Abstract
Background & Aims
Quantitative microarray analyses have shown increased expression of IL-15 mRNA in the esophagus of patients with eosinophilic esophagitis (EoE), a recently recognized allergic disorder with poorly understood pathogenesis.
Methods
Quantitative PCR and ELISA analyses were performed to examine protein and transcript levels in tissue samples from patients with EoE. Tissues from IL-15Ra-deficient and wild-type (control) mice were also examined. Tissue eosinophilia was determined by immunostaining for major basic protein (MBP) and flow cytometry for cell-surface receptors.
Results
Quantitative PCR analyses demonstrated that levels of IL-15 and its receptor IL-15Ra were increased ~6- and ~10-fold, respectively, in tissues from patients with EoE and ~3- and ~4-fold, respectively, in mice with allergen-induced EoE. A >2-fold increase in serum IL-15 protein was also detected in human EoE samples compared with those from healthy individuals. Human IL-15 mRNA levels correlated with esophageal eosinophilia (p<0.001). IL-15Ra-deficient mice were protected from allergen-induced esophageal eosinophilia compared with controls (p<0.001), even though similar levels of airway eosinophilia were observed in all mice. IL-15 activated STAT5 and CD4+ T cells to produce cytokines that act on eosinophils. Incubation of primary esophageal epithelial cells from mice and humans with IL-15 caused a dose-dependent increase in the mRNA expression and protein levels of eotaxins-1, -2 and -3.
Conclusion
IL-15 mediates in the pathogenesis of EoE. IL-15 activates CD4+ T cells to produce cytokines that act on eosinophils.
Keywords: Eosinophils, Esophagus, Interleukin (IL)-15, IL-15Ra, T cells, EoE Patients
Introduction
Eosinophilic esophagitis (EoE, formally referred as EE) is a clinicopathological disease associated with symptoms similar to those described in patients with gastroesophageal reflux disease (GERD) and an abundant accumulation of eosinophils in the esophageal epithelium.1–5 EoE is differentiated from GERD by the magnitude of intraepithelial eosinophils and the lack of response to acid suppressive therapy.1, 2 Animal modeling has established that Th2 cytokine signaling is required for induction of experimental EoE in mice.6, 7 Considerable evidence supports a critical role for the Th2 cytokines IL-5 and IL-13 in EoE pathogenesis.8, 9 The highest increases in gene expression in the esophagus of EoE patients is for eotaxin-3, an eosinophil chemoattractant and activator produced by esophageal epithelial cells.10 We recently reported increased expression of IL-15 mRNA in the esophagus of EoE patients using microarray gene analysis.10 IL-15 is similar in structure to IL-2 and both share a number of biological activities including the ability to stimulate the proliferation and differentiation of activated T cells.11, 12 In addition, IL-15 can target natural killer (NK) cells and induce their activation in an antigen-independent manner. 11, 12 This process is thought to contribute to intestinal inflammatory responses including those found in celiac disease, a disease that shares features with EoE including being triggered by food antigens, the involvement of epithelial cells (although squamous epithelium in EoE), and the overexpression of NK cell activation antigens such as the MHC like molecule MIC.10, 13–15 Notably, mice deficient in IL-15 or the IL-15 receptor (IL-15Rα) have defective naïve and memory CD8+ T cells, intestinal intraepithelial lymphocytes (IEL), and NK cells.16, 17
The present study defines a critical role for IL-15 in the pathogenesis of EoE. We demonstrate that IL-15 and its specific receptor IL-15Rα are increased in the esophagus of EoE patients. Notably, IL-15 transcript levels correlate significantly with esophageal eosinophilia in active and treated EoE patients. IL-15Rα-deficient mice are protected from the development of experimental EoE (but not respiratory allergic inflammation) following allergen exposure. Mechanistically, IL-15 primes CD4+ T cells to produce eosinophil-activating Th2 cytokines (e.g. IL-5, IL-13). Additionally, IL-15 induces primary esophageal epithelial cells to produce eosinophil-activating chemokines in mice (eotaxin-1, eotaxin-2) and in humans (eotaxin-3). Taken together, these findings define a novel role for IL-15 in Th2 responses and provide evidence that IL-15 has an essential role in EoE pathogenesis.
Materials and Methods
Patient biopsies
Formalin fixed-paraffin embedded biopsy samples were obtained from the esophagus of non-EoE or EoE patients following an Institutional Review Board-approved protocol. The non-EoE individuals and EoE patients were selected without regard for age, atopic status, or gender. The non-EoE biopsies were obtained from patients who presented with symptoms typical of GERD and EoE but were found to have completely normal esophageal endoscopic and microscopic analysis. Diagnosis was established based on the maximum esophageal eosinophil count per high-power fields (hpf) (x400). Non-EoE or “normal” patients were defined as having 0 eosinophils/hpf and no basal layer expansion. Typically, these patients had abdominal pain, and some had allergic diseases including asthma or rhinitis. EoE patients were defined as having ≥ 24 eosinophils/hpf. Patients included in this study commonly had EoE or other allergic diseases such as asthma or atopic dermatitis. Detailed patient characteristics including treatment and dietary restriction are listed in Supplementary Table 1.
Quantitative PCR
The RNA samples (500 ng) were subjected to reverse transcription using iScript reverse transcriptase (Bio-Rad) according to the manufacturer’s instructions. Human and murine IL-15, eotaxin-1, eotaxin-2, eotaxin-3, IL-5 and IL-13 were quantified by real-time PCR using IQ5 (Bio-Rad). Results were then normalized to human or mouse GAPDH or β-actin amplified from the same cDNA mix and expressed as relative gene expression. cDNA were amplified using the primers listed in Supplementary Table 2.
Enzyme-Linked Immunosorbent Assay (ELISA)
IL-15 protein concentrations in the serum of non-EoE and EoE patient samples and eotaxin-1, -2, -3 protein concentrations in the cell supernatant and lysate of cultured epithelial cells were quantified by using a DuoSet ELISA kit (R & D Systems).
Immunohistochemical (IHC) detection of IL-15 in EoE patient esophageal biopsies
Frozen, 5-μm esophageal biopsies sections of non-EoE and EoE patients were immunostained with biotinylated anti-hIL-15 (PeproTech) using IHC staining methods previously described. 18, 19 The specificity of anti-lL15 IHC staining was tested using biotinylated anti-rabbit IgG on EoE biopsies.
Mice
Specific pathogen-free BALB/c, IL-15Rα-deficient mice with matched control wild-type mice were obtained from the Jackson Laboratory (Bar Harbor, ME). All of the experiments were performed on age- and gender-matched mice 6–8 weeks of age. The mice were maintained in a pathogen-free barrier facility, and animals were handled according to Institutional Animal Care and Use Committee and National Institute of Health guidelines.
Induction of experimental allergic EoE in mice
A mouse model of allergic EE was established using methods described previously with a few modifications.6 The detailed procedure is provided in the supplementary materials and methods.
Airway eosinophil analysis
The mouse lungs were lavaged. Recovered BALF was centrifuged, resuspended, and measured for total cell numbers. Cytospin cell preparations were made for differential cell counts. The BALF eosinophil counts were used as an indication of lung eosinophilia. Details are provided in the supplementary materials and methods.
