This review highlights the role of microRNAs (miRNAs) in tissue regeneration, discusses key challenges in translating this knowledge to the clinic, and outlines recent technological advances that aim to address these issues. By combining a comprehensive knowledge of miRNA biology with cutting-edge delivery technologies, it is clear that miRNAs hold significant promise for tissue regenerative therapies in the future.
Keywords: Tissue regeneration, MicroRNAs, Stem cells, Delivery systems
Abstract
In recent years, the role of miRNAs in post-transcriptional gene regulation has come to the fore with strong evidence to indicate an important role for microRNAs (miRNAs) in the regulation of a wide range of fundamental biological processes. Notably, this includes the regulation of both endogenous tissue repair mechanisms and the growth and differentiation of stem cells (both adult and pluripotent). As a result, manipulation of miRNA signaling holds great promise for regenerative medicine, which aims to harness either endogenous or implanted cells to promote tissue repair. However, to fully realize this potential, it will be necessary to combine advances in our biological understanding with new technologies that allow precise spatiotemporal modulation of specific miRNA candidates. In this review, we highlight the role of miRNAs in tissue regeneration, discuss key challenges in translating this knowledge to the clinic, and outline recent technological advances that aim to address these issues. By combining a comprehensive knowledge of miRNA biology with cutting-edge delivery technologies, it is clear that miRNAs hold significant promise for tissue regenerative therapies in the future.
Introduction
MicroRNAs (miRNAs) are a class of small noncoding RNAs that play an important role in post-transcriptional gene regulation. Initially synthesized as longer precursors (pri-miRNA), miRNAs are processed through a series of stages to mature, cytoplasmic miRNA duplexes of ∼22 nucleotides in length. Their regulatory activity is conferred upon loading into the miRNA-induced silencing complex (miRISC) in which one strand of the RNA duplex guides the miRISC to its target mRNA. Gene expression is downregulated by inhibiting translation, targeting the mRNA for degradation, or a combination of both [1].
miRNAs usually act by binding to recognition sites in the 3′-untranslated region (3′-UTR) of target mRNAs via complementary base pairing. Such pairing does not have to be perfect across the whole miRNA sequence but instead differs between miRNA-mRNA pairs with the primary requirement being the matching of a 6–8-nucleotide seed sequence at the 5′ end of the miRNA [2]. The prevalence of miRNAs (miRBase [v20] contains more than 2,500 mature human miRNAs [3]), coupled with their known ability to regulate many target genes and the strong evolutionary conservation of miRNA recognition sites, suggests that miRNA regulation influences a large proportion of the gene regulatory network and thereby a wide range of fundamental biological processes.
This review focuses on the potential of miRNAs in regenerative medicine. It will discuss the important role of miRNAs in endogenous tissue repair and stem cell-mediated regeneration, address some of the strategies being developed to translate miRNA technology to the clinic, and finally highlight novel applications of miRNAs that may present future possibilities for tissue regeneration.
miRNAs in Tissue Regeneration
Endogenous Tissue Regeneration
The cellular mechanisms that underlie organ regeneration are diverse, with cellular differentiation, dedifferentiation, transdifferentiation, and proliferation all contributing to regenerative phenomena [4]. These complex alterations in cellular plasticity involve context-dependent regulation of gene expression networks to reconstruct tissues in a spatiotemporally controlled manner. Although the reactivation of developmental signaling pathways appears to be an important evolutionarily conserved mechanism for tissue regeneration [5], the molecular processes that trigger and coordinate these signaling networks are largely unknown. To this end, several recent studies in model systems spanning invertebrates, lower vertebrates, and mammals have revealed a previously unappreciated role for miRNAs in regenerative processes. Given their capacity to modulate genes involved in common biological signaling pathways [6], miRNAs are emerging as potential candidates for therapeutic modulation of endogenous tissue regeneration.
Among invertebrates, hydra and planarians are the champions of organ regeneration and can regrow an entire organism from amputated body fragments. This involves the activation of pluripotent stem cells known as neoblasts, and deep-sequencing studies have revealed that scores of miRNAs are differentially expressed in normal and regenerating planarian tissues, including neoblasts [7, 8]. However, the functions of individual miRNAs during tissue regeneration in hydra and planarians remain largely unknown.
Certain lower vertebrates such as teleost fish and urodele amphibians can also mount a robust regenerative response following traumatic tissue injury, including appendage amputation. Studies in adult zebrafish suggest that miRNAs functionally regulate regeneration of complex tissues including the retina, tail fin, spinal cord, and heart [9–12]. For example, depletion of miRNA (miR)-133 is an important upstream signal for induction of regeneration following tail fin amputation or cardiac resection in the zebrafish. Inhibition of miR-133 using morpholinos in this model accelerated blastemal cell proliferation, an effect mediated, at least in part, through repression of the monopolar spindle 1 (mps1) kinase [13]. Similarly, gain- and loss-of-function studies in the zebrafish have established an important role for miR-133 in the regulation of adult heart regeneration in which overexpression of miR-133 inhibited cardiomyocyte proliferation and abolished the regenerative response to cardiac injury, whereas inhibition of miR-133 augmented the cardiomyocyte proliferative response [11]. These studies have established an important role for miR-133 in the regulation of proliferative signaling during vertebrate organ regeneration.
