ER quality control factors GRP94 and OS-9 associate not for the disposal of ERAD substrates but instead because OS-9 sequesters and degrades aberrant forms of GRP94, which are hyperglycosylated at cryptic acceptor sites and have altered structure and activity. This highlights a novel mechanism of quality control of an ER-resident chaperone.
Abstract
The tight coupling of protein folding pathways with disposal mechanisms promotes the efficacy of protein production in the endoplasmic reticulum (ER). It has been hypothesized that the ER-resident molecular chaperone glucose-regulated protein 94 (GRP94) is part of this quality control coupling because it supports folding of select client proteins yet also robustly associates with the lectin osteosarcoma amplified 9 (OS-9), a component involved in ER-associated degradation (ERAD). To explore this possibility, we investigated potential functions for the GRP94/OS-9 complex in ER quality control. Unexpectedly, GRP94 does not collaborate with OS-9 in ERAD of misfolded substrates, nor is the chaperone required directly for OS-9 folding. Instead, OS-9 binds preferentially to a subpopulation of GRP94 that is hyperglycosylated on cryptic N-linked glycan acceptor sites. Hyperglycosylated GRP94 forms have nonnative conformations and are less active. As a result, these species are degraded much faster than the major, monoglycosylated form of GRP94 in an OS-9–mediated, ERAD-independent, lysosomal-like mechanism. This study therefore clarifies the role of the GRP94/OS-9 complex and describes a novel pathway by which glycosylation of cryptic acceptor sites influences the function and fate of an ER-resident chaperone.
INTRODUCTION
Protein quality control (QC) in the endoplasmic reticulum (ER) is defined as the coordination of major pathways that have evolved to fold nascent polypeptides and transport properly folded proteins to their destinations. Molecular chaperones and QC enzymes contribute to protein maturation by binding to folding intermediates, preventing aggregation, adding posttranslational modifications, and inducing conformational changes (Braakman and Hebert, 2013; Gidalevitz et al., 2013). Nonetheless, a significant percentage of proteins still fail to fold, owing to the complexity of folding pathways, molecular crowding in the ER, and protein-modifying mutations and external factors such as cellular stress. Maturation-incompetent secretory proteins are routinely culled by ER-associated degradation (ERAD), a remediation pathway that facilitates protein retrotranslocation across the ER lipid bilayer to the cytosol, where substrates are targeted for proteasomal degradation (Smith et al., 2011; Brodsky, 2012; Olzmann et al., 2012). Although some signals that can initiate ERAD are understood, such as the rate-limiting mannose trimming of oligosaccharides on substrate proteins (Aebi et al., 2010), the targeting of proteins from folding/maturation to degradation pathways and the molecular components that oversee such triage decisions remain enigmatic aspects of ER QC.
An intriguing link between protein folding and ERAD arose from the report of a complex between glucose-regulated protein 94 (GRP94, gp96, HSP90B1) and osteosarcoma amplified 9 (OS-9; Christianson et al., 2008). GRP94 is an ER-resident, metazoan-specific member of the HSP90 family of ATP-dependent molecular chaperones, as well as a major ER calcium buffer (Macer and Koch, 1988; Melnick et al., 1994; Randow and Seed, 2001; Biswas et al., 2007; Yang et al., 2007). Unlike other ER chaperones, such as BiP, calnexin, and calreticulin, GRP94 is required to assist in the folding of a select subset of client proteins that includes immunoglobulins (Melnick et al., 1994), insulin-like growth factors (Wanderling et al., 2007), some toll-like receptors (TLRs) and integrins (Randow and Seed, 2001; Yang et al., 2007), and select members of the Wnt pathway (Liu et al., 2013). However, little is known about how GRP94 promotes folding of its clients, except that it is believed to bind late-stage folding intermediates (Melnick et al., 1994), its ATPase activity is essential (Ostrovsky et al., 2009), and it uses a C-terminal binding region for the chaperoning of integrins and TLRs (Wu et al., 2012).
OS-9 is a more recently characterized ER-resident lectin that participates in the substrate recognition and escort phases of ERAD (Bernasconi et al., 2008, 2010; Christianson et al., 2008). OS-9 is subject to alternative splicing, with at least two isoforms (OS-9.1 and OS-9.2) detected at the protein level under normal growth conditions (Bernasconi et al., 2008; Christianson et al., 2008). Two additional variants (OS-9.3 and OS-9.4) are also predicted, although it is unclear whether or when these isoforms are expressed (Kimura et al., 1998). OS-9 and a related lectin, XTP3-B/Erlectin, bind preferentially to the trimmed forms of high-mannose oligosaccharides found on many ERAD substrates through mannose-6-phosphate receptor homology (MRH) domains (Hosokawa et al., 2009, 2010; Satoh et al., 2010). These lectins also recognize and bind substrates in a glycan-independent manner (Bhamidipati et al., 2005; Bernasconi et al., 2008; Christianson et al., 2008), although how this is accomplished is poorly understood. OS-9 is able to assemble with the ubiquitin ligase Hrd1, through the scaffolding protein SEL1L, to form a complex central to mammalian ERAD (Christianson et al., 2008, 2012; Hosokawa et al., 2008; Mueller et al., 2008).
Because GRP94 has been characterized primarily as a profolding component of the ER, whereas OS-9’s known activity is in protein disposal, the biological implication of their association is not obvious. Given these roles, such a complex could have the following possible functions: 1) OS-9 may be an obligate client of GRP94; 2) GRP94 may be degraded via OS-9; or 3) the complex could participate in a particular step of ER QC. Specifically, we wondered whether GRP94 and OS-9 coordinate to identify proteins with stalled folding pathways, perhaps by associating with substrates and assaying their glycan-trimming status. Indeed, evidence exists for a mutual function of GRP94 and OS-9 in ERAD, since cells depleted of GRP94 exhibited an impairment in the disposal of a soluble glycoprotein, the null Hong Kong mutant of α1-antitrypsin (NHK), similar to the phenotype of OS-9 silencing (Christianson et al., 2008).
In this article, we provide a detailed functional analysis of the GRP94/OS-9 complex, through which we show that OS-9 preferentially associates with aberrant conformers of GRP94 marked by hyperglycosylation of cryptic N-linked glycan acceptor sites. These species are subject to enhanced degradation in an OS-9-mediated, ERAD-independent, lysosomal-like pathway. This work therefore provides a clear example of regulated turnover of an ER-resident chaperone and expands our limited knowledge of how the ER can degrade not just misfolded secretory proteins, but also ER-resident QC factors themselves.
RESULTS
GRP94 does not play a direct role in ERAD
A physical and functional interaction with OS-9 suggests that GRP94 actively participates in ERAD (Christianson et al., 2008), yet any specific role that it may play remains poorly defined. GPR94’s established status as a molecular chaperone makes it an attractive candidate for the task of substrate recognition, in which it could identify, bind, and transfer select misfolded protein intermediates to ERAD machinery, including OS-9. To investigate such a role, we began by testing the interactions between GRP94, OS-9, and several model ERAD substrates. To reflect the variety of proteins cleared through ERAD, we used a diverse panel of misfolded α1-antitrypsin (α1AT) mutants, including the NHK truncation (Sifers et al., 1988; Liu et al., 1997; Christianson et al., 2008), the aggregation-prone PI Z mutant (Lomas et al., 1992; Teckman and Perlmutter, 2000; Cabral et al., 2002; Hidvegi et al., 2010), and an NHK variant in which all three N-glycosylation acceptor sites were mutated to render it nonglycosylated (AAA or QQQ; Cormier et al., 2009; Christianson et al., 2012; Ushioda et al., 2013).
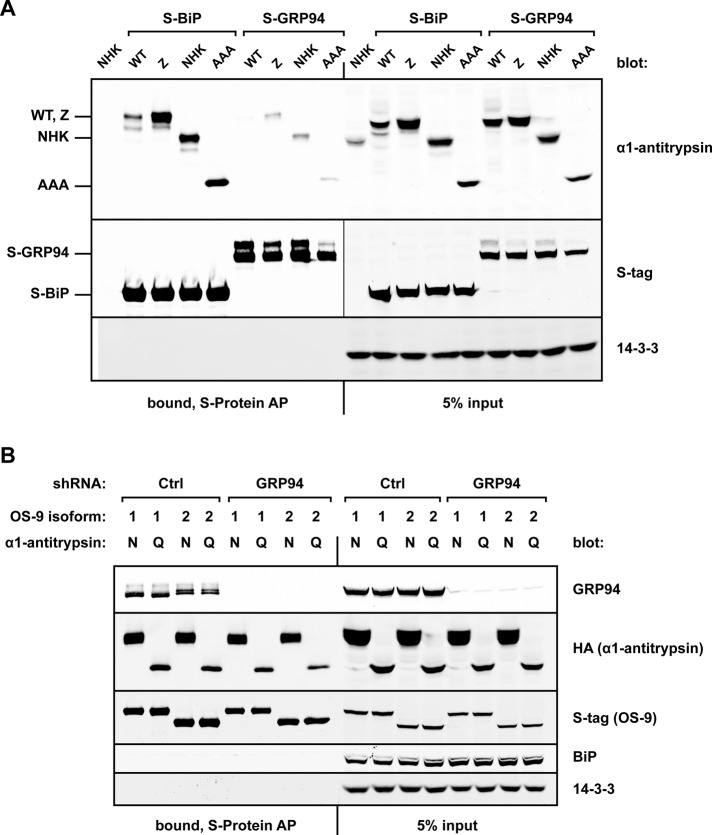
When these α1AT constructs were coexpressed with either S-tagged BiP (S-BiP) or S-tagged GRP94 (S-GRP94) in HEK293T cells, affinity purifications (AP) with S-protein agarose revealed that the α1AT variants associated poorly with GRP94 (Figure 1A). In contrast, BiP bound robustly to the α1AT substrates, with preference for the misfolded species. This is in agreement with previous reports of BiP's role in binding nonnative proteins, as well as with its role in ERAD (Kabani et al., 2003; Ushioda et al., 2008, 2013).
FIGURE 1.