Esophageal eosinophil analysis
Esophageal, 5-μm paraffin tissue sections were immunostained with antiserum against mouse eosinophil major basic protein (anti-MBP, a kind gift of Drs. James and Nancy Lee, Mayo Clinic, Scottsdale, AZ) as previously described. 18, 19 Positively-stained cells were quantified with digital morphometry using the Metamorph Imaging System (Universal Imaging Corp) and expressed as eosinophils/mm2 as previously described.9, 20 Eosinophils in esophageal biopsies of non-EoE and EoE patients were identified and quantified in hematoxylin- and eosin-stained tissue sections as eosinophils/hpf (400X). Details of eosinophil staining and quantification are provided in the supplementary materials and methods.
Isolation of human and mouse primary esophageal epithelial cells
The human primary esophageal epithelial cells (hPEEC) were isolated from the esophageal biopsies. 10 Detailed descriptions of human and mouse primary epithelial esophageal cell isolation and culture are provided in the supplementary materials and methods.
Esophageal single-cell isolation and flow cytometric analysis
Esophageal single-cell isolation was performed as described recently21 and is detailed in the supplementary materials and methods. Esophageal cells were stained with cell surface molecule-specific antibodies for flow cytometric analysis using a fluorescence activated cell sorter (FACS). The following reagents were used for specific antigen analysis: anti- MHCII, CD11c, CD11b, and CD45 (pan marker) to identify macrophages and dendritic cells in the esophageal total leukocyte (CD45+) population; mouse and human anti-cytokeratin to validate epithelial cell characteristics; anti-mIL-15Rα and anti-hIL-15Rα to identify the IL-15 receptor on respective cell types; and anti- pSTAT3, pSTAT5, and pSTAT6 to examine specific STAT phosporylation and respective isotype controls obtained from BD Biosciences. FcR block (anti-CD16/anti-CD32) was added to all surface staining mixtures. 7ADD was used to exclude dead cells. The cells were incubated for the specific antigens with the required combination of antibodies at 4 °C for 45 min followed by two washes. The intracellular IL-15 was detected using anti-IL-15 antibody obtained from PeproTech, Inc. FACS analysis was performed using a FACSCalibur (BD Biosciences) and analyzed using CellQuest software (BD Biosciences).
Isolation of CD4+ T cells
Total splenic lymphocytes from BALB/c mice were aseptically harvested by gently mashing isolated spleens through a 70-μm nylon mesh cell strainer. A single-cell suspension was prepared and RBCs were lysed. Total splenic lymphocytes were incubated with a magnetically-labeled cocktail of biotin-conjugated antibodies against CD8a (Ly-2), CD45R (B220), DX5, CD11b (Mac-1) and Ter-119, followed by anti-biotin microbeads provided in the kit (Miltenyi Biotec Inc) to exclude non-CD4+ T cells. The purity of the CD4+ T cells was analyzed by FACSCalibur (BD Immunocytometry Systems) and was ≥95%.
Cytokine analysis
The cytokine mRNA and protein was determined by using real-time PCR and ELISA analysis. Details are provided in supplementary materials and methods.
CD4+ T cell proliferation assay
Freshly isolated splenic CD4+ T cells from wild-type BALB/c mice were cultured in triplicate wells (2×105 cell/well) of 96-well plates in complete RPMI-1640 medium alone (negative control), anti-CD3/anti-CD28 (positive control; 2 μg/ml of each, BD Pharmingen,), or in the presence of 10, 100, or 500 ng/ml mIL-15 (PeproTech Inc). Cells were incubated for 3 days at 37 °C with 5% CO2. Eighteen hours before the end of incubation, 1 μCi of 3H-thymidine was added per well. The cells were harvested onto glass fiber filter paper, and 3H-thymidine incorporation was determined by a beta counter (Topcount NXT)
Western analysis
STAT phosphorylation (pSTAT5) in CD4+ T cells treated with 100 ng/ml IL-15 for 0, 15, 30, 60, 120 and 240 min was examined by western blot analysis. Cell lysate from 2×106 cells was electrophoresed in 4–12% SDS-polyacrylamide gels (PAGE) and transferred to nitrocellulose membranes for total STAT and pSTAT detection. The detailed procedure is provided in the supplementary materials and methods.
Statistical analysis
Details of statistical analysis are provided in the supplementary materials and methods.
Results
IL-15 expression in human and experimental EoE
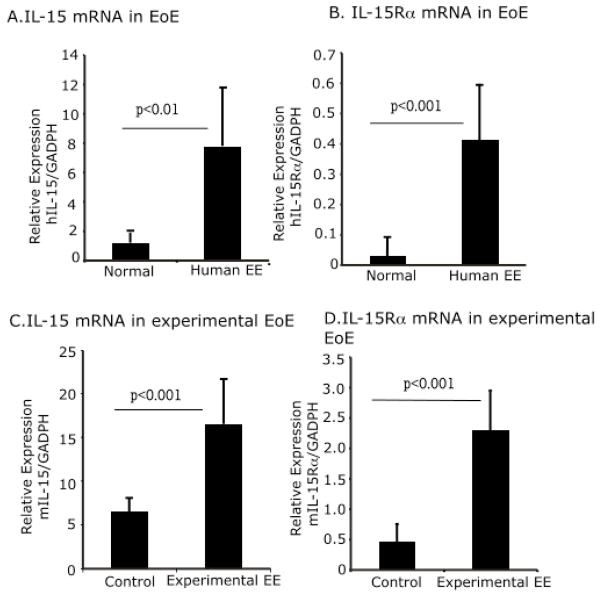
We previously reported that IL-15 mRNA is significantly increased in the esophagus of patients with EoE compared with non-EoE individuals.10 We validated these preliminary findings by quantifying IL-15 and IL-15Rα mRNA expression in the esophagus of EoE patients compared to non-EOE individuals. There was a ~6-fold increase in IL-15 and 10-fold increase in IL-15Rα mRNA levels in esophageal biopsies of EoE patients compared to non-EoE individuals (Fig 1 A, B). In a murine model of allergen-induced EoE, a ~3-fold increase in IL-15 and 4-fold increase in IL-15Rα mRNA levels was observed in allergen-challenged compared to saline-challenged mice (Fig 1 C, D).
Figure 1. Induced expression of IL-15 and IL-15Rα transcript in the esophagus of experimental and human EoE.
Esophageal mRNA expression of IL-15 and IL-15Rα was measured in non-EoE and EoE patient esophageal biopsies performing quantitative real-time PCR analysis. The relative expression compared to respective controls are shown, n=14–16 esophageal biopsies/group (A–B) and saline- and allergen-challenged mice, n = 12 mice/group (C–D). The data is expressed as mean ± SD.