In mammals, only a limited number of tissues harbor a robust regenerative capacity throughout life, including the liver and skeletal muscle. A number of miRNAs have been implicated in the regulation of endogenous tissue regeneration in mammals following damage to the liver or skeletal muscle including miR-206, miR-128, and miR-26 [14–16].
The Heart: An Example of miRNAs in Endogenous Regeneration
An area of intensive research in regenerative medicine is the heart, which has a much more restricted endogenous regenerative potential than liver or skeletal muscle. Interestingly, in contrast to the adult heart, the neonatal mouse heart can regenerate following myocardial infarction at postnatal day 1, but this regenerative potential is lost within the first 2 weeks after birth [17, 18]. Postnatal cardiac regenerative arrest coincides with alterations in the expression of several miRNAs including the developmental induction of a large miRNA family known as the miR-15 family [18, 19]. Gain- and loss-of-function studies have established an important role for the miR-15 family in the regulation of postnatal cardiomyocyte mitotic arrest and cardiac regenerative capacity. Overexpression of miR-195, a member of the miR-15 family, impairs the cardiac regenerative response of neonatal mice to myocardial infarction and is associated with a marked reduction in the incidence of cardiomyocyte proliferation. Conversely, postnatal inhibition of the miR-15 family prolongs cardiomyocyte proliferative capacity and extends the developmental window for cardiac regeneration following ischemic injury [18, 19]. These effects appear to be mediated through miR-15-dependent repression of a number of cell cycle-associated genes including the checkpoint kinase 1 (Chek1) [18, 19]. Recent studies have also identified a number of other miRNAs that govern cardiomyocyte proliferation, including the miR-17–92 cluster [20] as well as miR-199a and miR-590 [21]. In addition, miRNAs have been implicated in almost every aspect of cardiac regenerative biology including cardiac lineage commitment and differentiation of stem cells, as well as direct reprogramming of cardiac fibroblasts into cardiomyocytes (reviewed in [22]).
Collectively, these studies suggest that miRNAs form critical components of proliferative cell signaling networks and could potentially be harnessed for activation of endogenous cardiac regenerative mechanisms in adult tissues. Furthermore, the evidence implicating miRNAs in the regeneration of other organs also suggests that a similar approach could be applied to numerous body tissues.
miRNAs in Stem Cell-Mediated Regeneration