GRP94 binds poorly to ERAD substrates and is not required for OS-9/substrate interactions. (A) HEK293T cells were cotransfected with either S-tagged GRP94 or S-tagged BiP and a panel of untagged α1-antitrypsin constructs: WT, Z, NHK, or a nonglycosylated form of NHK, AAA. Cells were harvested 20 h posttransfection and lysed, and the postnuclear fraction was subjected to affinity purification (AP) with S-protein agarose. Whole-cell lysate inputs and AP bound fractions were examined by Western blot analysis to monitor the coprecipitation of α1-antitrypsin proteins with GRP94 and BiP. 14-3-3 served as a cytosolic loading control and to ensure stringency of the AP. (B) HEK293T cells were infected with lentivirus containing scrambled shRNA (Ctrl) or shRNA targeting GRP94, followed by selection in puromycin. In shCtrl and shGRP94 cells, S-tagged splice isoforms of OS-9 (OS-9.1 or OS-9.2, designated 1 or 2, respectively) were cotransfected with either HA-tagged NHK or nonglycosylated QQQ. AP of S-OS-9 was performed with S-protein agarose. Whole-cell lysate inputs and AP bound fractions were examined via Western blot analysis to observe OS-9 interactions. N, NHK; Q, QQQ.
Although GRP94 bound misfolded α1AT mutants poorly, we queried whether it was required to initiate or stabilize interactions between OS-9 and ERAD substrates. In this scenario, a GRP94-induced conformational change in OS-9 might facilitate association with misfolded proteins. To test this, we investigated whether depletion of endogenous GRP94 affected the ability of OS-9 to recognize and bind substrates. The S-tagged OS-9 splice isoforms OS-9.1 and OS-9.2 were coexpressed with either NHK or QQQ in cells sufficient or deficient for GRP94 (Eletto et al., 2012). Immunoblot analysis revealed that both NHK and QQQ were still robustly copurified with OS-9 via an interaction that occurred independently of GRP94 (Figure 1B). GRP94 coprecipitated with OS-9 in scrambled short hairpin RNA (shRNA)-expressing cells (shCtrl), confirming the interaction between these ER proteins. OS-9 failed to coprecipitate BiP, indicating that the GRP94 interaction is specific and does not occur with any abundant ER-resident protein. Taken together, our data show that GRP94 does not participate directly in binding to model α1AT ERAD substrates, nor does it affect OS-9’s capacity to do so.
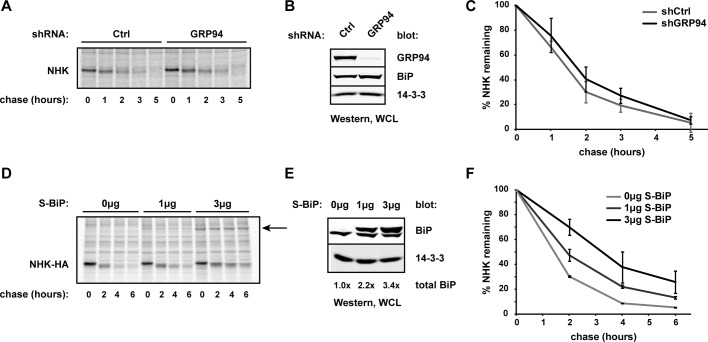
We next asked whether GRP94 was required for turnover of misfolded substrates by monitoring the degradation rates of transiently expressed NHK or misfolded nonsecreted 1 immunoglobulin light chain (NS1; Dul et al., 1992; Chillaron and Haas, 2000; Okuda-Shimizu and Hendershot, 2007). Using HEK293T cells sufficient or deficient for GRP94, pulse-chase analysis revealed that the absence of GRP94 did not alter the rates of disposal for either the glycosylated NHK (Figure 2, A and C) or the nonglycosylated NS1 substrate (Supplemental Figure S1). These data are in contrast to a previous study demonstrating that NHK degradation was impaired upon silencing of GRP94 (Christianson et al., 2008).
FIGURE 2.
GRP94 is not required for ERAD of NHK. (A) Untagged NHK was transiently expressed in HEK293T shCtrl and shGRP94 cells. At 44 h posttransfection, cells were pulse labeled with [35S]methionine/cysteine and chased over the indicated time course, as described in Materials and Methods. At each time point, cells were lysed and NHK isolated with anti–α1-antitrypsin and protein G Sepharose. Samples were separated by SDS–PAGE and the resulting gel scanned by phosphorimager. (B) Western blot of lysates from A, showing depletion of endogenous GRP94. BiP levels in shGRP94 cells are only 1.2 times those of shCtrl cells. WCL, whole-cell lysate. (C) Quantification of three independent pulse-chase experiments as described in A. Means and SDs are plotted. (D) Pulse-chase assays were carried out in parental HEK293T cells cotransfected with NHK-HA and either 0, 1, or 3 μg of S-BiP plasmid. At each time point, NHK was isolated by anti–HA-agarose. The arrow indicates S-BiP coimmunoprecipitating with NHK. (E) Western blot analysis of lysates from D, probed for BiP. Total BiP refers to the entire amount of BiP in a sample relative to the level of endogenous BiP in each sample. WCL, whole-cell lysate. (F) Quantification of two independent pulse-chase experiments as described in D. Means and SDs are plotted.
In an effort to resolve this discrepancy of functional participation of GRP94 in ERAD, we asked whether GRP94 ablation could affect ERAD indirectly. Up-regulation of BiP in response to either inhibition or depletion of GRP94 has been reported (Eletto et al., 2012), and indeed, elevated BiP levels were observed with transient knockdown of GRP94 (Supplemental Figure S2A). In contrast, BiP up-regulation was minimal in our ERAD assays because cells had been maintained under puromycin selection for several weeks after GRP94 depletion, allowing BiP levels to return to baseline (Figure 2B). These data suggest that transient GRP94 knockdowns used in previous studies would have likely resulted in elevated levels of BiP. Because BiP binds nonnative proteins robustly (Figure 1A), high levels of BiP could serve to stall substrate processing.
To ask directly whether BiP up-regulation altered the kinetics of ERAD, we monitored NHK turnover in HEK293T cells transiently expressing increasing amounts of BiP. We observed an impairment of substrate degradation that was proportional to the elevated steady-state levels of BiP (Figure 2, D–F), recapitulating the initial observation of inefficient NHK disposal in GRP94-depleted cells. Thus the coordinate up-regulation of BiP responding to transient loss of GRP94 may explain why impairment to NHK turnover was misconstrued as a direct role for GRP94 in ERAD. It also underscores the importance of considering compensatory cellular mechanisms during gene silencing. Of importance, allowing BiP levels to relax to baseline, as with our puromycin-selected cells, allowed us to demonstrate that GRP94 does not participate directly in ERAD of either glycosylated or nonglycosylated misfolded substrates.
Unique regions mediate the interaction between OS-9 and GRP94
Because our data indicated that GRP94 is dispensable for ERAD, we examined and mapped the physical association between GRP94 and OS-9 in order to gain novel functional insights. Both GRP94 and OS-9 have relatively few known interactions in the ER lumen, and we reasoned that elucidating the domains/surfaces necessary for complex formation might shed light on a distinct function.
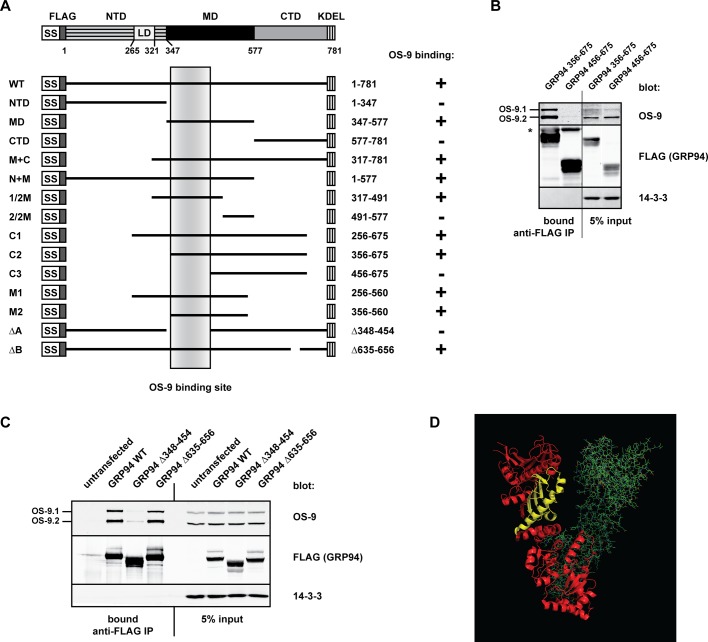
To identify the region of GRP94 necessary to bind OS-9, we designed a panel of GRP94 truncation and deletion mutants to include or exclude prominent structural features (Figure 3A). Crystallographic studies have demonstrated that individual domain structures align closely with those of full-length GRP94 (Dollins et al., 2005, 2007), indicating that the protein has a modular fold and rationalizing the use of truncations to probe interactions. Each FLAG-tagged GRP94 mutant was evaluated for its ability to associate with endogenous OS-9 in HEK293T cells. Systematic analysis of the truncation panel demonstrated that full-length GRP94, as well as constructs containing the middle domain (MD), were sufficient to coimmunoprecipitate OS-9 (summarized in Figure 3A). Specifically, amino acids (aa) 356–456 within the MD of GRP94 were required for OS-9 association (Figure 3B). These residues form a small β-sheet and a long, exposed α-helix within the N-terminal half of the MD (Figure 3D, yellow).
FIGURE 3.
The OS-9–binding site is located in the middle domain of GRP94. (A) Scheme of the domain architecture of GRP94 and the truncation and deletion mutants created to test the OS-9 interaction. OS-9 binding is indicated with a plus or minus sign to the right of each construct. The shaded vertical box refers to residues 356–456 in the middle domain of GRP94, which are common to all the truncations that interact with OS-9 and form the OS-9–binding region. NTD, N-terminal domain; LD, acidic linker domain; MD, middle domain; CTD, C-terminal domain. (B) Coimmunoprecipitation of endogenous OS-9 with the indicated FLAG-tagged GRP94 truncation mutants transiently expressed in HEK293T cells. Samples were harvested and lysed 20 h posttransfection, and GRP94 truncations were immunoprecipitated by anti–FLAG M1 agarose. Whole-cell lysate inputs and the immunoprecipitate bound fractions were examined via immunoblot analysis. 14-3-3, cytosolic control to ensure stringency of immunoprecipitation. Gel lanes were removed in this panel to show relevant samples. (C) FLAG-tagged WT GRP94 or the indicated deletion mutants of GRP94 were expressed in HEK293T cells, and coimmunoprecipitations were conducted as in B. (D) The OS-9–binding surface, indicated in yellow, contains a small β-sheet and a long α-helix in the MD. One GRP94 monomer is shown in ribbon diagram (red/yellow) and one in stick diagram (green); adapted from crystal structure 2O1U.