Human esophageal eosinophilia correlates with IL-15 mRNA expression levels
Next, we tested the hypothesis that esophageal eosinophilia may be associated with IL-15 mRNA expression in human EoE. The IL-15 mRNA expression in the esophagus positively correlated with peak tissue eosinophil counts (r=0.67, p<0.001) in esophageal biopsies of EoE patients (both active and treated) (Fig 2A). Additionally, we compared eosinophil peak numbers and IL-15 mRNA expression in patients with improved EoE (patients treated with swallowed fluticasone or dietary treatment). Patients with improved EoE following treatment had a significantly reduced expression of IL-15 mRNA (Fig 2B). Furthermore, a similar trend of IL-15 protein levels in serum samples was also observed (Fig 2C). No IL-15-positive cells were detected by IHC in the epithelial mucosa of non-EoE patient esophageal biopsies [Figure 2D (a, b)]. In contrast, there were IL-15-positive inflammatory cells in the elongated esophageal papillae as well as in the epithelial mucosa of EoE patient’s esophageal biopsies. [Figure 2 D (c, d)]. Interestingly, no anti-IL-15 immunoreactivity was detected on esophageal epithelial cells. Similarly, no immunopositive staining was detected in the negative control samples of EoE patients (data not shown). Additionally, supplementary figure 3 is included to show better visualization of ant-IL-15 immunopositive cells in EoE patient’s biopsies.
Figure 2. Analysis of esophageal eosinophilia and IL-15 in non-EoE, active EoE, and treated EoE patients.
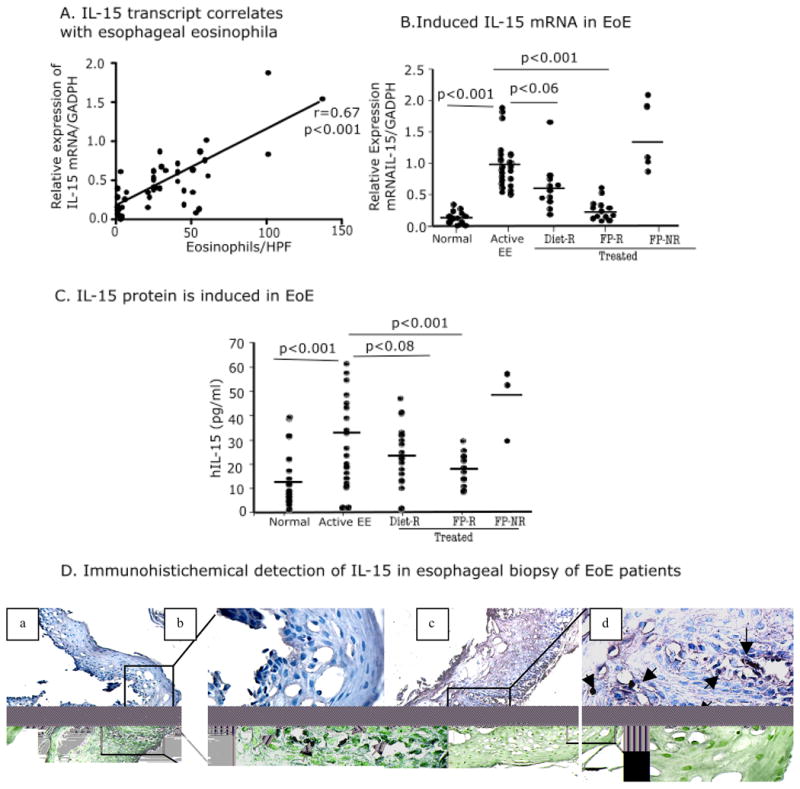
Correlation between the maximum eosinophil number/hpf and IL-15 mRNA expression in human EoE is shown (A). P -value and r were calculated using Spearman correlation test. IL-15 mRNA expression in non-EoE individuals, active EoE patients (> 24 eosinophils/hpf), patients on elemental diet, and patients responsive and not responsive to Flovent are shown (B). Serum IL-15 protein levels in non-EoE individuals, active EoE patients (> 24 eosinophils/hpf), patients on elemental diet, and patients treated and responsive to Flovent are shown (C). Immunoreactivity of IL-15 was tested on esophageal biopsies on non-EoE and EoE patients by performing IHC. No IL-15-positive cells were detected in non-EOE patient esophageal biopsies, marked with arrows, original magnification 10x [D, (a)], and inset magnification in 400x [D, (b)]. A number of infiltrating cells were detected positive for IL-15 in the esophageal biopsies of EoE patient, original magnification 10x [D, (c)] and inset magnification in 400x [D, (d)]. P-values were calculated using Kruskal–Wallis test followed by Dunn’s multiple comparison tests. Diet-R (diet treatment responder), FP-R (Flovent responders) and FP-NR (Flovent non-responders).
The role of IL-15 in the induction of experimental EoE
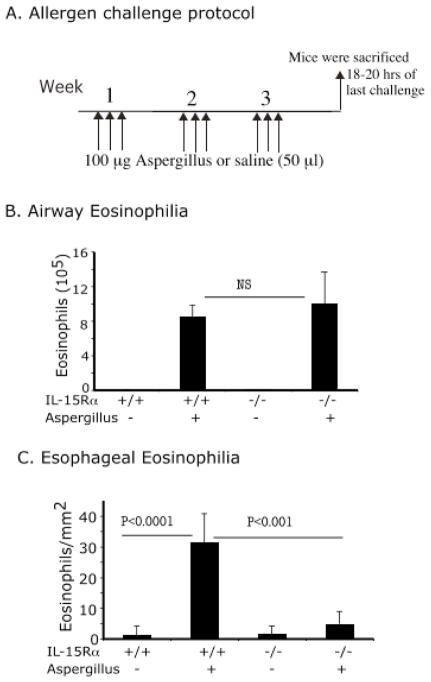
We were next interested in determining whether induction of esophageal eosinophilia was dependent upon IL-15. Accordingly, we induced experimental EoE by repeated intranasal allergen or saline exposure (Fig 3A). The eosinophil levels in the airways of allergen-treated IL-15Rα-deficient mice were comparable to wild-type mice (Fig 3 B). In contrast, eosinophil levels in the esophagus of allergen-challenged wild type and IL-15Rα-deficient mice were 31.5 ± 9.2/mm2 and 4.7 ± 3.9/mm2 respectively (mean ± S.D., n = 9–11, p<0.001) compared to 1.4 ± 2.8/mm2 and 1.7 ± 2.6/mm2 (mean ± S.D., n = 10–12) in saline-treated mice (Fig 3C).
Figure 3. IL-15Rα-deficient mice are protected from allergen-induced EoE.
Mice were intranasally challenged three times a week for 3 weeks with Aspergillus fumigatus (Asp) extract or saline as per the protocol shown (A). Bronchoalveolar lavage fluid was collected and analyzed 18–20 hrs after the last intranasal saline or allergen exposure, and BALF eosinophil numbers were counted and are shown (B). Eosinophils in the esophagus were counted by performing morphometric analysis and are expressed as eosinophils/mm2 (C). Data are expressed as mean ± S.D.; n = 12 mice/group. NS (non significant).