In addition to enhancing endogenous tissue regeneration, modulation of miRNAs for stem cell-based tissue genesis also holds significant promise. A growing body of evidence is revealing critical roles for miRNAs in the regulation of stem cell maintenance and differentiation, indicating that modulation of miRNA signaling in endogenous stem cell populations, or in vitro as part of a tissue-engineering strategy, may provide a useful tool with which to direct tissue regeneration (Table 1).
Table 1.
miRNAs involved in the regulation of pluripotent and mesenchymal stem cell properties
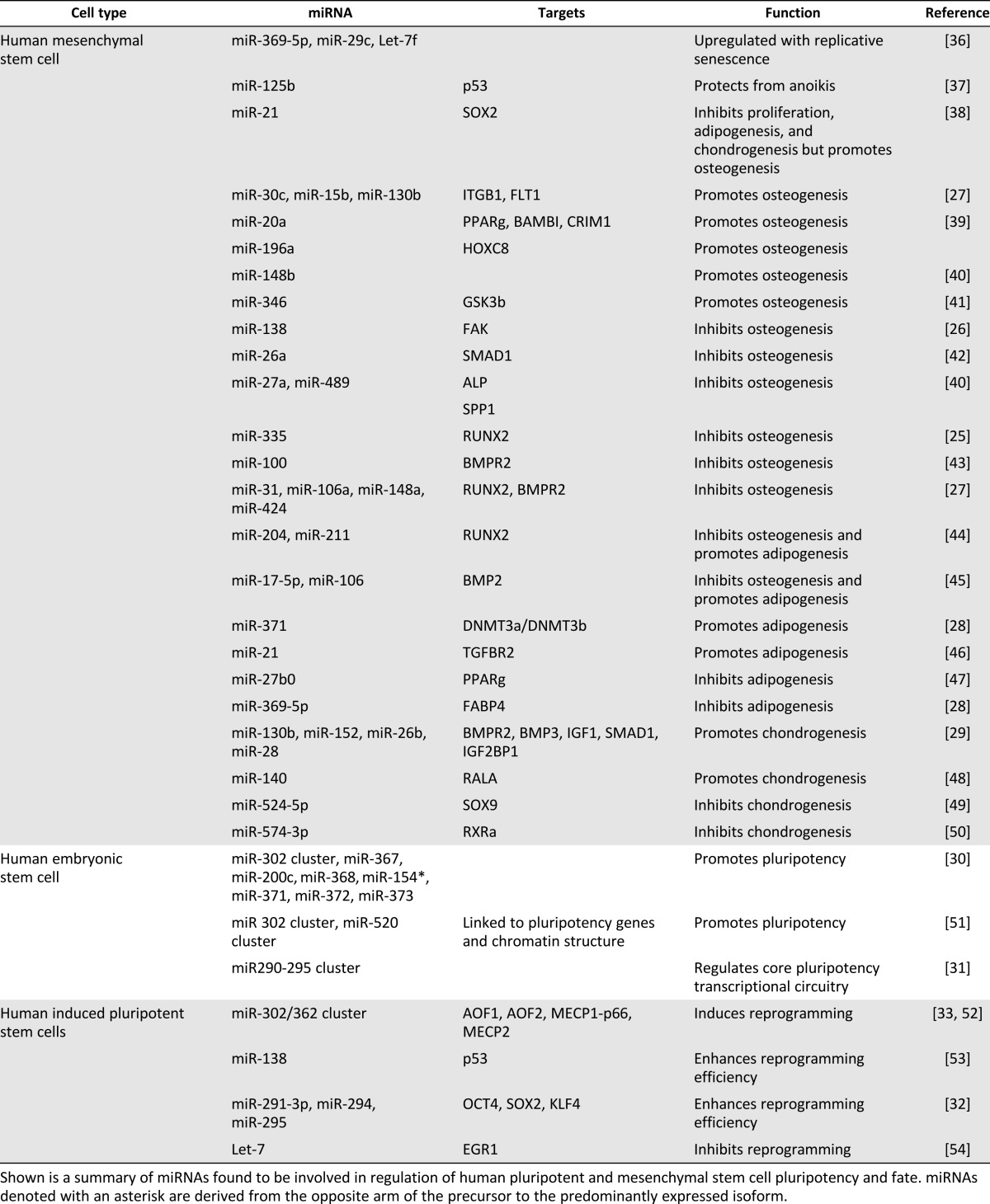
The importance of miRNA signaling has already been established for many adult stem cell types [23–25], although much remains to be understood. The potential for modulators of miRNA signaling to be used to direct stem cell-based regeneration is exemplified by studies using mesenchymal stem cells (MSCs), in which specific miRNAs are differentially expressed when MSCs switch from a proliferative to differentiating state [25], or during maturation along the osteogenic [26, 27], adipogenic [28], and chondrogenic [29] lineages. To highlight the utility of such candidates in modulating tissue formation as part of a stem cell-based therapy, Eskildsen et al. [26] showed not only that miR-138 inhibition enhanced the expression of osteogenic genes and overexpression inhibited them, but also that when MSCs were implanted into NOD/SCID mice, inhibition of miR-138 enhanced ectopic bone formation, whereas overexpression of premiR-138 inhibited bone formation.
Future utility of miRNAs in stem cell-mediated regeneration also relates to the role of miRNAs in pluripotent stem cells (embryonic and induced pluripotent stem cells). Human embryonic stem cells express a characteristic panel of miRNAs [30], many of which are linked to the core pluripotency network [31]. Advancing on work showing that miRNAs can be used to enhance reprogramming efficiency [32], the miR-302/362 cluster is also capable of initiating reprogramming in its own right [33]. As with adult stem cells, miRNAs also play an important role in cell commitment, with roles already established for miRNA candidates in the regulation of numerous lineages (see recent reviews [34, 35]). Together, these studies indicate enormous future potential for the modulation of miRNA signaling to direct stem cell-based tissue regeneration.
Strategies for miRNA Modulation
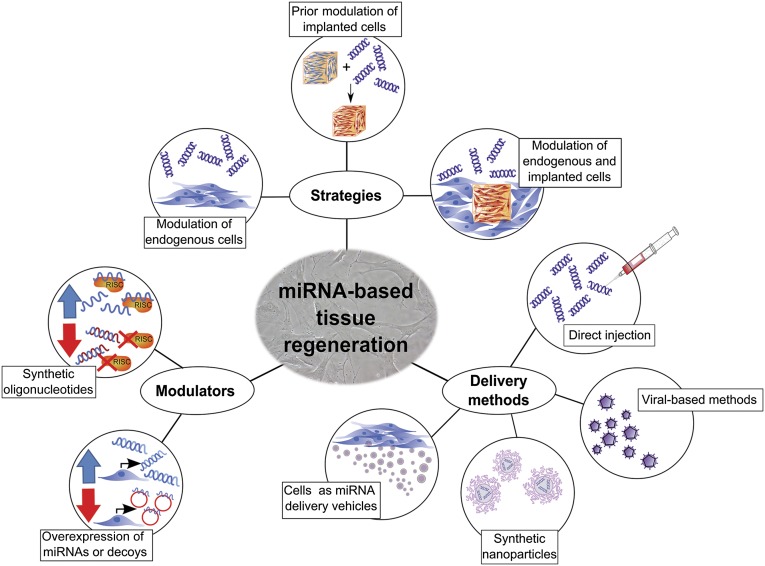
Modulation of miRNAs has clear potential for tissue regeneration, both in studies that advance our understanding of regenerative mechanisms and in those for clinical application. Opportunities for novel treatments include directing tissue formation by altering endogenous cellular activity, directing the behavior of (stem) cells incorporated into tissue-engineered constructs, or by manipulating the activities of a combination of both endogenous and implanted cells (Fig. 1).