One functional insight was provided by the deletion of aa 635–656 in the C-terminal domain of GRP94, previously assigned as a client binding site for at least TLRs and integrins (Wu et al., 2012). Deletion of this region had no effect on the OS-9 interaction (Figure 3C), suggesting that OS-9 does not bind as a client to GRP94. To confirm this, we monitored OS-9 stability in cells depleted for endogenous GRP94. Silencing of GRP94 did not affect the steady-state levels of OS-9 (Supplemental Figure S2A), nor did it alter the kinetic turnover of individual OS-9 isoforms (Supplemental Figure S2B). These data reveal that OS-9 folding is not reliant on GRP94 and that OS-9 does not interact with the chaperone as an obligate client.
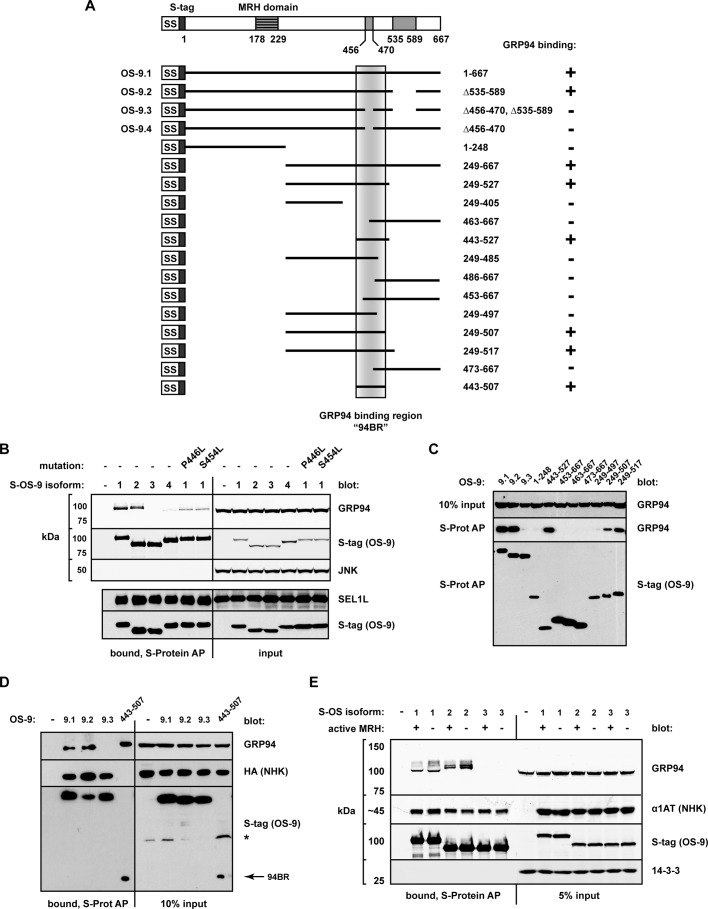
In parallel, we sought to identify the region of OS-9 required to bind GRP94. OS-9 can be readily detected at the protein level as two prominent isoforms (OS-9.1 and OS-9.2; Figure 3, B and C). Both isoforms were recovered in GRP94 immunoprecipitates at the same ratio as their relative cellular expression (Figure 3, B and C), indicating that neither isoform associates with GRP94 preferentially. Additional isoforms (OS-9.3, OS-9.4) are detectable as transcripts but have yet to be fully characterized (Kimura et al., 1998; Supplemental Figure S3; diagrammed in Figure 4A). OS-9.1 and OS-9.2 bind SEL1L and GRP94 through interactions that appear to be mutually exclusive (Christianson et al., 2008). We observed that while SEL1L was coprecipitated equally by all isoforms, GRP94 was bound only by OS-9.1 and OS-9.2 but not by OS-9.3 or OS-9.4 (Figure 4B). The loss of GRP94 binding that resulted from the excision of 15 aa (Δ456–470, in OS-9.3 and OS-9.4) suggested this OS-9 segment contributes directly to the interaction. Further, a naturally occurring somatic mutation (P446L; Vigneron et al., 2002) and a reported single-nucleotide polymorphism (S454L) in this vicinity lead to diminished GRP94 binding as well (Figure 4B), indicating a region sensitive to structural perturbations.
FIGURE 4.
OS-9 binds GRP94 through a region distinct from its MRH domain. (A) Scheme of the known architecture of OS-9, including its splice isoforms (OS-9.1–9.4) and the truncations used to identify the GRP94-binding site. GRP94 binding is indicated with a plus or minus sign to the right of each construct. The shaded vertical box demarcates the GRP94-binding region (94BR, aa 443–507). (B) Western blot analysis monitoring coprecipitation of endogenous GRP94 and SEL1L with OS-9 isoforms and point mutants (P446L, S454L). Each S-tagged OS-9 construct was expressed in HEK293 cells for 20 h, and samples were lysed and subjected to affinity purification by S-protein agarose. Whole-cell lysate inputs and AP-bound fractions were examined by immunoblotting. The blots were developed separately at the indicated lines. JNK, cytosolic control to ensure stringency of the AP. (C) Western blot monitoring coprecipitation of GRP94 with the OS-9 truncations; complexes were isolated and detected as in B. (D) Coprecipitation of NHK-HA and GRP94 by the indicated OS-9 splice isoforms and 94BR (aa 443–507). HEK293 cells were cotransfected with NHK-HA and the indicated S-OS-9 constructs, followed by AP with S-protein agarose and immunoblot analysis. Arrow, 94BR. (E) Coprecipitation of NHK and endogenous GRP94 by WT or MRH-domain mutants (R188A) of S-tagged OS-9 isoforms. AP and analysis as in D.
To define more precisely this GRP94-binding region, we generated a sliding window series of S-tagged truncations of OS-9.1 and monitored coprecipitation of endogenous GRP94 (summarized in Figure 4A). The analysis identified a minimal OS-9 region (aa 443–507) that was both necessary and sufficient for GRP94 interaction at levels comparable to those of full-length OS-9.1 (Figure 4, C and D). This region has been designated the GRP94-binding region (94BR). Secondary structure algorithms (Jpred [Cuff and Barton, 2000]; and PsiPred [Jones, 1999]) predict two extended α-helices within the 94BR (aa 448–471, 485–502) linked by a short, unstructured segment. Fragments bisecting the 94BR through the short linker (e.g., OS-9.1249-497, OS-9.1463-667) failed to support GRP94 binding (Figure 4C). The inability to bisect, mutate, or reduce the 94BR without compromising interaction argues for the mutual contribution of these two helices to form the surface needed to support GRP94 binding. Although the 94BR was sufficient to coprecipitate GRP94, it was not able to bind NHK (Figure 4D). This observation strongly suggests that OS-9 has multiple sites of interaction and indicates that OS-9 recognizes GRP94 and ERAD substrates via distinct regions. Together these data identify the 94BR as the OS-9 region responsible for binding GRP94 (but not for NHK) and raise the possibility that alternative splicing could alter the interaction profile and function of OS-9.
OS-9 does not require its lectin activity to bind GRP94
A prominent feature of OS-9 is its MRH domain, through which OS-9 selectively recognizes trimmed oligosaccharides during ERAD (Szathmary et al., 2005; Hosokawa et al., 2009; Satoh et al., 2010). Given that GRP94 is itself a glycoprotein, we asked whether the MRH domain contributed to the formation or stabilization of the OS-9/GRP94 complex. Mutating conserved residues in the MRH domain (e.g., R188) abolishes oligosaccharide binding by OS-9 (Hosokawa et al., 2009; Satoh et al., 2010). The persistence of GRP94 interaction with OS-9R188A demonstrated that the MRH domain is not required for complex formation (Figure 4E). Consequently, OS-9 recognizes the GRP94 polypeptide in a manner that is not dependent on binding to oligosaccharides. This is consistent with reports of GRP94’s typical glycan structure being mannose8 (Lewis et al., 1985), a form poorly recognized by the OS-9 MRH domain (Hosokawa et al., 2009). The ability of OS-9 to coprecipitate nonglycosylated ERAD substrates (e.g., QQQ; Figure 1B) and the association of OS-9R188A with NHK (Figure 4E) clearly demonstrate a polypeptide-binding capacity for OS-9 (Bernasconi et al., 2008), but one that is likely to be distinct from its interaction with GRP94.