IL-15 cellular source in EoE
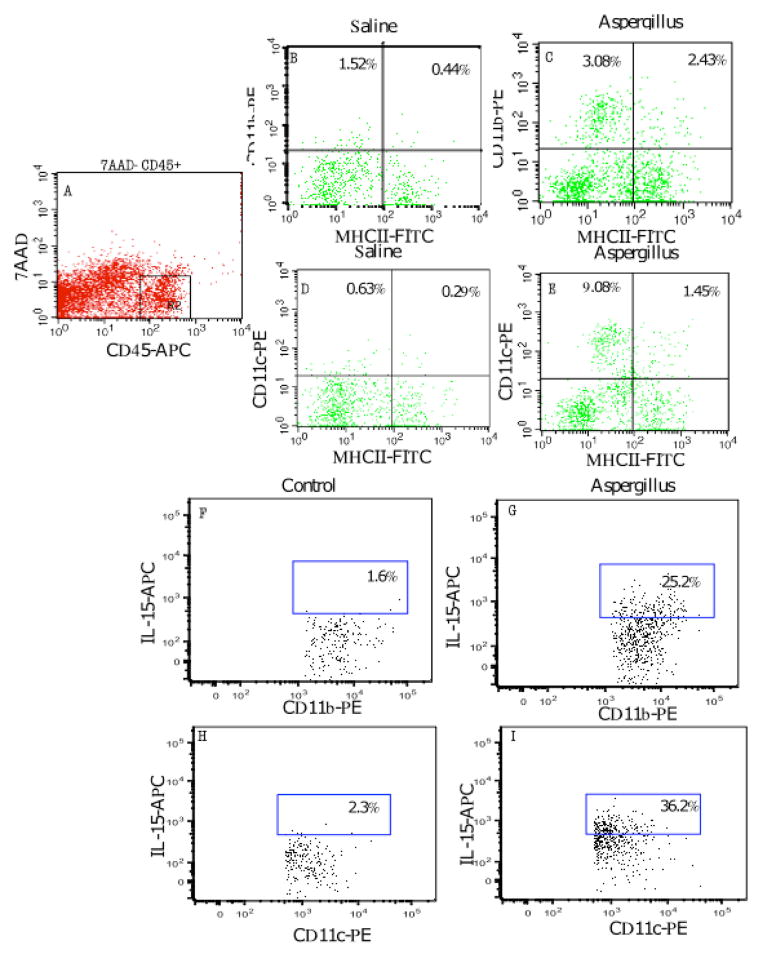
Next, we examined whether IL-15-producing macrophages and dendritic cell populations are increased in the esophagus. In order to test this, total esophageal cells were isolated from the saline- and allergen-challenged mice, and the number of macrophages and dendritic cells were examined in the total leukocyte (anti-CD45+) cell population by FACS. We observed a ~6-fold increase in the frequency of macrophages and a ~ 3-fold increase in the frequency of dendritic cells in the esophagus of allergen-challenged mice compared to saline-challenged wild-type mice (Fig 4A–E). We then examined whether these cells were a source of IL-15. Intracellular cytokine detection analysis revealed that both macrophages and dendritic cells produced IL-15 following allergen challenge. Intracellular IL-15 was detected in gated MHCII positive, CD11b or CD11c positive cells isolated from allergen-challenged mice. Intracellular IL-15 was detected in ~ 25% MHCII+/CD11b+ cells (Fig 4 G) compared to ~1.6% in control cells (Fig 4 F) and ~36% MHCII+/CD11c+ cells (Fig 4 I) compared to 2.3% in control cells (Fig 4 H).
Figure 4. Macrophages and dendritic cells are increased following experimental EoE induction.
The total esophageal cells from saline- and Aspergillus-challenged mice were analyzed for macrophages and dendritic cells using FACS analysis. The 7AAD− CD45+ (pan marker) cells were gated (A) and CD11b/MHC class II double positive cells were analyzed for macrophages in saline- (B) and Aspergillus-challenged mice (C). Similarly, CD11c/MHC class II double positive cells were analyzed for dendritic cells in 7AAD− CD45+ (pan marker) gated cells in saline- (D) and Aspergillus-challenged mice (E). Further, intracellular IL-15 was detected in MHCII-CD11b double positive cells (G) and MHCII-CD11c double positive cells (I) isolated from allergen-challenged mice. Some baseline IL-15-positive cells were also detected in control cells (F, H). These experiments are representative of three independent experiments performed in triplicate. Data are expressed as percent change in cell populations.
IL-15 activates CD4+ T cells
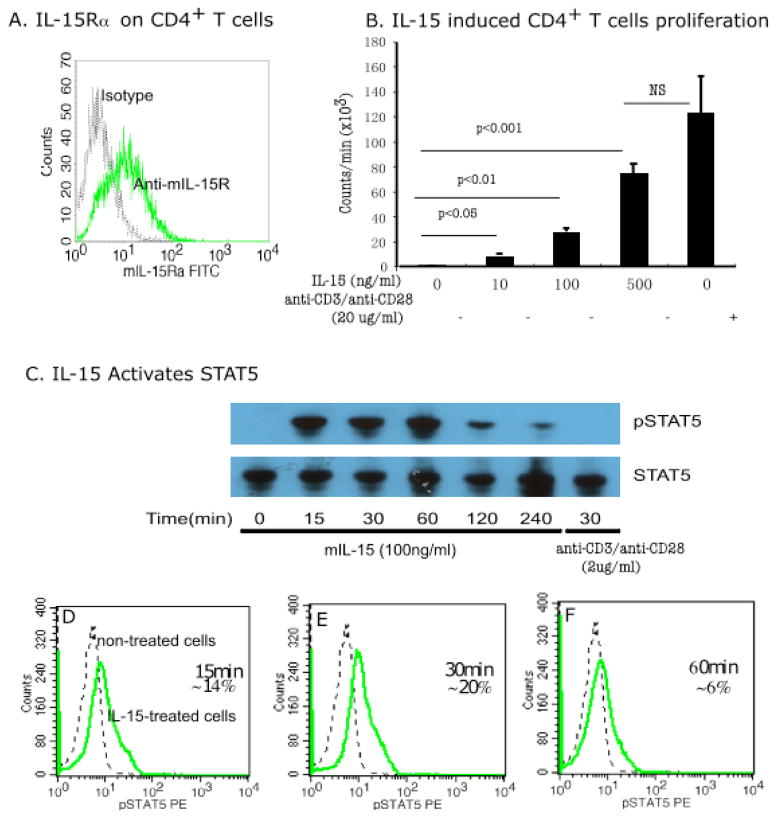
We tested the hypothesis that IL-15 may activate CD4+ T lymphocytes. We first examined whether IL-15 receptor was expressed on CD4+ T cells. Indeed, our observation found that ~30% of freshly isolated murine splenic CD4 T cells expressed the IL-15 specific receptor IL-15Rα (Fig 5A). Second, we tested the hypothesis that IL-15 activates and proliferates CD4+ T cells. A dose-dependent increase in cell proliferation was observed following IL-15 treatment; the cell proliferation at a dose of 500 ng/ml was comparable to the positive control used in the experiment (2 μg each of anti-CD3/anti-CD28) (Fig 5B). In addition, we also examined IL-15-induced CD4+ T cell proliferation that was regulated by a family member of signal transducer and activator of transcription (STAT). Both IL-15-treated and non-treated cells were tested for STAT5 activation by performing western blot analysis. We observed STAT5 phosphorylation after treating the cells with IL-15 for 15–60 min compared to non-treated cells (Fig 5C). Further, we tested the hypothesis that STAT5 is activated in specific subsets of CD4+ T cells or in all CD4+ T cells in response to IL-15. In order to test this we performed flow cytometric analysis using anti-pSTAT5 antibody and only 20% of 100 ng/ml of IL-15 treated CD4+ T cells showed maximum activation 30 min post-treatment compared to non-treated cells (Fig 5 D, E, F). The other STAT family of molecules, STAT3 and STAT6, showed no response to IL-15 exposure at any time point tested; representative data is shown (Supplementary Fig 1 A, B).