Figure 1.
MicroRNA-based tissue regeneration. Strategies using miRNAs for tissue regeneration therapies include altering endogenous cellular activity, directing the behavior of (stem) cells incorporated into tissue-engineered constructs, or targeting both implanted and endogenous cells. Modulation of miRNAs can be achieved by the delivery of miRNA mimics or anti-miRs and by overexpressing miRNAs or miRNA sponges in the cell type of interest. Methods for delivery of such modulators include simple injection, viral overexpression, delivery via synthetic nanoparticles, or delivery from cells via extracellular vesicles. Abbreviations: miRNA, microRNA; RISC, miRNA-induced silencing complex.
Synthetic Oligonucleotides
Short oligonucleotides are a popular option for miRNA modulation; levels of a specific miRNA can be increased by delivering synthetic miRNA mimics or knocked down by the action of short complementary oligonucleotides (anti-miRs or antago-miRs) that anneal to the guide strand of the mature miRNA, blocking miRISC activity [36]. The stability and efficiency of these synthetic molecules can be improved by modifications including functionalization of ribose units in the passenger stand with moieties such as 2′-O-methyl, 2′-O-methoxyoxyethyl or 2′-fluoro groups [37]. Alternatively locked nucleic acids (LNAs) incorporate the formation of a 2′,4′-methylene bridge in the ribose unit to produce a bicyclic nucleotide [38].
Further, regarding the wide use of such synthetic oligonucleotides to study cell behavior in vitro, these methods have shown some success for tissue regeneration in vivo. For example, the inclusion of short double-stranded miR mimics of miR-1, miR-133, and miR-206 with atelocollagen injections was shown to enhance the generation of muscle defects in rats [39]. However, the longevity of these molecules is limited, and so overexpression strategies are of interest for longer-term modulation of miRNA expression levels.
Overexpression Strategies
miRNAs can also be upregulated by overexpression at the level of the pri-miRNA, pre-miRNA, or fully mature miRNA sequence. Overexpression of either pre-miRNAs or pre-miRNA with some flanking sequence may be more biologically relevant than simply using the mature miRNA sequence, because the transcript interacts with the endogenous cellular machinery ensuring correct processing and final function [40]. Overexpression strategies can also be used to knock down miRNA signaling. For example, Ebert et al. [41] developed DNA-based sponge transcripts, containing multiple miRNA-binding sites in the 3′-UTR and a bulge at the position usually cleaved by Ago2 (to force a stable interaction between the miRNA and sponge sequence). These sponge transcripts were shown to inhibit the activity of all miRNAs with a specific seed sequence and thus can be used to functionally inhibit entire miRNA families. The further development of tough decoy RNAs, expressed from lentiviral vectors, used a similar principle and successfully inhibited miRNAs for more than 1 month [42]. This concept of a sponge transcript has also recently been found to exist in nature, with the discovery of naturally occurring circular RNAs [43].
Key Challenges in the Modulation of miRNA Signaling for Tissue Regeneration
Candidate Selection and Kinetics
A key advantage of miRNAs as therapeutic agents is that one miRNA can modulate numerous genes [44], thus providing the potential to influence targets at several levels in a regulatory network and provide an extremely robust biological output. However, this promiscuity of miRNAs also makes selection of therapeutic candidates particularly challenging to ensure miRNA modulation results in the expected biological response. To achieve this, a thorough understanding of exquisitely complex regulatory networks, as aided by bioinformatic and systems biology approaches, will be essential.
In silico algorithms are commonly used to predict miRNA-mRNA interactions (e.g., Targetscan [45], PicTar [46], and miRANDA [47]) and when combined with gene ontology analysis (e.g., DAVID [48] or Ingenuity) are often used to infer a functional response. Target prediction algorithms are constantly evolving to improve their accuracy (each provides varying outputs and are known to produce false positive results with a relatively high occurrence), but with a deeper knowledge of miRNA-mRNA interactions, the complexity of this task is becoming more evident. For example, it is now clear that miRNAs do not only bind to partner mRNAs via the canonical seed sequence (nucleotides 2–8), but that sites in the center of the miRNA can also facilitate miRNA-mRNA interactions [49].
To aid in narrowing down the specific effects, high-throughput methods, such as Ago HITS-CLIP (Argonaute high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation) now allow the actual miRNA-mRNA interactions in any particular biological context to be probed. With these advances, the ability to determine direct miRNA targets and subsequently evaluate the effect of a miRNA on specific biological pathways will undoubtedly aid in the identification of appropriate candidates for therapeutic intervention. miRNA therapies must also account for the fact that tissue regeneration is a multistage process and provide the required kinetics of modulation. On a whole tissue level, this could mean coordinating the behavior of multiple cell types, whereas regenerative strategies using stem cells will need to consider the differential requirements of the developmental program through which a stem cell will progress. As an example, Zhang et al. [50] identified miRNAs that were upregulated at different stages of osteogenesis, suggesting specific functions at different stages of differentiation. Consequently, the best outcomes will occur when the temporal provision of miRNA modulators is matched to these biological requirements.