OS-9 preferentially binds to hyperglycosylated forms of GRP94
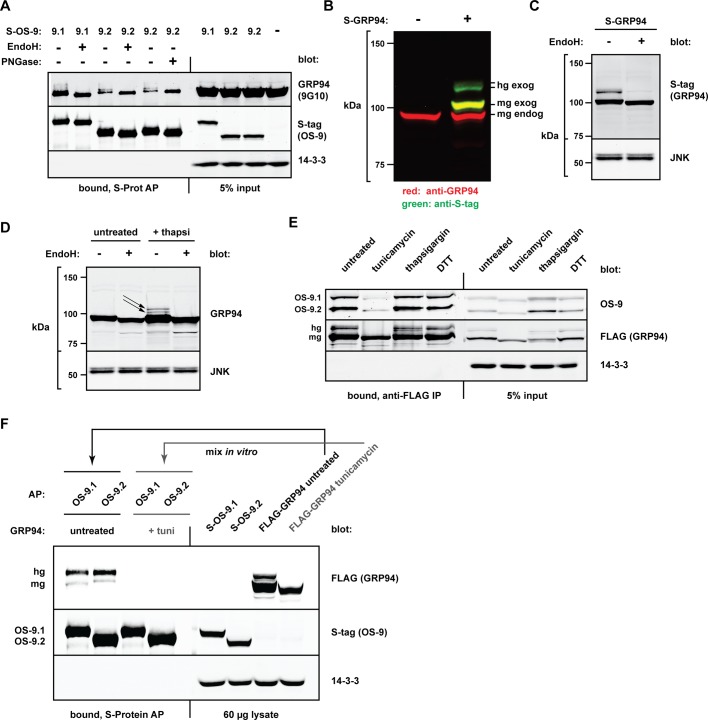
During our investigation, we consistently observed that OS-9 pull downs were enriched in higher–molecular weight forms of endogenous GRP94 (Figures 1B and 4E). Detection of these species was dependent on the sensitivity of immunoblotting, as well as on sufficient electrophoretic resolution. Owing to the ∼3- to 12-kDa shifts of these forms, we hypothesized that they represented a minor cellular population of hyperglycosylated GRP94 (hgGRP94). Mammalian GRP94 contains six canonical N-glycan acceptor sites (N41, N86, N196, N424, N460, N481; residue number starting from the first amino acid after the signal sequence) but is predominantly monoglycosylated at N196 (mgGRP94; Qu et al., 1994). Treatment of GRP94 coprecipitated by S-tagged OS-9 with either endoglycosidase H (EndoH) or peptide-N-glycosidase F (PNGase F) caused the higher–molecular weight species to collapse into a single band corresponding to the unglycosylated form (Figure 5A). This confirms that a small population of endogenous GRP94 is hyperglycosylated under normal growth conditions and can be distinguished by preferential binding to OS-9. Because OS-9R188A still bound robustly to hgGRP94 species (Figure 4E), enrichment of these forms cannot be attributed simply to an increase in available glycans binding more molecules of OS-9. Although the band shifts of GRP94 appeared different between pull downs with OS-9.1 and OS-9.2 (Figures 1B and 4E), we ascribe this to the fact that S-OS-9.1 runs at the same mobility as GRP94 species and alters their apparent size (Supplemental Figure S4). The faster mobility of S-OS-9.2 in SDS–PAGE permits resolution of the true size shifts of hgGRP94.
FIGURE 5.
OS-9 preferentially associates with hyperglycosylated forms of GRP94. (A) HEK293T cells expressing S-OS-9.1 or S-OS-9.2 were harvested 20 h posttransfection and lysed, and complexes were affinity purified with S-protein agarose. Samples were split equally and treated with or without EndoH or PNGase F as described in Materials and Methods. Whole-cell lysates and AP-bound fractions were analyzed via immunoblotting to examine the coprecipitating endogenous GRP94 and visualized with the 9G10 anti-GRP94 antibody. (B) Differential detection of mgGRP94 and hgGRP94 by the monoclonal 9G10 antibody. Lysates from untransfected HEK293T cells or cells expressing S-tagged GRP94 were subjected to immunoblotting with the 9G10 antibody (red) and anti–S-tag (green). hg exog, hyperglycosylated S-GRP94; mg exog, monoglycosylated S-GRP94; mg-endog, monoglycosylated endogenous GRP94. Lanes were removed from this blot to show relevant samples. (C) Whole-cell lysates from cells expressing S-GRP94 were subjected to EndoH (or mock) treatment to observe the size shift of both mgGRP94 and hgGRP94 species. (D) Formation of endogenous hgGRP94 forms (black arrows) in HEK293T cells treated with thapsigargin (0.3 μM, 19 h) with the 9G10 monoclonal anti-GRP94 antibody. Cell lysates from untreated and thapsigargin-treated cells were subjected to EndoH treatment to observe size shifts of both mgGRP94 and hgGRP94 species. (E) FLAG-GRP94 was transiently expressed in HEK293T cells, which were left untreated or treated with tunicamycin (10 μg/ml, 20 h), thapsigargin (3 μM, 20 h), or DTT (1 mM, 1 h). Cells were harvested and lysed, and equal protein amounts were immunoprecipitated by anti–FLAG M1 agarose. Coprecipitating endogenous OS-9 was examined via Western blot analysis. 14-3-3, control to ensure stringency of the immunoprecipitation. hg, hyperglycosylated FLAG-GRP94; mg, monoglycosylated FLAG-GRP94. (F) In vitro coprecipitation of GRP94 by OS-9 from mixed lysates. S-OS-9.1, S-OS-9.2, and FLAG-GRP94 were individually expressed in separate plates of HEK293T cells. FLAG-GRP94–expressing cells were left untreated or treated with tunicamycin (1 μg/ml, 20 h) to prevent glycosylation. S-OS-9.1 and S-OS-9.2 lysates were each mixed with equal amounts of total protein from each FLAG-GRP94 lysate (untreated or tunicamycin treated), and the resulting complexes were affinity purified by S-protein agarose. GRP94 and OS-9 were detected by immunoblot with anti-FLAG and anti–S-tag, respectively.
The conformation-specific, monoclonal antibody 9G10 recognizes an epitope within the first acidic domain of GRP94 (aa 290–350; Edwards et al., 1984; Vogen et al., 2002). Of note, detection of OS-9-bound GRP94 by 9G10 increased dramatically when samples were treated with either EndoH or PNGase F (Figure 5A), suggesting that one or more N-linked glycans sterically hinder access of the antibody to the GRP94 epitope. To explore this, we expressed S-tagged GRP94 (S-GRP94) in HEK293T cells and probed the resulting Western blot with the 9G10 antibody. Two bands corresponding to GRP94 were detected: endogenous mgGRP94 and exogenous monoglycosylated S-GRP94 (Figure 5B, red channel). Probing the same membrane with anti-S-tag revealed the presence of monoglycosylated S-GRP94, as well as a significant population of hyperglycosylated S-GRP94 (Figure 5B, green channel). Thus, even though 9G10 can recognize hgGRP94 (Figures 1B and 4E), it is far more effective in detecting mgGR94. Because 9G10 is widely used to detect GRP94, the minor population of hgGRP94 may have often been overlooked in other studies.
A significant fraction of ectopically expressed GRP94 exists as hyperglycosylated forms (Figure 5B), which are observed regardless of the epitope tag's type or location (unpublished data). Two hgGRP94 bands are typically resolved by SDS–PAGE, but treatment with kifunensine, a mannosidase I inhibitor, distinguishes them into three clear species, ∼9–15 kDa larger than mgGRP94, which corresponds to GRP94 glycosylated on three to five sites (Supplemental Figure S5). Like the endogenous hgGRP94, these species are sensitive to EndoH (Figure 5C). Of importance, these species are not formed due to disrupted ER homeostasis (Supplemental Figure S6, A and B). In addition, ectopic expression does not result in gross overexpression of the chaperone, as exogenous GRP94 is expressed less than endogenous GRP94 at all expression conditions tested (Supplemental Figure S6B). These levels are well within the normal range of GRP94 induction during the unfolded protein response (UPR). To determine whether excessive synthesis induces the formation or stabilization of hgGRP94, we treated HEK293T cells with thapsigargin overnight to activate the UPR and induce transcription and translation of endogenous GRP94. Indeed, EndoH-sensitive hgGRP94 species were formed during the response to ER stress (Figure 5D). Together these data reveal that small populations of GRP94 containing more than one N-linked glycan are present during normal growth conditions but are also created or stabilized when excess GRP94 is produced or when the ER environment is disturbed.
Because OS-9 preferentially binds hgGRP94 forms, yet does so independently of its MRH domain, we hypothesized that glycosylation of GRP94 alters its polypeptide conformation to expose the binding site for OS-9. If this is the case, a nonglycosylated GRP94 should be poorly recognized by OS-9, and in fact, treatment with the glycosylation inhibitor tunicamycin markedly reduced coimmunoprecipitation of OS-9 with GRP94 (Figure 5E). This was not due to UPR induction per se, because other ER stress inducers, such as thapsigargin and dithiothreitol (DTT), did not have the same effect. Tunicamycin also prevented glycosylation of OS-9 (N177), however, which could be contributing to complex formation. To rule this out, we expressed S-OS-9 and FLAG-GRP94 separately in HEK293T cells, treated the latter with or without tunicamycin, mixed the resulting lysates in vitro, and affinity purified complexes via OS-9. Whereas GRP94 from untreated cell lysates bound robustly to OS-9 (principally as hyperglycosylated forms), nonglycosylated GRP94 could not be coprecipitated (Figure 5F).
In contrast to the effects of tunicamycin, which completely inhibits glycosylation, treatment of cells with the mannosidase inhibitor kifunensine did not prevent the GRP94/OS-9 interaction (Supplemental Figure S7A). In fact, more hgGRP94 species were coprecipitated by OS-9, presumably because more molecules of OS-9 were available to sequester GRP94 in the absence of trimmed glycans on other substrates and ERAD components. Conversely, kifunensine reduced the interaction of OS-9 with its substrate NHK (Supplemental Figure S7B), further confirming that OS-9 recognizes substrates and GRP94 in fundamentally distinct manners. Collectively these data support a model in which glycosylation of GRP94 causes a surface in the MD to be exposed, which can then be recognized and bound by the 94BR of OS-9.
hgGRP94 species have altered conformations and decreased ATP binding
N-linked glycans are large, hydrophilic moieties that can facilitate protein folding through chaperone interactions (Michalak et al., 2009), promoting hydrophobic collapse (Jitsuhara et al., 2002), and preventing off-target folding pathways (Gidalevitz et al., 2013). However, oligosaccharides placed in regions of amino acid packing or within active sites can disrupt protein conformation and activity. Based on the crystal structure of GRP94 (Supplemental Figure S8; Dollins et al., 2007), only N424 and the constitutively modified N196 map to loop regions, the preferred location for oligosaccharide attachment (Zielinska et al., 2010). Other visible glycan acceptor sites map to α-helices, with one site (N86) clearly predicted to disrupt the ATP-binding pocket. This suggested that hgGRP94 forms might represent a pool of nonnative and possibly nonfunctional molecules.