Figure 5. IL-15 treatment of purified murine CD4+T cells.
FACS analysis was performed to examine IL-15 receptor on purified splenic CD4+ T cells. Approximately 30% of the purified population expressed IL-15-specfic receptor IL-15Rα (A). IL-15 dose dependently increased proliferation as observed by performing thymidine incorporation analysis as shown (B). Western blot analysis indicates that 100 ng/ml IL-15-induced STAT5 phosphorylation was observed between 15–60 min (C), and flow cytometric analysis indicates that only 20% of CD4+ T cells responded to 100 ng/ml of IL-15 for STAT5 phosphorylation at 30 min. The regular line in the histogram represents 100 ng/ml IL-15 treated, and the dotted line represents non-treated CD4+T cells (D–F). Data is representative of three independent experiments and are expressed as mean ± S.D.
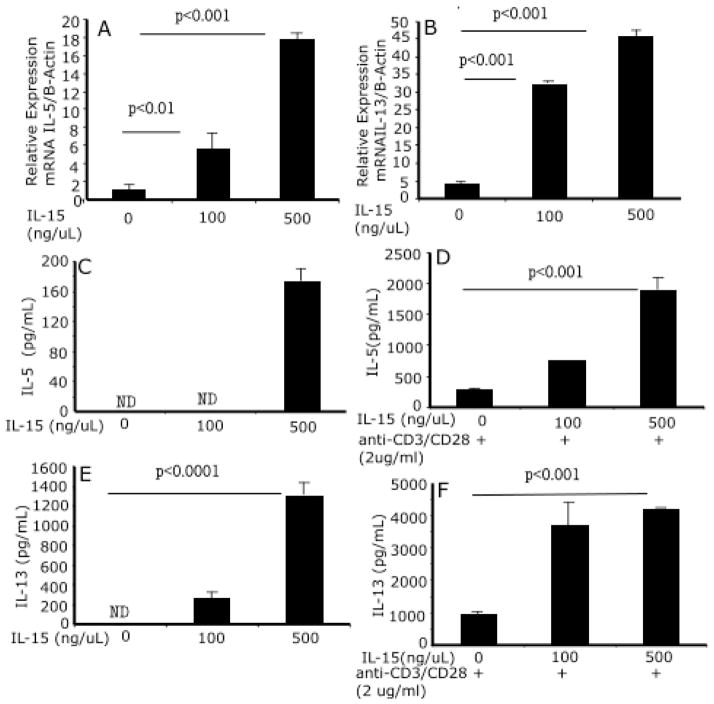
CD4+ T cells produce eosinophil-selective Th2 cytokines in response to IL-15
We next tested the hypothesis that IL-15 induces a Th2-type cytokine profile by CD4+ T cells. Of note, IL-5 and IL-13 transcripts (Fig 6 A, B) and proteins were increased following IL-15 exposure (Fig 6 C, D). Further, we also stimulated IL-15-treated cells with anti-CD3/anti-CD28 to examine whether IL-15 priming and anti-CD3/anti-CD28 stimulation enhances Th1 and Th2 cytokine production by CD4+ T cells. IL-15-primed CD4+ T cells, followed by anti-CD3/anti-CD28 stimulation, had enhanced production of both IL-5 and IL-13 (Fig 6 E, F). Additionally, IL-15-primed CD4+ T cells upon activation with anti-CD3/anti-CD28 also produced interferon-γ (data not shown).
Figure 6. CD4+ T cell and Th2 cytokine profiles following IL-15 exposure.
A dose response analysis indicates that IL-15 priming to CD4+ T cells induces Th2 cytokine (IL-5 and IL-13) mRNA (A, B) and proteins (C, E). The IL-15-primed CD4+ T cells following stimulation with anti-CD3/anti-CD28 enhance the production of IL-5 and IL-13 (D, F). These are representative of three independent experiments performed in triplicate. Data are expressed as mean ± S.D.
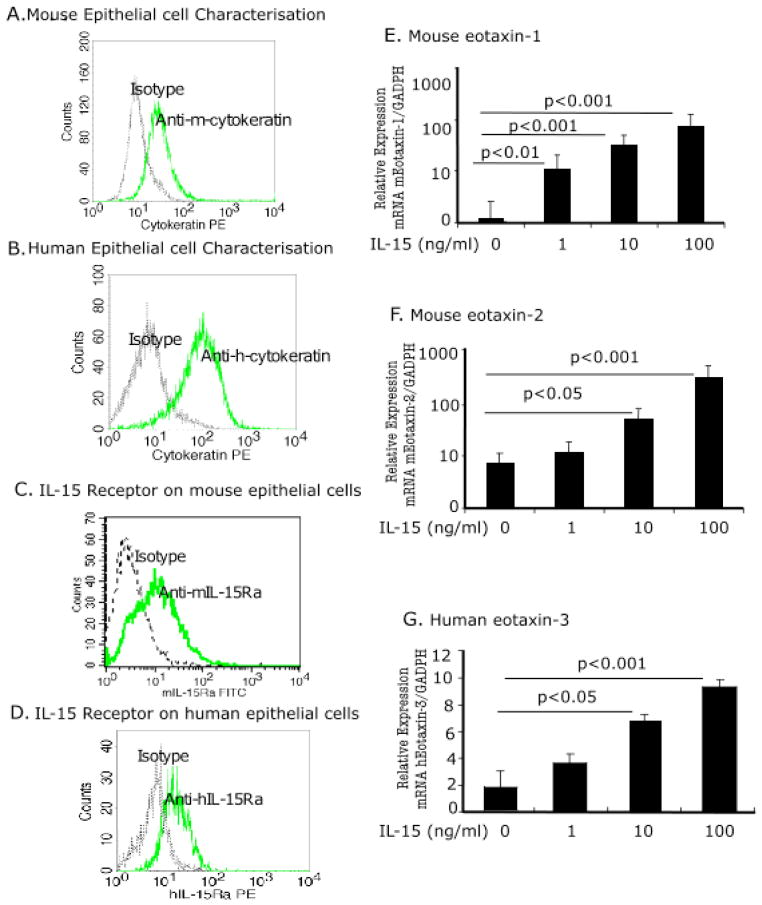
Eosinophil active chemokines (Eotaxin-1, -2 and -3) are induced in human and mouse primary esophageal epithelial cells following IL-15 treatment
Next, we tested the hypothesis that IL-15 induces eotaxins in the esophagus. We first, examined the characteristics of isolated cultured human and mouse epithelial cells by flow cytometric analysis using wide spectrum screening anti-cytokeratin subunits of 58, 56 and 52 kD (DakoCytomation, Denmark), a characteristic marker of epithelial cells. Isolated and cultured esophageal cells were positive for mouse (Fig 7A) and human cytokeratin (Fig 7B). Next, we examined IL-15 receptor on primary esophageal epithelial cells. Both mouse and human primary epithelial cells expressed IL-15 specific receptor IL-15Rα (Fig 7C and D). Further, we exposed mouse and human primary esophageal epithelial cells to IL-15 (0, 10, 1, 100 ng/ml) for 48 hours and tested eosinophil-specific chemokine induction in response to IL-15 exposure. Notably, IL-15 increased eotaxin-1 and eotaxin-2 mRNA in mouse primary esophageal epithelial cells (Fig 6E and F) and eotaxin-3 mRNA in human primary esophageal epithelial cells (Fig 6 G), yet eotaxin-3 protein was not detected in the supernatant. No significant change in eotaxin-1 and eotaxin-2 was observed in human primary esophageal epithelial cells following IL-15 treatment (Supplementary Fig 2 A, B). Additionally, we observed an increase of eotaxin-1 protein in mouse esophageal epithelial cells and eotaxin-3 protein in human esophageal epithelial cell lysate following 100ng/ml IL-15 treatment for 48 hours. The mouse eotaxin-1 levels following 100ng IL-15 treatment was 57.5 ± 8.4 pg/mg protein (mean ± SD) compared to 26.8 ± 2.6 pg/mg protein (mean ± SD, p<0.01) in non-treated cells, and eotaxin-3 protein level in human esophageal epithelial cell lysate following 100 ng/ml IL-15 treatment was 2.1 ± 1.3 ng/mg protein (mean ± SD) compared to 1.08 ± 0.03 ng/mg protein (mean ± SD) in non-treated cell lysate (mean ± SD, p<0.05). The mouse eotaxin-2 and human eotaxin-1 and eotaxin-2 protein levels were comparable in IL-15-treated and non-treated cell lysate (data not shown). Eotaxin-1, -2 and -3 levels were undetectable in the supernatant of IL-15-treated and non-treated human esophageal epithelial cells.
Figure 7. Eotaxin gene induction in human and mouse PEEC following IL-15 treatment.
