs
Understanding the normal cell precursors of CSCs and how these cells are transformed into CSCs is of great importance for elucidating the mechanisms regulating CSC survival, self-renewal, and differentiation. During mammary gland development, multipotent mammary stem cells (MaSCs) produce luminal- or basal-specific progenitor cells, which in turn differentiate into mature luminal or basal cells [11, 12]. Although these multipotent MaSCs are required for the long-term maintenance of mammary gland homeostasis, recent studies show that in postnatal glands, luminal and basal unipotent progenitor cells can independently sustain luminal and basal lineages, respectively, for an extended period of time [13–15]. This finding suggests that multiple mammary cell types have long-term self-renewal abilities and that BCSCs may originate from various precursors. Indeed, comparisons of the global gene expression profiles of tumor samples and specific mammary cell types suggest that different breast cancer subtypes may originate from distinct mammary cell types [8, 10]. For example, basal-like breast cancer is likely to originate from luminal progenitor cells, whereas multipotent MaSCs are likely the precursor of the claudin-low subtype [7, 10]. However, the gene expression profile of tumor bulk may not precisely reflect those of the cell of origin. Therefore, future studies using lineage-tracing strategies or introducing oncogenic mutations into specific cell types are needed to determine the exact cellular precursors of CSCs.
Identifying BCSCs: Markers and Functional Assays
The development of stem cell markers has been instrumental for studying normal and cancer stem cells. Clarke and colleagues [16] identified the first set of markers that can serve to highly enrich human BCSCs; they found that as few as 100 CD44+/CD24−/low cancer cells were able to form tumors when transplanted into immunodeficient mice, whereas tens of thousands of other cancer cells failed to do so. Non-cell-surface markers have also been identified for BCSCs. These markers include elevated aldehyde dehydrogenase (ALDH) activity, which allows cells to be labeled by an ALDH-activated fluorescence probe [17], and the tendency to proliferate more slowly than the rest of the cancer cells, which allows CSCs to retain a PKH26 label [18]. In addition, certain CSCs can be identified as the so-called side population because of their ability to exclude Hoechst dye [19]. The combination of some of these markers, such as CD44+/CD24−/low and ALDHhigh, has been shown to further enrich BCSCs when compared with individual markers [17].
Genetically engineered mouse models (GEMMs) provide highly tractable in vivo systems for studying tumor biology. Distinct CSC markers have been found for various breast cancer GEMMs as well. In a p53-null tumor model, CSCs are enriched in the Lin−CD29highCD24high population [20], whereas in the MMTV-Wnt1 model, CSCs are CD61high or Thy1+CD24+ [21, 22]. Interestingly, these markers do not enrich CSCs from MMTV-ErbB2 tumors [21]. This raises the possibility that CSCs from different subtypes of breast cancer may express distinct markers. Thus, understanding the subtype-specific properties of CSCs is important for developing therapeutic strategies to target these cells.
Because current CSC markers are not directly connected to the intrinsic properties of CSCs and their expression can fluctuate widely in different tumors, CSC markers must be combined with functional assays to convincingly identify CSCs. In addition to measuring CSC activity with in vivo tumor transplantation assays, BCSCs are also identified by sphere assays. Both normal MaSCs and BCSCs can grow in anchorage-independent suspension culture as spheroid structures (so called mammospheres or tumor spheres) [23]. Although the ability to form spheres in vitro is correlated with in vivo tumorigenicity in some cancers [24], discordance between sphere-forming and tumorigenic abilities have also been reported in certain tumors [25]. Therefore, careful evaluation of the tumor-initiating ability of sphere-forming cells is needed for individual tumor models. It is important to distinguish the spheroid structures formed by clonal expansion versus cell aggregation. This may be achieved by seeding cells at low density and adding methylcellulose to prevent aggregation [26]. Interestingly, supplementing Matrigel (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com) in sphere cultures facilitates the specific expansion of bona fide MaSCs [27, 28]. This new approach may also improve the reliability of sphere cultures for expanding BCSCs.