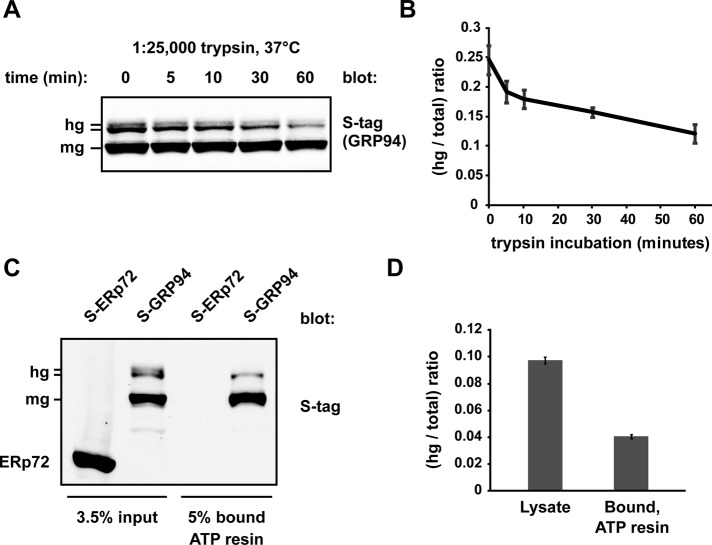
In results described earlier, we observed reduced reactivity of hgGRP94 with 9G10 (Figure 5B), an antibody whose binding reflects the state of the linker domain of GRP94 and its coordination with the N-terminal, nucleotide-binding domain (Vogen et al., 2002). To gain further evidence for abnormal hgGRP94 conformations, we performed limited trypsin proteolysis of lysates containing S-tagged GRP94. Bands corresponding to hgGRP94 were preferentially digested during a 1 h time course (Figure 6A), reflected by a decrease in the ratio of hg:total GRP94 determined from quantification of hgGRP94 and mgGRP94 band intensities (Figure 6B). This result suggests that hgGRP94 forms are folded less compactly than mgGRP94 in solution.
FIGURE 6.
hgGRP94 species exhibit altered conformation and lower ATP-binding activity. (A) S-GRP94–containing cell lysate was treated with dilute trypsin (1:25,000) for the indicated time course and then subjected to SDS–PAGE and immunoblotting with the anti–S-tag antibody. hg, hyperglycosylated S-GRP94; mg, monoglycosylated S-GRP94. (B) Quantification of three independent limited proteolysis experiments as in A. Bands corresponding to hgGRP94 were quantified as a fraction of total GRP94 and plotted over the 1-h time course. Means and SDs are plotted. (C) Affinity purification of S-ERp72 and S-GRP94 by γ-phosphate–linked ATP resin. Whole-cell lysate inputs and ATP-bound fractions were separated by SDS–PAGE and the resulting immunoblots probed with anti–S-tag. S-ERp72 was used as a control to ensure the ATP resin was specific for ATPases. (D) Quantification of three independent affinity purification experiments as in C; the hgGRP94 bands were quantified as a fraction of the total GRP94. Means and SDs are plotted.
The structural changes apparent in the hgGRP94 population prompted the question of whether hgGRP94 and mgGRP94 bound ATP with different capacities. Because ATP binding and hydrolysis are essential for activity (Ostrovsky et al., 2009), differential ATP binding would reflect altered functionality. We monitored the hg:total GRP94 ratio in S-GRP94–containing lysates and in the fraction bound to γ-phosphate–linked ATP resin (Haystead et al., 1993; Hughes et al., 2012). Though not completely excluded, hgGRP94 was preferentially lost during the enrichment of active mgGRP94 bound to ATP (Figure 6C). This suggests that hgGRP94 adopts the proper ATP-binding conformation less than half as well as mgGRP94 (Figure 6D). Moreover, the most heavily glycosylated form failed to bind ATP at all and likely represents a species of hgGRP94 modified at the N86 glycan acceptor site in the ATP-binding pocket. The fact that GRP94 functions as a dimer must also be considered, as the residual hgGRP94 bound to ATP may do so indirectly through a mgGRP94 partner.
GRP94 glycosylation state influences its rate of degradation
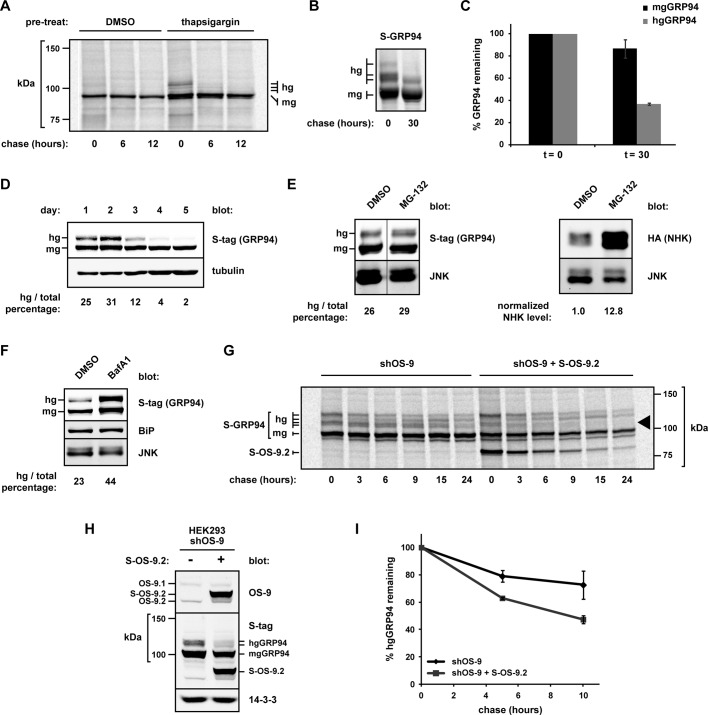
Because aberrant glycosylation alters both GRP94 conformation and ATP-binding activity, we hypothesized that hgGRP94 species may be subject to increased turnover in cells. We first examined the disposal of endogenous GRP94 after pretreatment with thapsigargin or vehicle control (dimethyl sulfoxide [DMSO]). hgGRP94 species were synthesized only after the thapsigargin treatment and were rapidly cleared from the cells, whereas mgGRP94 in both conditions was much longer lived (Figure 7A). In addition, we monitored the turnover of S-tagged GRP94 in HEK293T cells under no external stress. Whereas mgGRP94 was very stable, with ∼90% remaining after a 30 h chase, the hgGRP94 forms (quantified together) were degraded more rapidly, with half-life t1/2 < 20 h (Figure 7, B and C). The difference in half-life of endogenous and exogenous hgGRP94 is likely due to indirect effects of ER stress in Figure 7A, but in both cases hgGRP94 species were degraded significantly faster than mgGRP94. To explore differential cellular fates further, we examined S-GRP94 transiently expressed in HEK293T cells over 5 d. hgGRP94 forms accumulated during the first 2 d of expression but were selectively degraded as the expression plasmid was lost/silenced. In contrast, levels of exogenous mgGRP94 remained stable for the duration of the assay (Figure 7D). These data highlight the differential turnover of GRP94 populations and demonstrate that atypical glycosylation marks molecules with decreased biological stability.
FIGURE 7.
Hyperglycosylated GRP94 is subject to enhanced turnover via OS-9. (A) Pulse-chase analysis of endogenous GRP94 from untreated HEK293T cells or cells pretreated with thapsigargin (0.3 μM, 14 h). The 9G10 anti-GRP94 antibody was used to isolate GRP94 at the indicated time points. hg, hyperglycosylated GRP94; mg, monoglycosylated GRP94. (B) Pulse-chase analysis of S-GRP94 expressed in HEK293T cells. S-protein agarose was used to isolate GRP94 at the indicated time points. hg, hyperglycosylated S-GRP94; mg, monoglycosylated S-GRP94. (C) Quantification of hgGRP94 species (collectively) from three independent experiments as described in B. Means and SDs are plotted. (D) Western blot analysis of transiently expressed S-GRP94 in HEK293Ts over 5 d. hgGRP94 and mgGRP94 forms were detected by anti–S-tag, and the hg:total GRP94 percentage was calculated for each sample (indicated at bottom of blot). Increasing amounts of lysates were loaded for each sample due to cell growth. (E) S-GRP94–expressing HEK293T cells were treated with either DMSO or MG-132 (1 μM, 24 h). S-GRP94 levels were monitored and quantified via Western blot analysis. The percentage of hg:total GRP94 was calculated and is labeled under the respective lanes. Irrelevant gel lanes were digitally removed at the vertical line. To confirm proteasomal inhibition, NHK-HA–expressing cells were treated with DMSO or MG-132 (1 μM, 18 h), and lysates were evaluated for NHK levels by immunoblot. NHK was normalized to the JNK loading control. (F) HEK293T cells expressing S-GRP94 were treated with either DMSO or bafilomycin A1 (50 nM, 17 h). Samples were lysed and subjected to SDS–PAGE and immunoblot analysis for determination of GRP94 levels, as in E. The percentage of hg:total GRP94 was calculated and is indicated under the respective lanes. (G) HEK293 cells stably depleted of OS-9 (shOS-9) were transiently transfected with either S-GRP94 alone or with S-GRP94 and S-OS-9.2. The rate of degradation of GRP94 was determined by pulse-chase analysis; GRP94 was isolated at each time point with S-protein agarose. Black arrowhead, two hgGRP94 species with enhanced degradation upon OS-9 complementation. Note that OS-9.2 is also isolated at each time point because it shares the same S-tag epitope as GRP94. (H) Immunoblot of lysates from the cells used in G, showing the increase in OS-9 protein levels upon S-OS-9.2 expression. (I) Quantification of two independent experiments as in G (other than G). Total hgGRP94 is plotted relative to the pulse. Means and SDs are plotted.
hgGRP94 is degraded via OS-9 in a lysosomal-like pathway
Disposal of a less stable hgGRP94 population from the ER lumen could occur via the proteasome-dependent ERAD process or through an autophagy/lysosomal pathway. Given OS-9’s established role in ERAD, we first measured hgGRP94 stability in cells treated with the proteasome inhibitor MG-132 or expressing a dominant-negative form of the ubiquitin ligase Hrd1 (Hrd1C294A). Neither condition was sufficient to stabilize the hgGRP94 population, suggesting that ERAD was not responsible for their clearance (Figure 7E; unpublished data). Because OS-9 does not interact with GRP94 and ERAD substrates through the same region (Figure 4D), this result is not entirely unexpected. In contrast, cells treated with the vacuolar-type H+-ATPase inhibitor bafilomycin A1 exhibited significantly higher steady-state levels of hgGRP94, as well as an increased hg:total GRP94 ratio (Figure 7F). These data point toward the clearance of hgGRP94 from cells in a proteasome-independent manner, through bafilomycin A1-sensitive acidic compartments.
