New optogenetic tools are introduced that provide spatial and temporal control over the activity of heterotrimeric G protein subunits inside single cells. They are used to identify dynamic roles for G protein subunits in immune cell migration. They can be applied to study the mechanistic basis of other GPCR-regulated cellular functions.
Abstract
Cells sense gradients of extracellular cues and generate polarized responses such as cell migration and neurite initiation. There is static information on the intracellular signaling molecules involved in these responses, but how they dynamically orchestrate polarized cell behaviors is not well understood. A limitation has been the lack of methods to exert spatial and temporal control over specific signaling molecules inside a living cell. Here we introduce optogenetic tools that act downstream of native G protein–coupled receptor (GPCRs) and provide direct control over the activity of endogenous heterotrimeric G protein subunits. Light-triggered recruitment of a truncated regulator of G protein signaling (RGS) protein or a Gβγ-sequestering domain to a selected region on the plasma membrane results in localized inhibition of G protein signaling. In immune cells exposed to spatially uniform chemoattractants, these optogenetic tools allow us to create reversible gradients of signaling activity. Migratory responses generated by this approach show that a gradient of active G protein αi and βγ subunits is sufficient to generate directed cell migration. They also provide the most direct evidence so for a global inhibition pathway triggered by Gi signaling in directional sensing and adaptation. These optogenetic tools can be applied to interrogate the mechanistic basis of other GPCR-modulated cellular functions.
INTRODUCTION
A cell's function often depends on its ability to sense gradients of external cues and generate a polarized response such as directed migration or neurite initiation. There is a limited understanding of how dynamic networks of intracellular signaling molecules generate polarized cell behaviors. Network motifs have been proposed that can give rise to some of the features of cell migration, such as directional sensing, adaptation, and amplification of an external gradient (Xiong et al., 2011; Wang et al., 2012). However, existing experimental methods have provided mostly static information on the relevant signaling molecules, making it difficult to examine whether and how specific molecular interactions map onto these dynamic network motifs. In particular, there has been a lack of methods to exert spatial and temporal control over the activity of select signaling molecules inside a cell.
Optical manipulation of signaling presents an attractive approach for achieving such control (Toettcher et al., 2011). We recently used color opsins to spatially confine G protein–coupled receptor (GPCR) activity to a selected region of a single cell and gain optical control over immune cell migration (Karunarathne et al., 2013b) and neurite initiation and extension (Karunarathne et al., 2013a). The opsin approach optically activates an entire signaling pathway to orchestrate cell behavior, but new tools that provide optical control of downstream signaling molecules are required to dissect the network of dynamic interactions triggered inside a cell.
Here we create new optogenetic tools that enable light-triggered inhibition of endogenous G protein subunits in a selected region of a cell. We use them to generate reversible intracellular signaling gradients in cells exposed to a uniform extracellular stimulus. We apply this approach to study cell migration in a macrophage cell line, RAW 264.7.
GPCRs control migration of a wide variety of cell types, but the dynamic roles of the G protein α and βγ subunits in directing cell migration remain unclear. Signaling by βγ subunits is generally recognized as a requirement for GPCR-stimulated chemotaxis (Bagorda and Parent, 2008), and multiple βγ effectors have been implicated in cell migration (Li et al., 2003; Yan et al., 2012; Runne and Chen, 2013). However, it is unknown whether a gradient of active βγ is sufficient to trigger a directional response. Recent work in neutrophils suggests that βγ signaling may be primarily involved in controlling the motility rather than the directionality of a migrating cell (Kamakura et al., 2013). Meanwhile, there have been conflicting reports on the requirement of G protein αi subunit signaling in chemotaxis (Neptune et al., 1999; Kamakura et al., 2013), and there remains the possibility that GPCR activation of non–G protein pathways also contributes to chemotaxis (Neptune et al., 1999; Van Haastert and Devreotes, 2004).
Here we use our new optogenetic tools to address fundamental questions about chemotaxis: can a gradient of heterotrimeric G protein subunit activity stimulate all of the processes required for GPCR mediated chemotaxis, or is there an additional requirement for a gradient of G protein–independent signaling stimulated by the receptor? Is a gradient of βγ activity sufficient for directional sensing? Does G protein subunit activity at one end of a cell lead to inhibition of responses such as increased phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and lamellipodia formation at the opposite end?