Targeting and Dosage
Given knowledge of the correct miRNA candidate and timing, precise targeting will be necessary to provide control over the dose of miRNA modulator that reaches the target cell population; if too little of a modulator reaches its intended target, the efficiency of modulation may be insufficient to achieve the desired clinical outcome. Conversely, Mukherji et al. [51] showed that miRNAs establish a threshold mRNA level around which small changes in mRNA translate into much larger changes in protein synthesis. Therefore, simply swamping a system with a specific miRNA or anti-miR may not be an effective strategy. This effect is likely to be context-dependent because the characteristics of behavior around the threshold were also shown to be dependent upon the affinity of the miRNA to its target mRNA.
In mammalian cells, miRNA incorporation into the miRISC is random, meaning that the relative amount of a miRNA species determines the levels of the active miRNA [52]. Thus any change in the level of a particular miRNA may also affect the activity of others by competing for incorporation into the miRISC. This phenomenon was first noted when using siRNAs to target two genes at once, which resulted in only partial knockdown of both genes [53]. However, it has subsequently been shown that increasing siRNA or miRNA levels can also alter the activity of endogenous miRNAs [54].
In addition to controlling the level of miRNA in the intended target population, precise targeting will also be essential to ensure that modulators are delivered to the desired cell type at the right time. This is essential, because miRNAs can have different activities in different cell types. For example, members of the miR-125 family have a tumor-suppressing function in ovarian, bladder, and breast cancer but act as oncogenes in endometrial, pancreatic, and prostate cancer [55], further highlighting the requirement for specificity of delivery. Dysregulation of miRNAs has been linked to cancer [56], and so the potential for disruption of miRNA signaling networks to cause cancer is a very real risk if miRNA modulation is not targeted precisely to the cell or tissue of interest. This concern is particularly relevant to the field of regenerative medicine, in which the very nature of stem cells (especially pluripotent stem cells) is to be able to replicate indefinitely and develop into multiple tissue types. There is inevitably overlap between the key genes controlling both pluripotency and differentiation and those integral to cancer initiation and development [57]. Therefore the spatiotemporal modulation of key miRNAs must be well controlled in strategies that use stem cell populations for tissue regeneration to reduce the possibility of cancer as a potential side effect. Overall, in the development of strategies for miRNA-based tissue regeneration, it is clear that the challenges of intelligent target selection coupled with precision in the dosage and spatiotemporal modulation of miRNA targets must be addressed.
Technologies to Modulate miRNAs for Tissue Regeneration
Direct Injection
A range of existing methods to deliver miRNA exists including direct injection and viral and non-viral-based methodologies. The simplest method for delivery of miRNAs and inhibitors is by direct injection. While this is the simplest method, longer-term stability may be a problem because miRNAs will be rapidly cleared by the kidneys (because of their inability to bind to plasma proteins) and degraded by nucleases in serum. Despite this, some miRNA inhibitors have proven to be quite stable in the circulation following direct injection via various routes including intravenous and subcutaneous administration. For example, nonhuman primates given intravenous injections of LNA-anti-miR in phosphate-buffered saline (against miR-122) showed efficient silencing of miR-122 in liver and effects on plasma cholesterol [58]. A single bolus injection of LNA-anti-miR has also been shown to be active for several weeks [59].
Viral-Based Methods
Viral-based methods are the most prevalent for in vitro assessment of miRNA efficacy and are similar to candidates currently used for transducing any genetic material to cells [60]. Different viral vectors exist, each with their own advantages and disadvantages, but the most commonly used are adeno or adeno-associated viral vectors, as well as lentiviral and retroviral vectors. Viral vectors have the advantage of enabling long-term expression (or control of expression with inducible promoters); however, translation to the clinic is challenging because of their inherent toxicity and immunogenicity (reviewed by Liu and Berkhout [61]). Adeno-associated virus (AAV) currently offers the most promise for in vivo delivery of miRNAs, because of its observed lower levels of toxicity and lack of off-target effects. Different serotypes allow broad tissue targeting, which can then be combined with tissue-specific promoters. They inherently have limited cloning capacity, although for miRNAs, this is not an issue [62].
Although the current safest option for viral-based delivery, AAV, still results in both cell-specific and humoral immunity, there have been reports of recombination with WT virus, and large-scale viral production is laborious (and hence expensive). Their ultimate use within a regenerative medicine framework will depend on the tissue being targeted and the ability to engender the AAVs with ligand-based specificity at a reasonable cost.