Finally, we asked whether the turnover of hgGRP94 was dependent on OS-9. Using HEK293 cells stably depleted for OS-9 by a 3′ untranslated region–targeted shRNA (shOS-9; Bernasconi et al., 2008), we monitored clearance of S-tagged GRP94. When S-OS-9.2 was recomplemented in these cells, hgGRP94 turnover was markedly enhanced (Figure 7, G–I). In fact, certain glycosylated species (Figure 7G, black arrowhead) were chased with similar kinetics to OS-9 itself. We conclude that OS-9 does not simply sequester hgGRP94 species but can also facilitate their disposal.
DISCUSSION
To investigate the links between protein folding and disposal in the ER, we examined in detail the complex formed between GRP94 and OS-9. Despite the many potential roles for this interaction, our data demonstrate that OS-9 sequesters and degrades a hyperglycosylated population of nonnative GRP94 molecules.
GRP94-depleted cells were shown previously to be impaired in their ability to degrade the misfolded substrate NHK (Christianson et al., 2008), implicating GRP94 in ERAD. However, our analysis now shows that this observation cannot be attributed to GRP94 directly, since GRP94-deficient cells are able to degrade glycosylated NHK and nonglycosylated NS1 with the same kinetics as GRP94-sufficient cells. The likely explanation for the previous findings is that transient depletion of GRP94 induced the expression of BiP, which we show here can trap NHK in the ER and delay its degradation.
If it does not function in ERAD, what purpose would a GRP94/OS-9 complex serve? In considering alternative roles, we demonstrated that GRP94 binds OS-9 at a region distinct from its TLR and integrin-binding site (Yang et al., 2007) and that OS-9 half-life, steady-state levels, and substrate binding are all unaffected by the loss of GRP94. Thus OS-9 is not reliant upon GRP94 for its own folding, nor is GRP94 required for OS-9 recognition of ERAD substrates. Several lines of evidence suggest that the uniqueness of the interaction between GRP94 and OS-9 may underlie an alternative function for the complex.
First, the N-terminal half of the GRP94 MD is essential for interaction with OS-9. Little is known about the MD of GRP94, except that its alignment with the N-terminal domain (NTD) is required for formation of the split ATPase-active site (Meyer et al., 2003; Chu et al., 2006; Dollins et al., 2007). The corresponding MD of the cytosolic HSP90 paralogue contains multiple client- and cofactor-binding sites, including one for the cofactor Aha1, which accelerates the ATPase activity of HSP90 (Lotz et al., 2003; Meyer et al., 2003). However, since OS-9 shares no homology with Aha1, their modes of interaction are likely to differ. The only known cofactor of GRP94 is CNPY3, which binds to its NTD (Liu et al., 2010). Thus OS-9 is the first protein known to bind GRP94 via the MD.
Second, a region in the C-terminal portion of OS-9, which we designated the 94BR, is both necessary and sufficient for association with GRP94. Of note, this region is disrupted in two OS-9 splice variants that fail to bind GRP94, suggesting that alternative splicing may alter the functionality and specificity of OS-9. The 94BR is not sufficient to bind NHK, indicating that OS-9 does not bind GRP94 as an ERAD substrate. Glycosylation of GRP94 is required for OS-9 binding, yet OS-9’s MRH domain is dispensable. The resolution of this apparent paradox comes from the notion that oligosaccharide-induced conformation changes in the GRP94 polypeptide are necessary for OS-9 recognition.
Third, although its MRH domain is not required for GRP94 binding, OS-9 preferentially associates with a minor population of endogenous hyperglycosylated GRP94. Whereas the majority of GRP94 is monoglycosylated, hgGRP94 species are observed with N-glycans on additional one to four cryptic acceptor sites. An analysis of the potential glycan acceptor sites on the crystal structure of GRP94 hinted that extraneous glycans would disrupt its conformation (Zielinska et al., 2010). Indeed, hgGRP94 species are more sensitive to trypsin proteolysis, have a lower avidity to a conformation-specific antibody, and have impaired binding to ATP, all indications that these species are nonnative. Their altered structure enhances association with OS-9 and leads to a clear cellular fate: hgGRP94 forms are degraded much more rapidly than mgGRP94.
hgGRP94 species have been observed in both mammalian and insect cells (Mazzarella and Green, 1987; Kang and Welch, 1991; Qu et al., 1994; Feldweg and Srivastava, 1995), although it was not clear whether this was solely an overexpression artifact. More recently, Yang et al. (2007) showed that hgGRP94 forms exist endogenously and associate with clients TLR4 and TLR9. Clearly, whereas improper glycosylation may affect certain activities of GRP94, the C-terminal TLR-binding site appears to be unaffected by conformational changes elsewhere in the molecule. Note that the altered conformation of hgGRP94 might interfere with proper ATP hydrolysis, trapping complexes with a subset of the client pool. Such interactions would be unproductive for folding of clients and thus highlight the importance of clearing a malfolded chaperone from the ER.
How do hgGRP94 species form? Their presence at low abundance in unstressed cells demonstrates that these species are synthesized constitutively. However, even minor overproduction of GRP94 (via either the UPR or ectopic expression) can lead to an increased fraction of hyperglycosylated forms. Because cells have the ability to monitor the activity (rather than quantity) of GRP94 (Eletto et al., 2012), excess GRP94 could be regulated in an active process. However, production of correctly folded GRP94 could also depend on QC factors acting on the folding process of GRP94 itself. For instance, in addition to acting coordinately on clients (Melnick et al., 1994), BiP association may contribute to the fidelity of GRP94 folding as it enters the ER lumen, perhaps by associating with the NTD while the C-terminus of GRP94 is being synthesized (Melnick et al., 1992; Molinari and Helenius, 2000). More recently, cyclophilin B was shown to bind GRP94. This interaction may be mediated via the acidic linker domain of GRP94 and could assist in the isomerization of proline residues (Jansen et al., 2012). Such resources may become overwhelmed at times of increased protein load or decreased fidelity, leading to improper folding of GRP94 and exposure of cryptic glycosylation sites.
From our study, we cannot unequivocally distinguish whether aberrant glycosylation of hgGRP94 occurs cotranslationally or posttranslationally. However, it has been demonstrated that the STT3B isoform of the oligosaccharyltransferase (OST) complex is used to posttranslationally glycosylate proteins containing acceptor sites within the ∼55 most-C-terminal residues (Shrimal et al., 2013). Because GRP94’s last acceptor site lies >300 residues from the C-terminus, STT3B-mediated posttranslational glycosylation is not predicted. In addition, because hgGRP94 species are observed immediately after pulse labeling, we favor the interpretation that excessive glycosylation of GRP94 occurs cotranslationally.
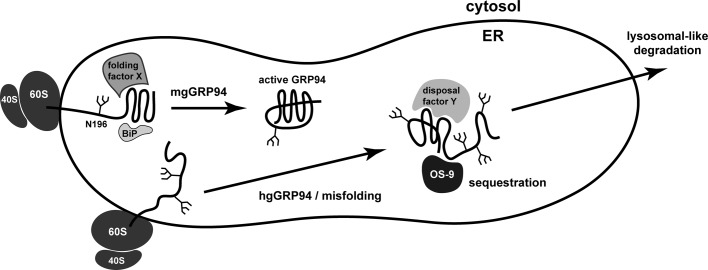
We envision a model in which the folding of the NTD dictates the fate of each GRP94 molecule (Figure 8). In unpublished observations, we confirmed the results of Qu et al. (1994), which demonstrate that alterations to the NTD of GRP94 force the use of downstream glycan acceptor sites, underlying the importance of this early domain to proper regulation of glycosylation. As GRP94 is inserted into the ER lumen during translation, proper folding of the NTD would result in the addition of a single glycan at N196, perhaps due to interactions with folding factors such as BiP. This monoglycosylated form is an active chaperone and represents the vast majority of GRP94 in the ER. Alternatively, the NTD of GRP94 may be inefficiently glycosylated at alternate positions or the NTD in a small percentage of molecules may misfold, exposing downstream cryptic glycosylation sites. Addition of extra glycans may be a stochastic process, governed principally by the accessibility and orientation of downstream acceptor sites to the OST complex. Once the aberrant modifications occur, hgGRP94 forms are then subject to specialized recognition by OS-9. OS-9 may sequester these species away from client proteins, perhaps by removing them toward the ERQC compartment, where OS-9 is reportedly localized (Leitman et al., 2014). OS-9 then facilitates the disposal of the hgGRP94 species, although our model does not exclude the possibility that other factors may also contribute.
FIGURE 8.
Model for the formation and disposal of hgGRP94. Our data support a model in which the glycosylation fate of GRP94 is determined as the chaperone enters the ER lumen. If folded properly, the NTD of GRP94 receives a single oligosaccharide at N196 and enters the pool of active chaperone (mgGRP94). However, if the folding factors acting on GRP94 (e.g., BiP) are exhausted or if the NTD spontaneously misfolds, aberrant glycosylation on cryptic acceptor sites can result, leading to the production of hgGRP94 species. Modification of these cryptic sites subsequently alters both the conformation and activity of GRP94. The hgGRP94 pool is preferentially sequestered by the ER-resident lectin OS-9 through a specialized polypeptide recognition domain (94BR) and not direct oligosaccharide binding. GRP94 is cleared by OS-9 through an ERAD-independent, lysosomal-like process. Because hgGRP94 species form endogenously under normal growth conditions, this is a constitutive process; however, it is enhanced when GRP94 synthesis is induced ectopically or via the UPR.
Degradation of hgGRP94 occurs within acidic compartments and not by proteasomes via ERAD. Although autophagy could explain these observations, hgGRP94 clearance may also be mediated via ERAD tuning vesicles, the route of degradation for OS-9, SEL1L, and EDEM1 (Bernasconi et al., 2012). hgGRP94 preferentially associates with OS-9, and so these species may be packaged into vesicles together. Analysis of the cellular localization of GRP94 and OS-9 is required for further evidence of this hypothesis.
Protein glycosylation is clearly a useful mechanism for cellular control of protein structure, function, and fate. Oligosaccharides mediate chaperone interactions and can be beneficial for the biophysics of protein folding (Jitsuhara et al., 2002; Michalak et al., 2009; Gidalevitz et al., 2013). In addition, trimming of glycans provides key signals for the initiation and timing of ERAD (Tokunaga et al., 2000). Recently it was shown that posttranslational glycosylation of the transthyretin mutant D18G routed it to ERAD (Sato et al., 2012). Here we show that when GRP94 is synthesized in abundance, extraneous glycans are added to signal for faster degradation by a non-ERAD pathway. Thus hyperglycosylation of cryptic acceptor sites can be used to control protein function and turnover, in particular of a long-lived ER-resident chaperone.