RESULTS
Creating intracellular signaling gradients using uniform ligand stimulation and confined optical inhibition
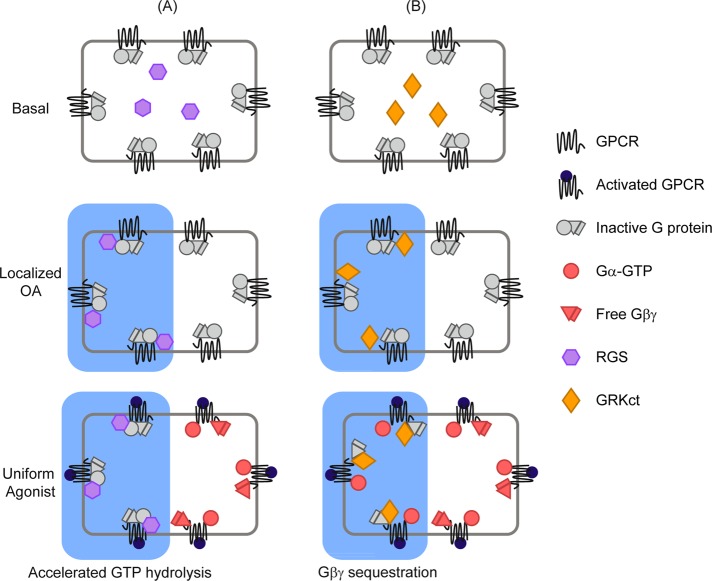
The general scheme used in our experiments is shown in Figure 1. We combined spatially uniform stimulation of GPCRs by a chemoattractant with confined optical inhibition of G protein signaling on one side of a cell. We applied two approaches: light-triggered acceleration of GTP hydrolysis on the α subunit, and optical recruitment of a βγ-sequestering domain.
FIGURE 1.
Generating intracellular signaling gradients by localized optical inhibition. (A) Optical recruitment of an RGS protein to a spatially confined region of the plasma membrane generates localized GAP activity, resulting in deactivation of the α subunit and the βγ complex. (B) Local inhibition of βγ signaling by optical recruitment of a βγ-sequestering peptide. Both approaches provide spatial control over G protein subunit activity downstream of uniform GPCR activation.
Design of an optically controlled GTPase-accelerating protein
GTPase-accelerating proteins (GAPs) act allosterically on G protein α subunits to accelerate the transition from active αGTP to inactive αGDP (Ross and Wilkie, 2000). Spatially localized acceleration of GTP hydrolysis at the α subunit can potentially reduce signaling by both the α and βγ subunits because deactivated αGDP rapidly rebinds the βγ complex and prevents its interaction with effectors (Lin and Smrcka, 2011). We sought to gain optical control over regulator of G protein signaling 4 (RGS4), which has GAP activity on both the αi and αq subunit types (Berman et al., 1996; Hepler et al., 1997). In yeast, exogenously expressed RGS4 has been shown to localize to the plasma membrane and inhibit the GPCR-regulated mating pathway (Srinivasa et al., 1998). A truncated mutant, RGS4(Δ1-33), did not localize to the plasma membrane and did not exhibit GAP activity. Its function, however, was rescued by addition of an alternative, C-terminal membrane–targeting domain (Srinivasa et al., 1998). These results suggested that it might be possible to gain optical control over the GAP activity of RGS4 by replacing its native membrane-targeting domain with a light-induced membrane-targeting domain.
The CRY2PHR and CIBN domains from Arabidopsis thaliana proteins cryptochrome 2 (CRY2) and CIB1 exhibit blue light–dependent binding and can be used for light-triggered recruitment of a CRY2-fused protein to the plasma membrane (Kennedy et al., 2010). We fused CRY2PHR-mCherry to RGS4(Δ1-33) to make CRY2-mCh-RGS4Δ. We then coexpressed this construct in HeLa or RAW 264.7 cells with a construct containing CIBN fused to the plasma membrane–targeting C-terminal domain from KRas (CIBN-CaaX; Idevall-Hagren et al., 2012). We found that CRY2-mCh-RGS4Δ translocated from the cytosol to the plasma membrane on photoactivation with 445-nm light (Figures 2 and 3).