The characteristics of isolated and cultured mouse and human PEEC was verified by immunostaining with mouse and human anti-cytokeratin antibody and tested by performing FACS analysis, mouse (A) and human (B). The cells were further examined for the expression of IL-15 receptor using IL-15Rα antibody against mouse and human and data is shown (C, D). Eosinophil-specific chemokines eotaxin-1, -2, and -3 expression following IL-15 stimulation in mouse and human PEEC was quantified using real-time PCR. Eotaxin-1 (E) eotaxin-2 (F) mRNA expression in mouse primary esophageal epithelial cells following 48 h of mIL-15 (0, 1, 10, 100 ng/ml) exposure is shown. Eotaxin-3 mRNA expression in human primary esophageal epithelial cells following 48 h of hIL-15 (0, 1, 10, 100 ng/ml) exposure is shown (G). These are representative of three independent experiments performed in triplicate. Data are expressed as mean ± S.D.
Discussion
Eosinophil accumulation in the esophagus is characteristic of a variety of clinical disorders including GERD, eosinophilic gastroenteritis, and EoE. 1, 2 Recent clinical studies have suggested that the prevalence of these disorders, especially EoE, is increasing. 22–24 Prior microarray gene chip analysis indicated induction of the IL-15 gene in EoE patients.10 The present study further analyzed the gene expression and functional role of IL-15 in the development of EoE with particular focus on the mechanistic induction of eosinophil-selective cytokines and chemokines of Th2 responses in the esophagus of experimental and human EoE.
Expression and protein levels of IL-15 are increased in human and experimental EoE. Using quantitative real-time PCR analysis, we confirmed our earlier microarray data10 and demonstrated that IL-15 and IL-15Rα mRNA levels were significantly increased in human and experimental EoE. Notably, transcript levels of IL-15 strongly correlated with esophageal eosinophils in active EoE patients and significantly decreased in improved treated EoE patients. Interestingly, both macrophages and dendritic cells have previously been shown to be a rich source of IL-15.25–28 These two cell types have a role in innate and adaptive immunity; under certain conditions, these cells produce a number of mediators that activate T cells to generate inflammatory cytokines. Both macrophages and dendritic cells were indeed increased in experimental EoE, and dendritic cell increases have been previously reported in human EoE.29 These findings and the induced serum levels of IL-15 suggest that IL-15 may be a non-invasive biomarker in the diagnosis of EoE.
IL-15 has a role in experimental EoE pathogenesis. In an in vivo model of wild-type and IL-15Rα gene-deficient mice exposed to intranasal allergen, 6, 7 IL-15Rα gene-deficient mice failed to develop EoE following allergen treatment despite having strong eosinophilic inflammation in the lungs comparable to wild type mice. These findings establish a role for IL-15 in allergen-induced EoE pathogenesis and implicate its contribution to human EoE.
IL-15 may contribute to EoE pathogenesis by directly inducing Th2 cytokine production. Mechanistic analysis demonstrated that IL-15 priming activates Th2 cytokine-producing CD4+ T cells. These data are in accordance with the previous reports that IL-15 is required for T cell proliferation and activation.11, 12, 16, 17 Further studies are needed to examine the role of specific CD4+ T cell subsets requiring IL-15 for growth and survival. The present study also identified a role for STAT5 in regulating IL-15-mediated CD4+ T cell activation, proliferation, and eosinophil active Th2 cytokine production. Our finding that IL-15 activates STAT5 is in accordance with prior studies.30, 31 In addition, STAT5 has been previously implicated in Th2 cytokine production in the absence of IL-2,32–34 and IL-2 production is impaired in mouse model of EoE following allergen challenge.21 Previous research has demonstrated that allergen-induced EoE is partially dependent on STAT6; whereas, IL-13-induced EoE is dependent on STAT6.20, 35 Notably, IL-15-induced Th2 cytokine release is not dependent on STAT6, as IL-15 exposure fails to activate STAT6 in CD4+ T cells. These findings further enhance our understanding of the mechanism of allergen-induced Th2 cytokine-mediated STAT6- dependent and independent induction of EoE, and the accumulating evidence suggests that EoE pathogenesis is regulated by both STAT5 and STAT6. We previously reported a contributory role for adaptive T cell immunity in allergen-induced experimental EE,7 and IL-15 is critical in the maintenance of innate immunity.28 We showed that IL-15 exposure to specific CD4+ T cells produces Th2 cytokines independent of STAT6 activation, and this finding is supported by our flow cytometric analysis that shows only ~20% of CD4+ T cells activate STAT5 in response to IL-15. However, further investigation is needed to establish the role of IL-15 in STAT5-mediated Th2 cytokine production. Taken together, these findings indicate that IL-15 may have an important role in bridging innate immunity to adaptive immunity.