In addition, targeting is also important at the subcellular level; the major site of miRNA activity is within the cytosol, making it important for mimics or inhibitors not to be sequestered to other intracellular compartments. Primarily, this concerns the escape of exogenous miRNAs/modulators from the endosome because cellular uptake is mediated by endocytotic pathways that lead, via the early and late endosome, to the lysosome, within which RNAs are degraded (reviewed in [63]). Therefore methods for miRNA/anti-miR delivery should incorporate mechanisms for endosomal escape, something that is beyond the capability of AAVs. Synthetic delivery systems can, however, be tailored to address this issue.
Synthetic Methods
Non-viral-based or “synthetic” methods for genetic material delivery have seen significant attention over the last decade, and the approaches are numerous. These delivery vehicles have recently been reviewed by Zhang et al. [62], and hence here only a brief summary will be provided, and their applicability within a regenerative medicine framework discussed. Current synthetic delivery systems being used with miRNA can be categorized into (a) lipid-based; (b) polyethyleimine (PEI)-based; (c) dendrimer-based; (d) poly(α-hydroxyacid) polymers (as particles or scaffolds); (e) naturally occurring biopolymers as particles (e.g., chitosan and protamine [64], atelocollagen, and protein translocation domain-derived peptides) or scaffolds; and (f) inorganic nanoparticles (gold, silica-based, or magnetic) and scaffolds [65].
Many of these methods rely on the self-assembly of synthetic materials (whether they be modified lipids (liposomes), prestructured polymers (dendrimers), or functionalized polymers with associative blocks or end groups (pluronics/tetronics)). It is this self-assembled nature that offers substantial advantages over viral-based methods, including controlled and tailored molecular composition, targeted ligation with desired cell surface antigens, tolerance of large (multiple plasmids) cargo sizes, controlled disassembly and release of payloads, simplified manufacturing, modification, scale up, ease of analysis and quality control, and low (tunable) immunogenicity [66].
In vitro, these synthetic systems have shown similar efficacy to viral-based methods, especially in the case of lipid-based and PEI-based systems, and their ability to display ligands for targeted ligation has been a major driver for their development and continual refinement. Use of synthetic systems in vivo are increasing, but challenges remain in terms of efficacy via intravenous versus local injection, sufficient delivery of miRNA to the site of injury without degradation or nonspecific uptake, targeted uptake by the appropriate cell type within complex tissues, and, thereafter, appropriate levels of overexpression or inhibition for appropriate time frames to invoke repair and regeneration.
Future Perspectives
Modulating miRNA Signaling to Overcome Limitations of the Cellular Microenvironment
In addition to the general concepts outlined above, the innovative use of miRs and technologies for modulation and delivery has the potential to bring about completely new approaches to tissue regeneration. For example, a major challenge in the field of regenerative medicine regards how to ensure the specificity of the regenerative response. This relates to the complexity of the cellular microenvironment where, in addition to soluble cues, cells (particularly stem cells) are sensitive to physical interactions (e.g., substrate stiffness, substrate creep and ECM composition and presentation [67–70]). When implanting cells into a damaged tissue, this has implications because the cells are introduced to a suboptimal microenvironment. Because of the sensitivity of cells to their surrounding microenvironment, these changes can influence the fate of the cells that are introduced as a cellular therapy and impede the regenerative response. Such effects have been noted with cells implanted into diseased heart [71] and bladder [72] tissue.
A similar theme also applies to tissue engineered therapies; although strategies aim to mimic the native tissue, it may not always be possible to create biomaterials with all of the desired properties. For example, there is significant interest in the use of hydrogel/stem cell composites for bone repair. However, the stiffness of hydrogels does not approach the high modulus of bone tissue [73] and may therefore limit the osteogenic potential of the cells encapsulated within them.
Situations such as these provide a new opportunity for miRNA therapies to modulate stem cell behavior and overcome some of the limitations of the extracellular environment. There is growing evidence that miRNAs have a significant role to play in regulating the response of cells to different extracellular stimuli [74]. With further knowledge of these events, it may be possible to override unwanted mechanical signals and promote tissue regeneration.
Cells as miRNA Delivery Vehicles
A further interesting avenue regards the use of cells as a delivery vehicle for regenerative miRNAs. Alternatively, this approach could be described as using miRNAs to enhance the paracrine effects of implanted cells and guide tissue regeneration. The concept of paracrine effects mediating the regenerative effect of cells used for cellular therapies is already well established. For example, it is widely accepted that many of the beneficial activities of MSCs are mediated by the secretion of proregenerative and anti-inflammatory growth factors and cytokines, for example in cardiac repair [71] or graft versus host disease [75]. Furthermore, it has been shown that miRNAs can be shuttled between cells packaged in small membrane vesicles (e.g., exosomes, shedding vesicles, apoptotic bodies), collectively referred to as extracellular vesicles (EVs) and that these can exert changes in gene expression of their target cells [76]. It is therefore a natural extension of this progression to suggest that cells could be used to deliver miRNAs to promote tissue regeneration. Alternatively, these EVs could be isolated and used to deliver a “pro-regenerative” miRNA payload. There has been some interest in the use of EVs for miRNA delivery. Guduric-Fuchs et al. [77] overexpressed miR-146a in HEK293T cells and showed that both the transfected cells and EVs released were enriched in this miRNA.