Intracellular Regulatory Networks of BCSCs
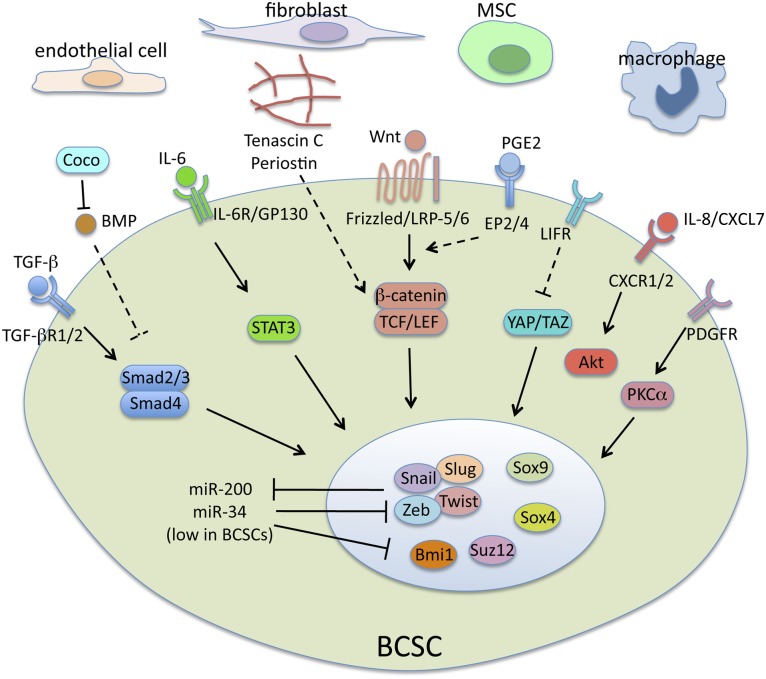
Multiple signaling pathways have been found to play a role in CSCs (Fig. 1). Interestingly, some of these pathways are associated with a developmental program called the epithelial-mesenchymal transition (EMT) and regulate the stem cell properties of both normal and cancer stem cells in the breast.
Figure 1.
Regulation of BCSCs. Stromal cells in the tumor microenvironment secrete various signaling molecules and deposit extracellular matrices. These molecules activate multiple stem cell-related signaling pathways, which lead to induction/activation of key stem cell nuclear factors, such as epithelial-mesenchymal transition-inducing transcription factors, Sox factors, and chromatin modifiers. Abbreviations: BMP, bone morphogenetic protein; BCSC, breast cancer stem cell; EP2/4, prostaglandin E receptors 2 and 4; IL, interleukin; LIFR, leukemia inhibitory factor receptor; MSC, mesenchymal stem cell; PDGFR, platelet-derived growth factor receptor; PGE2, prostaglandin E2; PKCα, protein kinase Cα; TGF-β, transforming growth factor β.
EMT and Acquisition of Normal and Cancer Stem Cell Properties
EMT was initially identified as a developmental program that enables polarized epithelial cells to acquire a motile mesenchymal phenotype. This allows stationary epithelial cells to gain the ability to migrate and invade during embryonic morphogenesis. In addition, EMT and its reverse process, the mesenchymal-epithelial transition, are integral steps of cell fate specification during gastrulation and organogenesis [29]. In carcinoma progression, reactivation of the EMT program promotes tumor metastasis by driving tumor cell invasion and enhancing tumor cell survival during the metastatic cascade [30].
In addition to its effect on promoting tumor cell dissemination, recent studies have shown that EMT confers stem-like properties on differentiated cells [26, 31]. The activation of EMT by multiple means, including transforming growth factor β (TGF-β) treatment and the expression of various EMT-inducing transcription factors, generates cells that express BCSC markers, such as CD44highCD24low, and form mammospheres [26, 31]. These cells also have greatly increased tumor-initiating ability, one of the defining characteristics of CSCs [26]. Studies in murine mammary glands have revealed that normal MaSCs similarly express mesenchymal traits [26, 27]. In particular, they highly express a key EMT transcription factor, Slug (also called Snail2), and Slug overexpression greatly increases stem cell activity, as demonstrated by an in vivo gland reconstitution assay [27]. These studies suggest a mechanistic link between normal and cancer stem cells.