IL-15 may stimulate esophageal epithelial cells to induce eosinophil-selective chemoattractants in the esophagus of experimental and human EoE. Eotaxins play a role in eosinophil chemoattraction,36, 37 and eotaxin-1 has been shown to have a role in allergen- and IL-13-induced experimental EoE.6, 20 Eotaxin-3 is the most highly expressed gene in human EoE, and epithelial cells are the major source of eotaxin-3 in the human esophagus.10 Our analysis indicated that the isolated primary cells retain the characteristics of epithelial cells and that they express specific receptors for IL-15. Furthermore, we demonstrated that mouse and human primary esophageal cells induces mRNA and protein levels of eotaxins in mouse and human following IL-15 exposure. It is important to note that we detected eotaxin protein in the cell lysate not in the supernatant of IL-15 treated epithelial cells indicating that eotaxin-3 protein is bound to the cell surface, as recently reported.38 Our observations are consistent with these findings, but needs more clarification to address whether eotaxin-3 is bound to cell surface or maintained in the epithelial cells to recruit eosinophils in the tissue.
In light of previous research and this study’s findings, we propose that IL-15 contributes to EoE induction by directly inducing Th2 cytokine production by CD4+ cells and eotaxins by esophageal epithelial cells. The identification of IL-15 in the pathogenesis of EoE suggests that targeting this molecule may be useful for the treatment of EoE. The therapeutic significance of our findings certainly deserves further attention. IL-15 is principally produced by macrophages and dendritic cells during innate immune response and subsequently profoundly influences adaptive immunity.39, 40 IL-15 is a growth factor for γδ T cells and NKT cells, and both these cells can play a role in innate and adaptive immunity under certain conditions.41, 42 Notably, we have previously reported increased IL-5+ NK cells in EoE patients.43 Therefore, a possible role of iNKT or γδ T cells should not be ruled out in EoE pathogenesis.
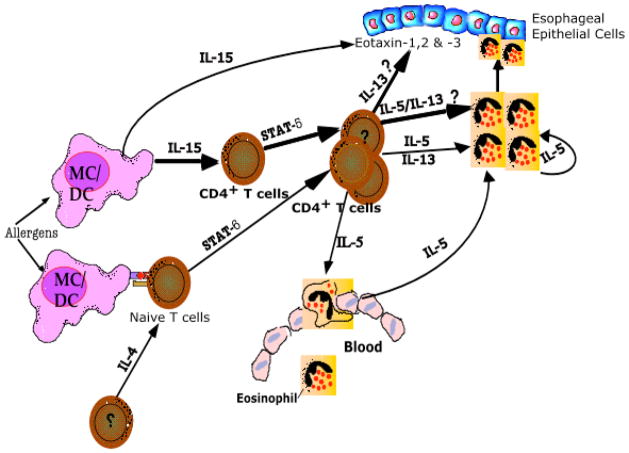
In conclusion, we demonstrate that esophageal biopsies of EoE patients and the esophagus of mice with experimental EoE have increased expression of IL-15 mRNA and protein. IL-15-induced CD4+ T cell proliferation and Th2 cytokine production, as well as the induction of eosinophil-specific chemokine mRNA by primary esophageal epithelial cells provides a mechanistic functional pathway for IL-15 in EoE. Importantly, mice deficient in the IL-15Rα gene are protected from the development of experimental EoE. Induced IL-15 expression significantly correlates with esophageal eosinophilia in humans and IL-15 levels were reduced following treatment in improved EoE patients. Collectively, these studies identify a novel role for IL-15 in regulating Th2 responses and provide evidence that IL-15 has a key role in EoE pathogenesis. Additionally, we include a prospective summarized pathway of IL-15-induced disease pathogenesis as a diagrammatic representation in Figure 8.
Figure 8. Diagrammatic proposed pathway representation of IL-15-induced EoE.
Allergen-induced IL-15 producing macrophages (MC) and dendritic cells (DC) are increased in the esophagus in experimental EoE. IL-15 activates and proliferates specific populations of CD4+ T cells. IL-15-induced CD4+ T cells activation in certain conditions produces eosinophil-active cytokines IL-5 and IL-13 that are regulated by signal transducer and activator of transcription (STAT) 5. In addition, IL-15 induces eosinophil active chemokines (eotaxin) in the esophageal epithelial cells that attract eosinophils into the esophageal epithelial mucosa from the blood. The STAT5 mediated pathway of Th2 cytokine induction support previous findings that allergen-induced Th2 responses in EoE are dependent and independent to STAT6.
Supplementary Material
Acknowledgments
This work was supported in part by the grants NIH RO1 DK067255 (AM), NIH R01 AI080581 (AM), AI45898 (MER), AI070235 (MER), the Digestive Health Center (DHC) grant DK078392, Campaign Urging Research for Eosinophilic Disease (CURED), Food Allergy Project, and the Buckeye Foundation. The authors also thank Andrea Lippelman and Shawna Hottinger for editorial assistance and Drs. James and Nancy Lee (Mayo Clinic, Scottsdale, AZ) for the generous supply of anti-MBP.
Footnotes
There are no conflicts of interest to disclose for all authors except Dr. Rothenberg who discloses consultancy relationships with Merck, Ception Therapueutics, Nycomed, Biocryst Pharmaceuticals, and Centocor.
Xiang Zhu and Meiqin Wang, Acquisition of data, and data statistical analysis.
Parm Mavi and Madhavi Rayapudi, Technical support.
Akhilesh Pandey, Technical support.
Anil Mishra, Study concept and design, analysis interpretation, study supervision and provided funding.
Marc Rothenberg, Critical revision of the manuscript.
Ajay Kaul and Philip E Putnam, Patient material support.