MATERIALS AND METHODS
Chemicals and antibodies
The following chemicals were used: thapsigargin (MP Biomedicals, Santa Ana, CA), tunicamycin and kifunensine (Calbiochem, Billerica, MA), puromycin (InvivoGen, San Diego, CA), and bafilomycin A1, MG-132, and DTT (Sigma-Aldrich, St. Louis, MO). For Western blot analyses, the following antibodies were used: anti-FLAG M1 and anti-tubulin (Sigma-Aldrich), anti–α1-antitrypsin (Dako, Glostrup, Denmark), anti–14-3-3z and anti-SEL1L (Santa Cruz Biotechnology, Dallas, TX), anti–S-tag and anti–OS-9 (Abcam, Cambridge, England), anti–GRP94 9G10 and anti-KDEL (Enzo, Farmingdale, NY), anti-GRP94 (provided by Ineke Braakman, Utrecht University, Utrecht, Netherlands), anti-HA (Covance, Princeton, NJ), anti-BiP (BD Biosciences, San Jose, CA), and anti-PERK and anti-JNK (Cell Signaling Technologies, Danvers, MA). Secondary antibodies were obtained from LI-COR (IRDye 680 and 800 nm, for use with the LI-COR Odyssey system [Lincoln, NE]) and Santa Cruz Biotechnology (horseradish peroxidase conjugated).
Plasmids and cloning
Untagged α1-antitrypsin constructs (WT, NHK, PIZ, and AAA) were gifts from Daniel Hebert (University of Massachusetts, Amherst, MA), and Oscar Burrone (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy) kindly provided V5-tagged NS1. S-tagged isoforms of OS-9 and NHK-hemagglutinin (HA) were provided by Ron Kopito (Stanford University, Stanford, CA) and were described previously (Christianson et al., 2008). Site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA) was used to generate all point mutants described. For the nonglycosylated version of NHK, the QQQ variant of α1-antitrypsin was created by mutating the three glycan acceptor Asn residues with Gln, whereas the AAA construct was provided by retaining the Asn but mutating the third amino acid in the N-X-S/T sequence to alanine. S-BiP and S-GRP94 were generated by subcloning the chaperones’ mature sequences (amplified from HeLa cells by reverse transcriptase [RT]-PCR) into the S-OS-9.1 vector, digested with AgeI and XbaI, containing the bovine preprolactin signal sequence followed by the S-tag. All S-tagged OS-9 truncations were created by PCR amplification of the indicated sequences and inserted into the S-OS-9.1 vector digested with AgeI and XbaI. FLAG-tagged GRP94 was constructed from a murine anionic-trypsin II signal sequence, followed by the FLAG peptide and the mature WT murine GRP94 sequence, cloned into the pcDNA3.1(+)Zeo vector (Invitrogen, Carlsbad, CA) using NheI and NotI restriction sites. This plasmid was modified to be resistant to shRNA as previously described (Eletto et al., 2012). FLAG-tagged GRP94 truncations were constructed by PCR amplification of restriction sites appended to the respective N- and C-terminal residues for each mutant and cloned into the WT FLAG-GRP94 vector digested with BamHI and NotI. GRP94 truncations lacking the natural C-terminus were engineered with the KDEL ER-retrieval sequence. Deletion mutants of FLAG-GRP94 were created by site-directed mutagenesis, which introduced two AgeI sites at either side of the sequence to be excised. Plasmids were then digested with AgeI and religated to remove the indicated regions.
Cell culture and immunoblotting
Stable shOS-9 HEK293 cells were a kind gift of Maurizio Molinari (Institute for Research in Biomedicine, Bellinzona, Switzerland). HEK293T, HEK293, HEK293 shOS-9, and NIH3T3 cells were maintained in DMEM (Life Technologies, Carlsbad, CA and Lonza, Basel, Switzerland) supplemented with PSG (100 U/ml penicillin, 100 μg/ml streptomycin, 2.92 mg/ml l-glutamine, Life Technologies), and 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) at 37°C in a 5% CO2 incubator. shRNAs (Sigma-Aldrich) were packaged into lentivirus in HEK293T cells, and infections were carried out as described previously (Eletto et al., 2012). Transient transfection experiments were conducted when cells reached 70–90% confluency, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were generally harvested 20 h posttransfection for immunoblot analysis. Briefly, cells were washed in phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 5 mM KCl, 5 mM MgCl2, 1% NP-40) supplemented with protease inhibitors (Roche, Basel, Switzerland) and 20 mM iodoacetamide (Sigma-Aldrich). After 20 min on ice, samples were spun at 13,000 × g, and postnuclear supernatant fractions were further analyzed. Protein concentration was determined using the Pierce bicinchoninic acid (BCA) assay (Thermo, Waltham, MA) and quantified on a Synergy HT plate reader (Bio-Tek, Winooski, VT). Samples were diluted in reducing Laemmli buffer and run on acrylamide gels. To observe the small size differences of hgGRP94 species, 7 or 8% acrylamide gels were typically used. Separated proteins were transferred to nitrocellulose or polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA) and blocked in 5% nonfat milk in Tris-buffered saline (TBS). Primary antibodies were diluted in 1% nonfat milk in TBS plus 0.1% Tween (TBST) and incubated with membranes overnight. Secondary antibodies were diluted 1:10,000 in 5% nonfat milk in TBST. Membranes were either probed with ECL reagent (Pierce) and exposed to film or scanned directly using the Odyssey scanner (LI-COR).
Pulse-chase and immunoprecipitation assays
Coprecipitation studies and pulse-chase analyses were conducted using the following purification reagents: anti–FLAG M1 agarose (Sigma-Aldrich), S-protein agarose (Novagen, Billerica, MA), anti–HA agarose (Sigma-Aldrich), anti–V5 agarose (Sigma-Aldrich), or protein G Sepharose (Invitrogen) for other antibodies. For coprecipitation experiments, lysates were incubated with the respective purification reagent and buffer TNNB (50 mM Tris, pH 7.5, 250 mM NaCl, 0.5% NP-40, 0.1% bovine serum albumin [BSA]) for 3–5 h. Beads were spun for 30 s at 1200 rpm, washed in buffer TNN (TNNB without BSA) four times, and finally resuspended in reducing Laemmli buffer. For samples treated with EndoH or PNGase F (New England Biolabs, Ipswich, MA), beads were instead resuspended in water and denaturing buffer or water and buffer G7, respectively, and treated according to the manufacturer's protocol. Samples were separated by SDS–PAGE, and Western blot analysis was performed as described. For consistency between figures, in some panels the immunoprecipitate bound fractions were digitally moved to the left side of the panel, with whole-cell lysate inputs shown on the right. Other digital alterations, such as the removal of irrelevant lanes, are indicated in the respective legends.
For pulse-chase experiments, 60-mm plates of cells were transiently transfected at 70–90% confluency with the plasmid(s) of interest. At 20 h posttransfection, cells were split to poly-d-lysine–coated plates. One day later, cells were washed with Hank's buffered saline solution (Life Technologies) and starved in labeling medium (DMEM without l-glutamine/l-methionine/l-cystine [Corning, Corning, NY] supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 2.92 mg/ml l-glutamine (Life Technologies), 10 mM HEPES, 1 mM sodium pyruvate [Mediatech, Corning, NY], and 3% dialyzed Hyclone [Little Chalfont, United Kingdom] serum) for 20 min. Cells were labeled for 20–30 min with [35S]methionine/cysteine diluted into labeling medium at ∼88 μCi/ml (Environmental Health and Radiation Safety, University of Pennsylvania, Philadelphia, PA). After labeling, cells were washed in ice-cold PBS supplemented with 5 mM each of methionine and cysteine (Calbiochem), followed by chase medium (growth medium with 5 mM methionine/cysteine), and finally incubated with fresh chase medium for the indicated times. At each time point, cells were harvested using the lysis procedure described. Labeled proteins of interest were isolated from lysates using the reagents described for 20 h, washed twice in TNN buffer, and resuspended in reducing Laemmli buffer. Samples were separated by SDS–PAGE with autoradiography of the dried gels carried out on a Typhoon 9200 (Amersham Biosciences, Little Chalfont, United Kingdom). Quantification was performed using ImageQuant software (GE Healthcare, Little Chalfont, United Kingdom).
RT-PCR
For the identification of OS-9 splicing isoforms, total RNA was extracted from cell pellets via an RNA extraction kit (Qiagen, Venlo, Netherlands). RNA concentration was determined by spectrophotometer and the quality assessed by conventional agarose gel electrophoresis. cDNA was synthesized using 1 μg of total RNA, Reverse Transcriptase AMV (Roche), and random hexamers p[dN]6 (Roche). Semiquantitative PCR was performed using KAPA2G Fast PCR Kit (Kapa Biosystems, Wilmington, MA). PCR products were separated by electrophoresis on a 1.5% agarose gel. Standard PCRs of individual OS-9–containing vectors were carried out as a direct comparison of band size. Primer sequences were as follows: forward, 5′-AGCCCGACCAAGGATGATACAGTAAG-3′, and reverse, 5′-AGTCAGCCAACGTGCACCCTC-3′.
RT-PCR of XBP-1 was conducted on RNA samples extracted by TRIzol (Life Technologies). cDNA was synthesized with SuperScript II Reverse Transcriptase (Life Technologies) using the manufacturer's protocol. XBP-1 was amplified with the following primers: forward, 5′-AAACAGAGTAGCAGCTCAGACTGC-3′, and reverse, 5′- TCCTTCTGGGTAGACCTCTGGGAG-3′. PCR products were separated by electrophoresis on a 2% agarose gel, and band intensities corresponding to spliced and unspliced XBP-1 were quantified using ImageJ (National Institutes of Health, Bethesda, MD).