In fact, Slug is a key target of several tumor and metastasis suppressors. For example, the loss of the tumor suppressor BRCA1 is linked to the formation of aggressive basal-like breast cancer and promotes mammary stem/progenitor cell expansion [10, 32]. Interestingly, BRCA1 has been shown to decrease Slug protein stability [33], whereas Slug, conversely, represses BRCA1 expression by recruiting a histone demethylase to its promoter [34]. These molecules form a reciprocal negative-feedback loop that maintains cells in either a stem or a differentiated cellular state. Such reciprocal negative feedback loops appear to be a common strategy in maintaining cells in metastable EMT and stem cell states, as further described below. Other suppressors of tumor progression and metastasis, such as Elf5, Simgleminded-2s, and miRNA-34, also repress Slug expression [35–37].
Cellular changes induced by EMT have direct impacts on stem cell signaling pathways. A cardinal feature of EMT is the disruption of adherens junctions and epithelial polarity mediated by E-cadherin and other cell-cell adhesion molecules. In addition to freeing cells from being restrained in the epithelial layer, this has a significant impact on several key signaling pathways. First, the loss of E-cadherin releases β-catenin from plasma membrane sequestration and facilitates its nuclear translocation to activate canonical Wnt signaling [38], which is a central pathway in normal and cancer stem cells [39, 40]. Second, the disruption of epithelial polarity leads to activation of the Hippo pathway effectors YAP/TAZ, which are involved in the regulation of organ size and tissue homeostasis [41]. In breast cancer, the activation of YAP/TAZ increases the in vitro mammosphere-forming ability, in vivo tumorigenicity, and metastatic capacity of cancer cells [42, 43].
MicroRNAs and BCSCs
MicroRNAs have emerged as a class of key regulators of CSCs and EMT [44]. Among them, the miR-200 family plays a particularly interesting role in integrating the EMT program and core stem cell pathways [45]. The miR-200 family includes five microRNAs sharing homologous seed sequences: miR-200a, miR-200b, miR-200c, miR-141, and miR-429. These microRNAs form a reciprocal negative feedback loop with EMT-inducing transcription factors ZEB1 and ZEB2. miR-200 binds to multiple recognition sites in the 3′-untranslated regions of ZEB1 and ZEB2 mRNAs, thereby suppressing their expression [46–51]. Conversely, ZEB1 and ZEB2 inhibit the expression of miR-200s by binding to their promoters [52]. Therefore, in epithelial cells, high levels of miR-200s suppress the accumulation of EMT transcription factors and maintain the cells in the epithelial state. Upon activation of the EMT program, the upregulation of ZEB1 and ZEB2 represses miR200 expression and relieves ZEB inhibition, thereby driving the cells into a stable mesenchymal state.
Consistent with their mesenchymal attributes, MaSCs and BCSCs show downregulated miR-200 levels compared with their differentiated progeny [53]. The overexpression of miR-200 suppresses the clonogenicity of BCSCs and the ability of MaSCs to regenerate mammary ductal trees [53]. Conversely, the inhibition of miR-200 increases the number of CSCs in breast and pancreatic cancers [52, 54]. Mechanistically, miR-200 targets many stem cell-associated genes, including members of polycomb-repressive complexes, such as Bmi1 and Suz12 [52–54], both of which are key players in CSCs [54, 55], and pluripotent factors such as Sox2 and Klf4 [52]. Interestingly, the upregulation of Suz12 by miR-200 inhibition leads to the repression of the E-cadherin gene [54]. Additionally, miR-200 members inhibit components of the Notch pathway, a key stem cell signaling pathway [56]. Thus, the miR-200 family integrates EMT-inducing transcription factors and core stem cell pathways to form a regulatory circuitry that sustains the stem cell and mesenchymal states.