References
- 1.Dahms BB. Reflux esophagitis: sequelae and differential diagnosis in infants and children including eosinophilic esophagitis. Pediatr Dev Pathol. 2004;7:5–16. doi: 10.1007/s10024-003-0203-5. [DOI] [PubMed] [Google Scholar]
- 2.Kirsch R, Bokhary R, Marcon MA, Cutz E. Activated mucosal mast cells differentiate eosinophilic (allergic) esophagitis from gastroesophageal reflux disease. J Pediatr Gastroenterol Nutr. 2007;44:20–6. doi: 10.1097/MPG.0b013e31802c0d06. [DOI] [PubMed] [Google Scholar]
- 3.Desai TK, Stecevic V, Chang CH, Goldstein NS, Badizadegan K, Furuta GT. Association of eosinophilic inflammation with esophageal food impaction in adults. Gastrointest Endosc. 2005;61:795–801. doi: 10.1016/s0016-5107(05)00313-5. [DOI] [PubMed] [Google Scholar]
- 4.Rothenberg ME, Mishra A, Collins MH, Putnam PE. Pathogenesis and clinical features of eosinophilic esophagitis. J Allergy Clin Immunol. 2001;108:891–894. doi: 10.1067/mai.2001.120095. [DOI] [PubMed] [Google Scholar]
- 5.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109:1503–12. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 6.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra A, Schlotman J, Wang M, Rothenberg ME. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2007;81:916–924. doi: 10.1189/jlb.1106653. [DOI] [PubMed] [Google Scholar]
- 8.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168:2464–9. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, Cohen MB, Akers R, Hogan SP, Assa’ad AH, Putnam PE, Aronow BJ, Rothenberg ME. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton JD, Bamford RN, Peters C, Grant AJ, Kurys G, Goldman CK, Brennan J, Roessler E, Waldmann TA. A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci U S A. 1994;91:4935–9. doi: 10.1073/pnas.91.11.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, et al. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science. 1994;264:965–8. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- 13.Sollid LM, Jabri B. Is celiac disease an autoimmune disorder? Curr Opin Immunol. 2005;17:595–600. doi: 10.1016/j.coi.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21:367–77. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, Ebert EC, Green PH, Jabri B. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–66. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Tough DF, Sprent J. Lifespan of lymphocytes. Immunol Res. 1995;14:1–12. doi: 10.1007/BF02918494. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, Peschon JJ. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–80. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews AN, Friend DS, Zimmermann N, Sarafi MN, Luster AD, Pearlman E, Wert SE, Rothenberg ME. Eotaxin is required for the baseline level of tissue eosinophils. Proc Natl Acad Sci U S A. 1998;95:6273–8. doi: 10.1073/pnas.95.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME. Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest. 1999;103:1719–27. doi: 10.1172/JCI6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125:1419–27. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X, Wang M, Crump CH, Mishra A. An imbalance of esophageal effector and regulatory T cell subsets in experimental eosinophilic esophagitis in mice. Am J Physiol Gastrointest Liver Physiol. 2009;297:G550–8. doi: 10.1152/ajpgi.00148.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra A. Mechanism of eosinophilic esophagitis. Immunol Allergy Clin North Am. 2009;29:29–40. doi: 10.1016/j.iac.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118:1054–9. doi: 10.1016/j.jaci.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 24.Pentiuk S, Putnam PE, Collins MH, Rothenberg ME. Dissociation between symptoms and histological severity in pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2009;48:152–60. doi: 10.1097/MPG.0b013e31817f0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welte T, Koch F, Schuler G, Lechner J, Doppler W, Heufler C. Granulocyte-macrophage colony-stimulating factor induces a unique set of STAT factors in murine dendritic cells. Eur J Immunol. 1997;27:2737–40. doi: 10.1002/eji.1830271038. [DOI] [PubMed] [Google Scholar]
- 26.Kirman I, Vainer B, Nielsen OH. Interleukin-15 and its role in chronic inflammatory diseases. Inflamm Res. 1998;47:285–9. doi: 10.1007/s000110050331. [DOI] [PubMed] [Google Scholar]
- 27.Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–17. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohteki T. Critical role for IL-15 in innate immunity. Curr Mol Med. 2002;2:371–80. doi: 10.2174/1566524023362519. [DOI] [PubMed] [Google Scholar]
- 29.Teitelbaum JE, Fox VL, Twarog FJ, Nurko S, Antonioli D, Gleich G, Badizadegan K, Furuta GT. Eosinophilic esophagitis in children: Immunopathological analysis and response to fluticasone propionate. Gastroenterology. 2002;122:1216–1225. doi: 10.1053/gast.2002.32998. [DOI] [PubMed] [Google Scholar]
- 30.Burchill MA, Goetz CA, Prlic M, O’Neil JJ, Harmon IR, Bensinger SJ, Turka LA, Brennan P, Jameson SC, Farrar MA. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol. 2003;171:5853–64. doi: 10.4049/jimmunol.171.11.5853. [DOI] [PubMed] [Google Scholar]
- 31.Gagnon J, Ramanathan S, Leblanc C, Cloutier A, McDonald PP, Ilangumaran S. IL-6, in synergy with IL-7 or IL-15, stimulates TCR-independent proliferation and functional differentiation of CD8+ T lymphocytes. J Immunol. 2008;180:7958–68. doi: 10.4049/jimmunol.180.12.7958. [DOI] [PubMed] [Google Scholar]
- 32.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–48. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 33.Kagami S, Nakajima H, Suto A, Hirose K, Suzuki K, Morita S, Kato I, Saito Y, Kitamura T, Iwamoto I. Stat5a regulates T helper cell differentiation by several distinct mechanisms. Blood. 2001;97:2358–65. doi: 10.1182/blood.v97.8.2358. [DOI] [PubMed] [Google Scholar]
- 34.Welte T, Leitenberg D, Dittel BN, al-Ramadi BK, Xie B, Chin YE, Janeway CA, Jr, Bothwell AL, Bottomly K, Fu XY. STAT5 interaction with the T cell receptor complex and stimulation of T cell proliferation. Science. 1999;283:222–5. doi: 10.1126/science.283.5399.222. [DOI] [PubMed] [Google Scholar]
- 35.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129:985–94. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Collins PD, Marleau S, Griffiths-Johnson DA, Jose PJ, Williams TJ. Cooperation between interleukin-5 and the chemokine eotaxin to induce eosinophil accumulation in vivo. Journal of Experimental Medicine. 1995;182:1169–1174. doi: 10.1084/jem.182.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elsner J, Petering H, Kluthe C, Kimmig D, Smolarski R, Ponath P, Kapp A. Eotaxin-2 activates chemotaxis-related events and release of reactive oxygen species via pertussis toxin-sensitive G proteins in human eosinophils. Eur J Immunol. 1998;28:2152–8. doi: 10.1002/(SICI)1521-4141(199807)28:07<2152::AID-IMMU2152>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 38.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, Buckmeier BK, Jameson SC, Greenberg A, Kaul A, Franciosi JP, Kushner JP, Martin LJ, Putnam PE, Abonia JP, Wells SI, Rothenberg ME. Coordinate Interaction between IL-13 and Epithelial Differentiation Cluster Genes in Eosinophilic Esophagitis. J Immunol. 2010 Mar 5; doi: 10.4049/jimmunol.0903069. On line publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liew FY. The role of innate cytokines in inflammatory response. Immunol Lett. 2003;85:131–4. doi: 10.1016/s0165-2478(02)00238-9. [DOI] [PubMed] [Google Scholar]
- 40.Liew FY, McInnes IB. The role of innate mediators in inflammatory response. Mol Immunol. 2002;38:887–90. doi: 10.1016/s0161-5890(02)00014-7. [DOI] [PubMed] [Google Scholar]
- 41.Wilson SB, Delovitch TL. Janus-like role of regulatory iNKT cells in autoimmune disease and tumour immunity. Nat Rev Immunol. 2003;3:211–22. doi: 10.1038/nri1028. [DOI] [PubMed] [Google Scholar]
- 42.Sakuishi K, Oki S, Araki M, Porcelli SA, Miyake S, Yamamura T. Invariant NKT Cells Biased for IL-5 Production Act as Crucial Regulators of Inflammation. J Immunol. 2007;179:3452–62. doi: 10.4049/jimmunol.179.6.3452. [DOI] [PubMed] [Google Scholar]
- 43.Bullock JZ, Villanueva JM, Blanchard C, Filipovich AH, Putnam PE, Collins MH, Risma KA, Akers RM, Kirby CL, Buckmeier BK, Assa’ad AH, Hogan SP, Rothenberg ME. Interplay of adaptive th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45:22–31. doi: 10.1097/MPG.0b013e318043c097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.