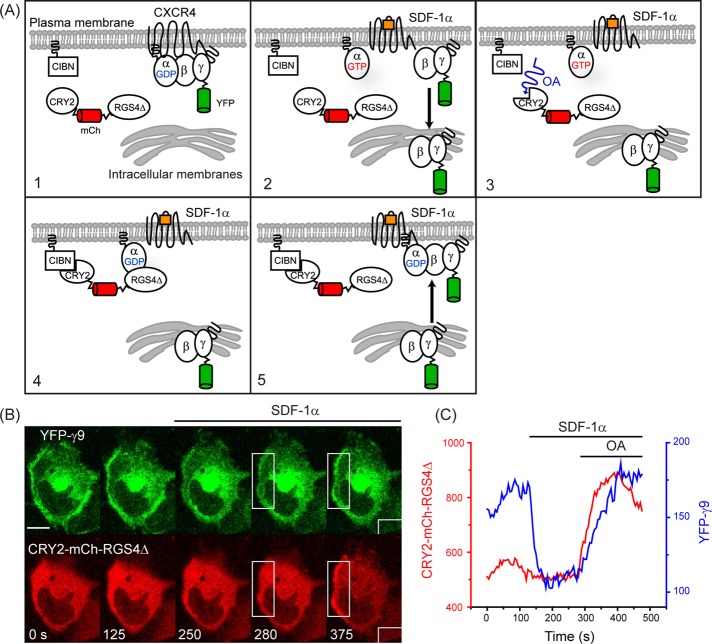
FIGURE 2.
Optical control of GTP hydrolysis with CRY2-mCh-RGS4Δ. (A) CXCR4 activation by SDF-1α triggers G protein activation, dissociation, and βγ translocation to intracellular membranes. OA of CRY2-mCh-RGS4Δ recruits it to the plasma membrane, where it can accelerate GTP hydrolysis on the α subunit. The increased concentration of αGDP at the plasma membrane results in reverse βγ translocation due to reformation of heterotrimers. (B) Live-cell imaging of a HeLa cell transiently transfected with CRY2-mCh-RGS4Δ, CIBN-CaaX, and YFP-γ9. Activation of endogenous CXCR4 receptors with 50 ng/ml SDF-1α triggered γ9 translocation from the plasma membrane to intracellular membranes. Photoactivation-stimulated translocation of CRY2-mCh-RGS4Δ to the plasma membrane. There it catalyzed hydrolysis of αGTP to αGDP, leading to reverse translocation of γ9 back to the plasma membrane due to the reformation of heterotrimers. Scale bar, 10 μm. (C) Time course of plasma membrane intensity for CRY2-mCh-RGS4Δ and YFP-γ9 in the photoactivated region.
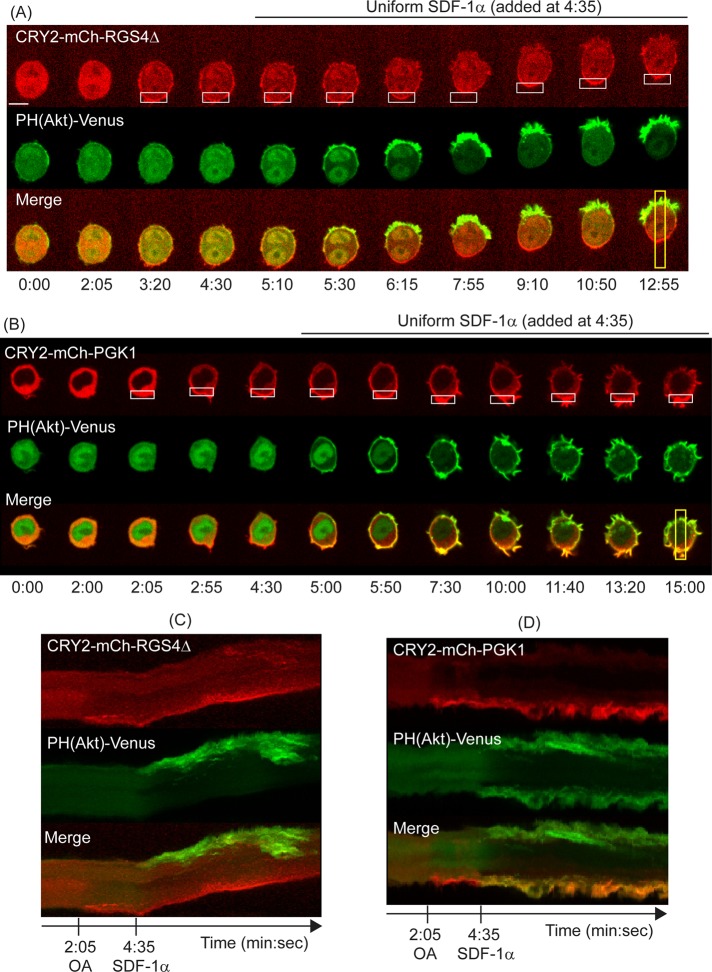
FIGURE 3.