CSC-inhibiting microRNAs are also important mediators of tumor suppressor functions. Loss of p53 decreases the miR-200c level and induces EMT and EMT-associated stem cell properties [57]. In fact, p53 induces the expression of multiple microRNAs that suppress EMT. For example, the activation of p53 downregulates Snail and other EMT-inducing transcription factors through the upregulation of the miR-34 family [37, 58]. Interestingly, Snail and ZEB1, conversely, repress miR-34 expression, forming yet another reciprocal negative feedback loop for regulating EMT. As such, loss of p53 tilts the balance of these feedback loops toward the accumulation of EMT transcription factors, hence promoting EMT.
Sox Family Transcription Factors
The Sox family includes 20 different transcription factors in mammals that share homologous high-mobility-group DNA-binding domains. Sox proteins play prominent roles in cell fate regulation during development, including the specification of embryonic and somatic stem and progenitor cells [59]. Several Sox family members have been found to play a role in the mammary gland and breast cancer.
In the normal mouse mammary gland, the coexpression of Sox9 and Slug can confer gland-reconstituting stem cell properties on mature luminal cells; conversely, the inhibition of Sox9 or Slug blocks stem cell activity [27]. Consistent with its role in stem cell self-renewal, Sox9 promotes the tumorigenicity and metastatic colonization of breast cancer cells. Interestingly, instead of inducing EMT, Sox9 activates a distinct program that acts synergistically with the EMT program to induce stem cells [27]. This suggests that the mammary stem cell state is controlled cooperatively by EMT and additional signaling pathways; thus, the mere acquisition of a mesenchymal state is not sufficient to endow full stem-cell potential on differentiated cells.
The pluripotency factor Sox2 is also frequently expressed in breast cancer, along with the activation of embryonic stem cell-like gene expression signatures; furthermore, the overexpression of Sox2 increases tumor sphere-forming efficiency [60, 61]. Sox4 is another Sox factor that is involved in breast cancer progression. Distinct from Sox9 and Sox2, Sox4 plays a direct role in activating EMT [62, 63]. It is required for TGF-β-induced EMT in breast cancer cells and is important for tumor growth and metastasis. Mechanistically, Sox4 upregulates Ezh2 expression, which introduces histone methylation on key EMT genes [62]. Ezh2 also promotes BCSC expansion through the activation of Raf1-β-catenin signaling [64].
Cancer Stem Cell Niche Signals
The stem cell niche, a specified tissue microenvironment in which stem cells reside, is critical for stem cell self-renewal, survival, and function [65]. Similar to normal stem cells, CSCs also rely on specific tumor microenvironments that provide paracrine and juxtacrine signals for maintaining CSC properties.
Developmental Signaling Pathways
Multiple developmental signaling pathways have been implicated in regulating BCSCs, including TGF-β, Wnt, and Notch. TGF-β is a potent EMT inducer that is secreted by multiple cell types in tumors [66]. Treating certain nonmalignant mammary epithelial cells or breast cancer cells with TGF-β efficiently activates EMT programs that are accompanied by the expression of BCSC markers, such as CD44highCD24low, and the increased ability to form mammospheres [26, 67, 68]. However, in normal human mammary epithelial cells, efficient activation of EMT requires the cooperation of the TGF-β and Wnt signaling pathways [67]. Interestingly, such cooperation is reminiscent of an early developmental program in which the TGF-β and Wnt interaction is critical for the induction of the Spemann organizer, in which EMT initially occurs during gastrulation [69]. The role of Wnt signaling in stem cells has been well documented [39, 40]. In adult mammary glands, MaSCs exhibit elevated Wnt signaling [15], and the overexpression of Wnt proteins or activation of canonical Wnt by Axin2 mutation or MMP3 overexpression promotes the expansion of MaSCs [70–72].