Determination of GRP94 conformation and activity
Limited proteolysis of S-GRP94–containing lysates was conducted as follows: lysates were prepared in lysis buffer without protease inhibitors and quantified via BCA assay. Equal amounts of lysate were aliquoted and subject to 1:25,000 dilution of trypsin (trypsin:lysate; Promega, Madison, WI) in triplicate. Samples were quenched with reducing Laemmli buffer at the indicated time points and subjected to SDS–PAGE and immunoblotting as described. To determine binding of GRP94 to ATP, S-ERp72– or S-GRP94–containing lysates were incubated with γ-phosphate–linked ATP resin (a kind gift of Timothy Haystead, Duke University, Durham, NC) as described previously (Hughes et al., 2012). Samples were then subjected to SDS–PAGE and immunoblotting procedures as described.
Supplementary Material
Acknowledgments
We thank Daniel Hebert, Oscar Burrone, and Ron Kopito for their kind contribution of plasmids; Timothy Haystead for the gift of γ-phosphate-linked ATP resin; and Maurizio Molinari for the shOS-9 HEK293 cells. We acknowledge the generosity of Ineke Braakman for use of antibodies and Stephen Seeholzer (The Children's Hospital of Philadelphia) and The Children's Hospital of Philadelphia Protein Core Facility for reagents. We also thank R. Kopito, J. Burkhardt, T. Gidalevitz, M. Marzec, D. Eletto, S. Boyle, V. Kulkarni, and J. Thomas for helpful discussions and comments. This work was funded by National Institutes of Health grants to Y.A. (AI-18001 and GM-77480) and by funding from Ludwig Cancer Research to J.C. D.D. was funded by National Institutes of Health Training Grants F31-NS084666-01 and T32-GM8275.
Abbreviations used:
- 94BR
GRP94-binding region of OS-9
- α1AT
α1-antitrypsin
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- hgGRP94
hyperglycosylated GRP94
- mgGRP94
monoglycosylated GRP94
- MRH
mannose-6-phosphate receptor homology
- NHK
null Hong Kong α1AT
- NS1
nonsecreted 1 immunoglobulin light chain
- QC
quality control
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0805) on June 4, 2014.
The authors declare no financial conflicts of interest.
REFERENCES
- Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, Molinari M. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol Cell. 2012;46:809–819. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Pertel T, Luban J, Molinari M. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J Biol Chem. 2008;283:16446–16454. doi: 10.1074/jbc.M802272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Biswas C, Ostrovsky O, Makarewich CA, Wanderling S, Gidalevitz T, Argon Y. The peptide-binding activity of GRP94 is regulated by calcium. Biochem J. 2007;405:233–241. doi: 10.1042/BJ20061867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harbor Perspect Biol. 2013;5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral CM, Liu Y, Moremen KW, Sifers RN. Organizational diversity among distinct glycoprotein endoplasmic reticulum-associated degradation programs. Mol Biol Cell. 2002;13:2639–2650. doi: 10.1091/mbc.E02-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillaron J, Haas IG. Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol Biol Cell. 2000;11:217–226. doi: 10.1091/mbc.11.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Maynard JC, Chiosis G, Nicchitta CV, Burlingame AL. Identification of novel quaternary domain interactions in the Hsp90 chaperone, GRP94. Protein Sci. 2006;15:1260–1269. doi: 10.1110/ps.052065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier JH, Tamura T, Sunryd JC, Hebert DN. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Dollins DE, Immormino RM, Gewirth DT. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. J Biol Chem. 2005;280:30438–30447. doi: 10.1074/jbc.M503761200. [DOI] [PubMed] [Google Scholar]
- Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul JL, Burrone OR, Argon Y. A conditional secretory mutant in an Ig L chain is caused by replacement of tyrosine/phenylalanine 87 with histidine. J Immunol. 1992;149:1927–1933. [PubMed] [Google Scholar]
- Edwards DP, Weigel NL, Schrader WT, O'Malley BW, McGuire WL. Structural analysis of chicken oviduct progesterone receptor using monoclonal antibodies to the subunit B protein. Biochemistry. 1984;23:4427–4435. doi: 10.1021/bi00314a029. [DOI] [PubMed] [Google Scholar]
- Eletto D, et al. Limitation of individual folding resources in the ER leads to outcomes distinct from the unfolded protein response. J Cell Sci. 2012;125:4865–4875. doi: 10.1242/jcs.108928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldweg AM, Srivastava PK. Molecular heterogeneity of tumor rejection antigen/heat shock protein GP96. Int J Cancer. 1995;63:310–314. doi: 10.1002/ijc.2910630227. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Stevens F, Argon Y. Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta. 2013;1833:2410–2424. doi: 10.1016/j.bbamcr.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haystead CM, Gregory P, Sturgill TW, Haystead TA. Gamma-phosphate-linked ATP-Sepharose for the affinity purification of protein kinases. Rapid purification to homogeneity of skeletal muscle mitogen-activated protein kinase kinase. Eur J Biochem. 1993;214:459–467. doi: 10.1111/j.1432-1033.1993.tb17942.x. [DOI] [PubMed] [Google Scholar]
- Hidvegi T, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284:17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kato K. The role of MRH domain-containing lectins in ERAD. Glycobiology. 2010;20:651–660. doi: 10.1093/glycob/cwq013. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PF, Barrott JJ, Carlson DA, Loiselle DR, Speer BL, Bodoor K, Rund LA, Haystead TA. A highly selective Hsp90 affinity chromatography resin with a cleavable linker. Bioorgan Med Chem. 2012;20:3298–3305. doi: 10.1016/j.bmc.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, et al. An interaction map of endoplasmic reticulum chaperones and foldases. Mol Cell Proteomics. 2012;11:710–723. doi: 10.1074/mcp.M111.016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitsuhara Y, Toyoda T, Itai T, Yamaguchi H. Chaperone-like functions of high-mannose type and complex-type N-glycans and their molecular basis. J Biochem. 2002;132:803–811. doi: 10.1093/oxfordjournals.jbchem.a003290. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Welch WJ. Characterization and purification of the 94-kDa glucose-regulated protein. J Biol Chem. 1991;266:5643–5649. [PubMed] [Google Scholar]
- Kimura Y, Nakazawa M, Yamada M. Cloning and characterization of three isoforms of OS-9 cDNA and expression of the OS-9 gene in various human tumor cell lines. J Biochem. 1998;123:876–882. doi: 10.1093/oxfordjournals.jbchem.a022019. [DOI] [PubMed] [Google Scholar]
- Leitman J, Shenkman M, Gofman Y, Shtern NO, Ben-Tal N, Hendershot LM, Lederkremer GZ. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol Biol Cell. 2014;25:1050–1060. doi: 10.1091/mbc.E13-06-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Turco SJ, Green M. Structure and assembly of the endoplasmic reticulum. Biosynthetic sorting of endoplasmic reticulum proteins. J Biol Chem. 1985;260:6926–6931. [PubMed] [Google Scholar]
- Liu B, et al. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1:79. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, et al. Essential roles of grp94 in gut homeostasis via chaperoning canonical Wnt pathway. Proc Natl Acad Sci USA. 2013;110:6877–6882. doi: 10.1073/pnas.1302933110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Intracellular disposal of incompletely folded human alpha1-antitrypsin involves release from calnexin and post-translational trimming of asparagine-linked oligosaccharides. J Biol Chem. 1997;272:7946–7951. doi: 10.1074/jbc.272.12.7946. [DOI] [PubMed] [Google Scholar]
- Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- Macer DR, Koch GL. Identification of a set of calcium-binding proteins in reticuloplasm, the luminal content of the endoplasmic reticulum. J Cell Sci. 1988;91 (Pt 1):61–70. doi: 10.1242/jcs.91.1.61. [DOI] [PubMed] [Google Scholar]
- Mazzarella RA, Green M. ERp99, an abundant, conserved glycoprotein of the endoplasmic reticulum, is homologous to the 90-kDa heat shock protein (hsp90) and the 94-kDa glucose regulated protein (GRP94) J Biol Chem. 1987;262:8875–8883. [PubMed] [Google Scholar]
- Melnick J, Aviel S, Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J Biol Chem. 1992;267:21303–21306. [PubMed] [Google Scholar]
- Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem J. 2009;417:651–666. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harbor Perspect Biol. 2012;5 doi: 10.1101/cshperspect.a013185. a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky O, Makarewich CA, Snapp EL, Argon Y. An essential role for ATP binding and hydrolysis in the chaperone activity of GRP94 in cells. Proc Natl Acad Sci USA. 2009;106:11600–11605. doi: 10.1073/pnas.0902626106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D, Mazzarella RA, Green M. Analysis of the structure and synthesis of GRP94, an abundant stress protein of the endoplasmic reticulum. DNA Cell Biol. 1994;13:117–124. doi: 10.1089/dna.1994.13.117. [DOI] [PubMed] [Google Scholar]
- Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- Sato T, et al. STT3B-dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol Cell. 2012;47:99–110. doi: 10.1016/j.molcel.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell. 2010;40:905–916. doi: 10.1016/j.molcel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Shrimal S, Trueman SF, Gilmore R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J Cell Biol. 2013;201:81–95. doi: 10.1083/jcb.201301031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279:G961–G974. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Brostrom C, Koide T, Arvan P. Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J Biol Chem. 2000;275:40757–40764. doi: 10.1074/jbc.M001073200. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Nagata K. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell. 2013;24:3155–3163. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron N, Ooms A, Morel S, Degiovanni G, Van Den Eynde BJ. Identification of a new peptide recognized by autologous cytolytic T lymphocytes on a human melanoma. Cancer Immun. 2002;2:9. [PubMed] [Google Scholar]
- Vogen S, Gidalevitz T, Biswas C, Simen BB, Stein E, Gulmen F, Argon Y. Radicicol-sensitive peptide binding to the N-terminal portion of GRP94. J Biol Chem. 2002;277:40742–40750. doi: 10.1074/jbc.M205323200. [DOI] [PubMed] [Google Scholar]
- Wanderling S, Simen BB, Ostrovsky O, Ahmed NT, Vogen SM, Gidalevitz T, Argon Y. GRP94 is essential for mesoderm induction and muscle development because it regulates insulin-like growth factor secretion. Mol Biol Cell. 2007;18:3764–3775. doi: 10.1091/mbc.E07-03-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Hong F, Gewirth D, Guo B, Liu B, Li Z. The molecular chaperone gp96/GRP94 interacts with Toll-like receptors and integrins via its C-terminal hydrophobic domain. J Biol Chem. 2012;287:6735–6742. doi: 10.1074/jbc.M111.309526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.