This approach using cells as delivery vehicles is related to previous work that has already used genetically engineered MSCs to deliver proteins and modulate an in vivo tissue response [78]. An advantage, particularly when using MSCs for this strategy, is that they naturally home to sites of disease or injury and so may provide a natural targeting mechanism to deliver miRNAs to the required site [79]. Although they did not directly look for evidence of EVs, Lee et al. [80] showed it is possible to use MSCs transfected with mimics of miR-124 and miR-145 to deliver them to glioma cells (both in vitro and in vivo) and that there were corresponding changes in gene expression and cellular behavior.
Conclusion
As our understanding of miRNA signaling improves, it is clear that there is significant potential to harness miRNAs to direct tissue regeneration, either via endogenous repair mechanisms or by directing the activity of implanted cells in cell therapies. With advances in our understanding, new opportunities to manipulate miRNAs for regenerative purposes also arise. The promise of miRNA-based regeneration is vast, because the right candidate holds the promise to target multiple levels of relevant cellular signaling pathway. Furthermore, the relative ease with which miRNAs can be pharmacologically targeted and their small size, compared with proteins, makes them attractive candidates for clinical use. However, many challenges are posed by the complexities of miRNA signaling networks and the precise spatiotemporal resolution that is required for therapies to be fully effective. Advances in the delivery of miRNA modulators are beginning to address some of these issues and will be critical for the successful translation of miRNA-based tissue regeneration strategies to the clinic. With parallel advances in both miRNA biology and delivery technologies, miRNAs hold significant promise for tissue regenerative therapies in the future.
Acknowledgments
We are grateful to Ernst Wolvetang for insightful discussions. J.E.F. is supported by an Australian Research Council Discovery Early Career Researcher Award Fellowship (DE13010098). E.R.P. is supported by grants and fellowships from the National Health and Medical Research Council and the National Heart Foundation of Australia (APP1033815 and 635530).
Author Contributions
J.E.F.: conception and design, manuscript writing, final approval of manuscript; E.R.P. and J.J.C.-W.: manuscript writing.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 5.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusev Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA. Methods. 2008;44:61–72. doi: 10.1016/j.ymeth.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Sasidharan V, Lu YC, Bansal D, et al. Identification of neoblast- and regeneration-specific miRNAs in the planarian Schmidtea mediterranea. RNA. 2013;19:1394–1404. doi: 10.1261/rna.038653.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu YC, Smielewska M, Palakodeti D, et al. Deep sequencing identifies new and regulated microRNAs in Schmidtea mediterranea. RNA. 2009;15:1483–1491. [Google Scholar]
- 9.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thatcher EJ, Paydar I, Anderson KK, et al. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci USA. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin VP, Lepilina A, Smith A, et al. Regulation of zebrafish heart regeneration by miR-133. Dev Biol. 2012;365:319–327. doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu YM, Gibbs KM, Davila J, et al. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur J Neurosci. 2011;33:1587–1597. doi: 10.1111/j.1460-9568.2011.07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin VP, Thomson JM, Thummel R, et al. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu N, Williams AH, Maxeiner JM, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122:2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motohashi N, Alexander MS, Shimizu-Motohashi Y, et al. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci. 2013;126:2678–2691. doi: 10.1242/jcs.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Ju W, Wang D, et al. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS One. 2012;7:e33577. doi: 10.1371/journal.pone.0033577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porrello ER, Mahmoud AI, Simpson E, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci USA. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porrello ER, Johnson BA, Aurora AB, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Huang ZP, Seok HY, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 22.Hudson JE, Porrello ER. The non-coding road towards cardiac regeneration. J Cardiovasc Transl Res. 2013;6:909–923. doi: 10.1007/s12265-013-9486-8. [DOI] [PubMed] [Google Scholar]
- 23.Cui Y, Xiao Z, Chen T, et al. The miR-7 identified from collagen biomaterial based 3-D cultured cells regulates neural stem cell differentiation. Stem Cells Dev. 2014;23:393–405. doi: 10.1089/scd.2013.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Li X, He Q, et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013;23:1356–1368. doi: 10.1038/cr.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomé M, López-Romero P, Albo C, et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18:985–995. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eskildsen T, Taipaleenmäki H, Stenvang J, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Yang T, Han J, et al. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem. 2011;112:1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- 28.Bork S, Horn P, Castoldi M, et al. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol. 2011;226:2226–2234. doi: 10.1002/jcp.22557. [DOI] [PubMed] [Google Scholar]
- 29.Han J, Yang T, Gao J, et al. Specific microRNA expression during chondrogenesis of human mesenchymal stem cells. Int J Mol Med. 2010;25:377–384. doi: 10.3892/ijmm_00000355. [DOI] [PubMed] [Google Scholar]