In contrast to Wnt, Notch induces the commitment of MaSCs to luminal-specific progenitors [73, 74]. Interestingly, certain aggressive breast cancers, including basal-like breast cancer, are likely to originate from luminal progenitor cells [7, 10]. Therefore, Notch may be particularly important for these breast cancer subtypes [75]. Interestingly, recent studies have also shown breast cancer subtype-specific effects for the TGF-β and Wnt pathways. Although TGF-β increases CSC numbers in claudinlow cancer cell lines, it suppresses CSCs in certain basal-like and luminal cell lines [68]. Similarly, Wnt-overexpressing fibroblasts promoted the growth of one patient-derived xenograft (PDX) model but inhibited another PDX [76]. Thus, future work is required to determine the tumor subtypes and molecular pathways that influence the response to these stem/progenitor cell pathways to properly target them for cancer therapy.
CSC-Associated Cytokine Networks
Stromal cells play a prominent role in tumor growth and progression and often interact with tumor cells through paracrine feedback loops [2]. For example, CSF-1 produced by carcinoma cells recruits macrophages and these macrophages then secrete epidermal growth factor, which promotes cancer cell invasion and intravasation [77, 78]. Cancer cells also stimulate carcinoma-associated fibroblasts to become myofibroblasts, which then stimulate tumor growth by activating the SDF1/CXCR4 pathway in cancer cells [79]. Furthermore, cancer cells recruit mesenchymal stem cells and stimulate their cytokine production, thereby promoting tumor growth and metastasis [80–82].
Tumor recruitment of stromal cells is facilitated by chronic inflammation mediated by inflammatory cytokines, such as interleukin 6 (IL-6), IL-8, and IL-1β [83, 84]. Interestingly, some of these cytokines also directly promote BCSC self-renewal and survival [83]. The activation of STAT3 by IL-6 through the IL-6 receptor/GP130 complex has been shown to induce BCSC expansion [85]. IL-6 also stimulates the recruitment of mesenchymal stem cells, which produce CXCL7 to increase the number of BCSCs in the tumor [82]. In addition, BCSCs express high levels of IL-8 receptor CXCR1, which prevents BCSC apoptosis [86]. Recent studies have also found RANK ligand (RANKL) to be an important stem cell-stimulating cytokine in the breast [87, 88]. The activation of the RANKL-RANK pathway induces EMT and increases the population of CD44highCD24low CSCs [89]. In mouse tumor models, RANKL accelerates tumor onset and promotes metastasis [90, 91]. In addition to cytokines, bioactive lipids such as prostaglandin E2 (PGE2) are also important mediators of inflammation. Multiple stromal cell types can produce PGE2, including carcinoma-associated fibroblasts and mesenchymal stem cells [81, 92]. PGE2 increases the number of ALDHhigh CSCs through the activation of Wnt/β-catenin signaling [81].
CSC Niche in Regulating Metastatic Colonization and Tumor Dormancy
Metastatic colonization, the outgrowth of disseminated cancer cells into overt metastases, is a major rate-limiting step of the metastatic cascade [93, 94]. Successful colonization requires the survival and self-renewal of tumor-initiating cells at metastatic sites. Clinical observations and mouse model studies suggest that disseminated cancer cells often remain dormant at metastatic sites, even if they survive the foreign microenvironment, and only some of them may reactivate to produce metastasis [95]. Recent studies have shown that establishing CSC niches at distant sites is crucial for the survival of CSCs and is required for the activation of their self-renewal ability for metastatic colonization [96].
Multiple cell-cell and cell-matrix interactions have been shown to be important for the survival and maintenance of CSCs at distant sites. The expression of VCAM-1 on cancer cells allows them to interact with macrophages and monocytic osteoclast progenitors via integrin α4β1. This interaction activates PI3K/Akt-mediated survival signals in cancer cells and promotes their osteolytic expansion [97, 98]. In addition, aggressive breast cancer cells contribute to their own CSC niche by secreting tenascin C at metastatic sites; tenascin C enhances Wnt and Notch signaling and promotes the survival and outgrowth of micrometastases [99]. Cancer cells also co-opt stromal cells at metastatic sites to establish a procolonization microenvironment [100]. For example, they stimulate the expression of periostin by stromal fibroblasts, which is required for maintaining disseminated CSCs by boosting Wnt signaling [101].
Unlike the active stroma in primary tumors, the distant tissue where disseminated tumor cells (DTCs) arrive tends to have a more quiescent microenvironment and these quiescent signals may force DTCs into dormancy. For example, abundant bone morphogenetic protein (BMP) ligands in the lung parenchyma inhibit CSC self-renewal, thereby causing metastatic dormancy. Expression of a BMP antagonist, Coco, promotes tumor-initiation ability and allows DTCs to reactivate and colonize [102]. Dormant DTCs were also found to reside in microvasculatures, sites where quiescent endothelial cell-derived thrombospondin-1 induces tumor dormancy. Upon the induction of neoangiogenesis, the sprouting vasculatures produce active TGF-β1 and periostin, two important CSC niche signals, to promote metastasis outgrowth [103]. Interestingly, BMPs have been shown to antagonize TGF-β signaling in part by competing for co-Smad [104]. However, although TGF-β signaling reactivates breast cancer cells from dormancy, TGF-β2 appears to induce dormancy in head and neck squamous cell carcinoma [105]. Future studies are needed to understand the contextual differences that contribute to these seemingly paradoxical effects.
Toward Cancer Stem Cell-Targeted Therapies
The most important clinical implication of the CSC model is that any effective cancer therapy has to be able to target CSCs. Indeed, evidence has shown that the current chemo- and radiotherapies fail to eliminate CSCs and instead mainly deplete the differentiated tumor cells comprising the tumor bulk. In culture, CSCs with mesenchymal attributes are more resistant to genotoxic agents than epithelial non-CSCs [106]. Similar findings were also observed by comparing tumor samples before and after neoadjuvant chemotherapy or endocrine therapy in patients. The tumor cells that survived these conventional therapies showed gene expression signatures similar to that of CD44highCD24−/low cells and mammospheres. These cells also express high levels of mesenchymal markers that are associated with BCSCs [107]. Furthermore, mammary stem/progenitor and BCSCs have a higher tolerance to radiation, which is mediated in part by Wnt signaling [108]. The radioresistance of CSCs is likely also mediated by their ability to reduce reactive oxygen species (ROSs), because ROSs are critical mediators of ionizing radiation-induced cell killing [109].
The resistance of CSCs to conventional therapies requires us to identify CSC-specific vulnerabilities. High-throughput screens in an experimentally engineered CSC model have identified a chemical compound, salinomycin, that selectively kills BCSCs [106]. Interestingly, salinomycin inhibits Wnt signaling [110], which is an important stem cell pathway in BCSCs, as discussed above. BCSCs with mesenchymal attributes also rely on protein kinase Cα (PKCα)-mediated signaling pathways, and inhibiting PKCα specifically targets CSCs [111]. Consistent with the role of TGF-β signaling in increasing CSCs [26, 67], TGF-β blockade prevents triple-negative breast cancer xenograft relapse after chemotherapy [112]. Furthermore, the radioresistance of CSCs can also be overcome by hyperthermia treatment mediated by optically activated gold nanoshells in vivo [113]. Understanding the mechanism by which hyperthermia sensitizes CSCs to radiation will allow for expansion of the usage of this new technology.
Conclusion
The studies of the past 10 years have led to a significant understanding of the biological properties and molecular pathways of breast CSCs. These efforts have also begun to shed light on potential CSC-targeted strategies that can be combined with the current standard of care for effective cancer treatment. However, significant and important knowledge gaps remain. In particular, current CSC studies mainly rely on tumor transplantation assays for determining CSC activity. These models do not completely recapitulate the spontaneous tumors that originate and progress in native tissue microenvironments. Indeed, the exact behaviors and properties of CSCs in these spontaneous tumors are still poorly understood. Approaches for labeling and manipulating CSCs in native tumor microenvironments, such as lineage tracing or genetic cell ablation, will help to yield novel insights into CSC biology. In addition, because CSCs share many regulatory machineries with normal SCs, we must develop strategies that selectively inhibit CSCs while sparing their normal counterparts.