- 30.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judson RL, Babiarz JE, Venere M, et al. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüningschrör P, Hauser S, Kaltschmidt B, et al. MicroRNAs in pluripotency, reprogramming and cell fate induction. Biochim Biophys Acta. 2013;1833:1894–1903. doi: 10.1016/j.bbamcr.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Choi E, Choi E, Hwang KC. MicroRNAs as novel regulators of stem cell fate. World J Stem Cells. 2013;5:172–187. doi: 10.4252/wjsc.v5.i4.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ameres SL, Horwich MD, Hung JH, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 38.Grünweller A, Hartmann RK. Locked nucleic acid oligonucleotides: The next generation of antisense agents? BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 39.Nakasa T, Ishikawa M, Shi M, et al. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park JE, Heo I, Tian Y, et al. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 44.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 45.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 46.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 47.John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Martin HC, Wani S, Steptoe AL, et al. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 2014;15:R51. doi: 10.1186/gb-2014-15-3-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci USA. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherji S, Ebert MS, Zheng GX, et al. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Zhang Z, O’Loughlin E, et al. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012;26:693–704. doi: 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McManus MT, Haines BB, Dillon CP, et al. Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol. 2002;169:5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- 54.Khan AA, Betel D, Miller ML, et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palanichamy JK, Rao DS. miRNA dysregulation in cancer: Towards a mechanistic understanding. Front Genet. 2014;5:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller FJ, Laurent LC, Kostka D, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 59.Hullinger TG, Montgomery RL, Seto AG, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 61.Liu YP, Berkhout B. miRNA cassettes in viral vectors: Problems and solutions. Biochim Biophys Acta. 2011;1809:732–745. doi: 10.1016/j.bbagrm.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Juliano RL, Ming X, Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suh JS, Lee JY, Choi YS, et al. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials. 2013;34:4347–4359. doi: 10.1016/j.biomaterials.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 65.Wu K, Song W, Zhao L, et al. MicroRNA functionalized microporous titanium oxide surface by lyophilization with enhanced osteogenic activity. ACS Appl Mater Interfaces. 2013;5:2733–2744. doi: 10.1021/am400374c. [DOI] [PubMed] [Google Scholar]
- 66.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cameron AR, Frith JE, Cooper-White JJ. The influence of substrate creep on mesenchymal stem cell behaviour and phenotype. Biomaterials. 2011;32:5979–5993. doi: 10.1016/j.biomaterials.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 68.Engler AJ, Sen S, Sweeney HL, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 69.Frith JE, Mills RJ, Cooper-White JJ. Lateral spacing of adhesion peptides influences human mesenchymal stem cell behaviour. J Cell Sci. 2012;125:317–327. doi: 10.1242/jcs.087916. [DOI] [PubMed] [Google Scholar]
- 70.Salasznyk RM, Klees RF, Hughlock MK, et al. ERK signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells on collagen I and vitronectin. Cell Commun Adhes. 2004;11:137–153. doi: 10.1080/15419060500242836. [DOI] [PubMed] [Google Scholar]
- 71.Leskelä HV, Satta J, Oiva J, et al. Calcification and cellularity in human aortic heart valve tissue determine the differentiation of bone-marrow-derived cells. J Mol Cell Cardiol. 2006;41:642–649. doi: 10.1016/j.yjmcc.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 72.Tolg C, Ahsan A, Dworski S, et al. Pathologic bladder microenvironment attenuates smooth muscle differentiation of skin derived precursor cells: Implications for tissue regeneration. PLoS One. 2013;8:e59413. doi: 10.1371/journal.pone.0059413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang LS, Du C, Chung JE, et al. Enzymatically cross-linked gelatin-phenol hydrogels with a broader stiffness range for osteogenic differentiation of human mesenchymal stem cells. Acta Biomater. 2012;8:1826–1837. doi: 10.1016/j.actbio.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 74.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoogduijn MJ, Popp F, Verbeek R, et al. The immunomodulatory properties of mesenchymal stem cells and their use for immunotherapy. Int Immunopharmacol. 2010;10:1496–1500. doi: 10.1016/j.intimp.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 77.Guduric-Fuchs J, O’Connor A, Camp B, et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haider HK, Jiang S, Idris NM, et al. IGF-1-overexpressing mesenchymal stem cells accelerate bone marrow stem cell mobilization via paracrine activation of SDF-1alpha/CXCR4 signaling to promote myocardial repair. Circ Res. 2008;103:1300–1308. doi: 10.1161/CIRCRESAHA.108.186742. [DOI] [PubMed] [Google Scholar]
- 79.Chapel A, Bertho JM, Bensidhoum M, et al. Mesenchymal stem cells home to injured tissues when co-infused with hematopoietic cells to treat a radiation-induced multi-organ failure syndrome. J Gene Med. 2003;5:1028–1038. doi: 10.1002/jgm.452. [DOI] [PubMed] [Google Scholar]
- 80.Lee HK, Finniss S, Cazacu S, et al. Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget. 2013;4:346–361. doi: 10.18632/oncotarget.868. [DOI] [PMC free article] [PubMed] [Google Scholar]