Acknowledgments
Research in the Guo laboratory is supported by Susan G. Komen for the Cure (CCR12224440), New York State Stem Cell Science (NYSTEM, C028109), and the Gottesman Institute for Stem Cell and Regenerative Medicine Research at Albert Einstein College of Medicine. I thank John Christin, Zheng Zhang, Almudena Bosch, and Chunhui Wang for their insightful comments on the manuscript.
Author Contributions
W.G.: manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The author indicates no potential conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 4.Lobo NA, Shimono Y, Qian D, et al. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 5.Shackleton M, Quintana E, Fearon ER, et al. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Visvader JE, Lindeman GJ. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Molyneux G, Geyer FC, Magnay FA, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 9.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 10.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 11.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visvader JE, Smith GH. Murine mammary epithelial stem cells: Discovery, function, and current status. Cold Spring Harb Perspect Biol. 2011;3:a004879. doi: 10.1101/cshperspect.a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rios AC, Fu NY, Lindeman GJ, et al. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 14.Van Keymeulen A, Rocha AS, Ousset M, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 15.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cicalese A, Bonizzi G, Pasi CE, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 19.Patrawala L, Calhoun T, Schneider-Broussard R, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 20.Zhang M, Behbod F, Atkinson RL, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaillant F, Asselin-Labat ML, Shackleton M, et al. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- 22.Cho RW, Wang X, Diehn M, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 23.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett LE, Granot Z, Coker C, et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 26.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Keckesova Z, Donaher JL, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spike BT, Engle DD, Lin JC, et al. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morel AP, Lièvre M, Thomas C, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proia TA, Keller PJ, Gupta PB, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu ZQ, Li XY, Hu CY, et al. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci USA. 2012;109:16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarti R, Hwang J, Andres Blanco M, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laffin B, Wellberg E, Kwak HI, et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siemens H, Jackstadt R, Hünten S, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 38.Onder TT, Gupta PB, Mani SA, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 39.Alexander CM, Goel S, Fakhraldeen SA, et al. Wnt signaling in mammary glands: Plastic cell fates and combinatorial signaling. Cold Spring Harb Perspect Biol. 2012;4:a008037. doi: 10.1101/cshperspect.a008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Yu FX, Guan KL. The Hippo pathway: Regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordenonsi M, Zanconato F, Azzolin L, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 43.Chen D, Sun Y, Wei Y, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamouille S, Subramanyam D, Blelloch R, et al. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop: A motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korpal M, Lee ES, Hu G, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 49.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurteau GJ, Carlson JA, Spivack SD, et al. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 51.Christoffersen NR, Silahtaroglu A, Orom UA, et al. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–1178. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 53.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iliopoulos D, Lindahl-Allen M, Polytarchou C, et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 56.Brabletz S, Bajdak K, Meidhof S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang CJ, Chao CH, Xia W, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim NH, Kim HS, Li XY, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarkar A, Hochedlinger K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leis O, Eguiara A, Lopez-Arribillaga E, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 62.Tiwari N, Tiwari VK, Waldmeier L, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Liang Q, Lei Y, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 64.Chang CJ, Yang JY, Xia W, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sneddon JB, Werb Z. Location, location, location: The cancer stem cell niche. Cell Stem Cell. 2007;1:607–611. doi: 10.1016/j.stem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheel C, Eaton EN, Li SH, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruna A, Greenwood W, Le Quesne J, et al. TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun. 2012;3:1055. doi: 10.1038/ncomms2039. [DOI] [PubMed] [Google Scholar]
- 69.Nishita M, Hashimoto MK, Ogata S, et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann’s organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 70.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 72.Kessenbrock K, Dijkgraaf GJ, Lawson DA, et al. A role for matrix metalloproteinases in regulating mammary stem cell function via the Wnt signaling pathway. Cell Stem Cell. 2013;13:300–313. doi: 10.1016/j.stem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Buono KD, Robinson GW, Martin C, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 75.Harrison H, Farnie G, Brennan KR, et al. Breast cancer stem cells: Something out of notching? Cancer Res. 2010;70:8973–8976. doi: 10.1158/0008-5472.CAN-10-1559. [DOI] [PubMed] [Google Scholar]
- 76.Green JL, La J, Yum KW, et al. Paracrine Wnt signaling both promotes and inhibits human breast tumor growth. Proc Natl Acad Sci USA. 2013;110:6991–6996. doi: 10.1073/pnas.1303671110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goswami S, Sahai E, Wyckoff JB, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 78.Patsialou A, Wyckoff J, Wang Y, et al. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–9506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 80.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 81.Li HJ, Reinhardt F, Herschman HR, et al. Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2012;2:840–855. doi: 10.1158/2159-8290.CD-12-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu S, Ginestier C, Ou SJ, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ginestier C, Liu S, Diebel ME, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120:485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Asselin-Labat ML, Vaillant F, Sheridan JM, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 88.Joshi PA, Jackson HW, Beristain AG, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 89.Palafox M, Ferrer I, Pellegrini P, et al. RANK induces epithelial-mesenchymal transition and stemness in human mammary epithelial cells and promotes tumorigenesis and metastasis. Cancer Res. 2012;72:2879–2888. doi: 10.1158/0008-5472.CAN-12-0044. [DOI] [PubMed] [Google Scholar]
- 90.Cao Y, Bonizzi G, Seagroves TN, et al. IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell. 2001;107:763–775. doi: 10.1016/s0092-8674(01)00599-2. [DOI] [PubMed] [Google Scholar]
- 91.Gonzalez-Suarez E, Jacob AP, Jones J, et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature. 2010;468:103–107. doi: 10.1038/nature09495. [DOI] [PubMed] [Google Scholar]
- 92.Rudnick JA, Arendt LM, Klebba I, et al. Functional heterogeneity of breast fibroblasts is defined by a prostaglandin secretory phenotype that promotes expansion of cancer-stem like cells. PLoS One. 2011;6:e24605. doi: 10.1371/journal.pone.0024605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 94.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu X, Mu E, Wei Y, et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging α4β1-positive osteoclast progenitors. Cancer Cell. 2011;20:701–714. doi: 10.1016/j.ccr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen Q, Zhang XH, Massagué J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011;20:538–549. doi: 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oskarsson T, Acharyya S, Zhang XH, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaplan RN, Riba RD, Zacharoulis S, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malanchi I, Santamaria-Martínez A, Susanto E, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 102.Gao H, Chakraborty G, Lee-Lim AP, et al. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ghajar CM, Peinado H, Mori H, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeisberg M, Hanai J, Sugimoto H, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 105.Bragado P, Estrada Y, Parikh F, et al. TGF-β2 dictates disseminated tumour cell fate in target organs through TGF-β-RIII and p38α/β signalling. Nat Cell Biol. 2013;15:1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci USA. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu D, Choi MY, Yu J, et al. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc Natl Acad Sci USA. 2011;108:13253–13257. doi: 10.1073/pnas.1110431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tam WL, Lu H, Buikhuisen J, et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhola NE, Balko JM, Dugger TC, et al. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest. 2013;123:1348–1358. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Atkinson RL, Zhang M, Diagaradjane P, et al. Thermal enhancement with optically activated gold nanoshells sensitizes breast cancer stem cells to radiation therapy. Sci Transl Med. 2010;2:55ra79. doi: 10.1126/scitranslmed.3001447. [DOI] [PMC free article] [PubMed] [Google Scholar]