Cell migration driven by localized Gi protein inhibition. (A) Image sequence of a live RAW 264.7 cell transiently transfected with CRY2-mCh-RGS4Δ, CIBN-CaaX, PH(Akt)-Venus, and CXCR4. Local OA was applied to generate a CRY2-mCh-RGS4Δ gradient before uniform addition of SDF-1α. Scale bar, 10 μm. (B) Negative control expressing CRY2-mCh-PGK1 instead of CRY2-mCh-RGS4Δ. (C, D) The t-stacks corresponding to the data in A and B. Localization of the RGS construct, but not the PGK construct, results in a PIP3 gradient, directional cell protrusions, and migration. White boxes correspond to OA regions. Yellow boxes show regions selected for generating the corresponding t-stacks.
Optical control over the GAP activity of an RGS protein can be demonstrated using a G protein βγ subunit translocation assay
We used a βγ subunit translocation assay to test whether light-activated recruitment of CRY2-mCh-RGS4Δ to the plasma membrane could regulate its GAP activity in a living cell. This assay leverages the αGDP-dependent plasma membrane targeting of βγ subunits to detect changes in the relative amounts of αGTP and αGDP in a living cell.
We previously showed that βγ subunits translocate reversibly from the plasma membrane to intracellular membranes upon GPCR activation (Akgoz et al., 2004; Azpiazu et al., 2006; Saini et al., 2007; Karunarathne et al., 2012). In unstimulated cells, G protein α and βγ subunits are primarily found as heterotrimers anchored to the plasma membrane by the lipid modifications on the α and γ subunits (Wedegaertner et al., 1995). GPCR activation triggers nucleotide exchange on the α subunit, resulting in dissociation of the αGTP and βγ subunits (Bondar and Lazar, 2014). The prenylated C-terminal domain of the γ subunits provides βγ subunits some membrane affinity, but it is insufficient for permanent anchoring in a membrane (O'Neill et al., 2012). As a result, free βγ subunits diffusively translocate to intracellular membranes (Saini et al., 2007; O'Neill et al., 2012). When receptors are deactivated, rebinding of βγ to αGDP results in their return to the plasma membrane.
Because reverse translocation of βγ subunits to the plasma membrane occurs through rebinding to αGDP, accelerating GTP hydrolysis on the α subunit should be capable of triggering reverse βγ translocation even if the receptors remain activated. We leveraged this feature of βγ translocation to test whether optical recruitment of CRY2-mCh-RGS4Δ to the plasma membrane can control its GAP activity.
We measured βγ translocation in HeLa cells by imaging a yellow fluorescent protein (YFP)–tagged version of γ9, a fast-translocating subunit (Figure 2). Consistent with previous observations (Karunarathne et al., 2012; O'Neill et al., 2012), activation of endogenous CXCR4 receptors with 50 ng/ml SDF-1α triggered βγ9 translocation from the plasma membrane to intracellular membranes, which was detected as a loss of YFP fluorescence from the plasma membrane (Figure 2, B and C). Localized optical activation (OA) of CRY2 resulted in localized accumulation of CRY2-mCh-RGS4Δ at the plasma membrane. This was accompanied by an increase of YFP-γ9 at the region proximal but not distal to the optically activated area. The light-triggered reverse βγ translocation occurred in the presence of continued receptor activity. No reverse βγ translocation was observed when CRY2-mCh lacking the RGS4 domain was optically recruited to the plasma membrane (Supplemental Figure S1). Thus the spatially confined reversal of βγ translocation detected here is consistent with optical recruitment of CRY2-mCh-RGS4Δ to the plasma membrane being able to locally trigger its GAP activity on α-GTP and thereby increase the concentration of α-GDP in that region.
Optically generated Gi protein signaling gradients direct migration of RAW 264.7 macrophage cells
The foregoing results showed that local optical activation of CRY2-mCh-RGS4Δ can trigger deactivation of αi-GTP and Gβγ in a selected area of a cell. This capability provides a way to create an intracellular G protein subunit activity gradient and examine polarized cell behaviors. We used it to examine the migratory response in RAW 264.7 macrophage cells. RAW cells are known to exhibit GPCR stimulated chemotaxis (Wiege et al., 2012), and we found that their low basal motility compared with commonly studied HL-60 neutrophils and Dictyostelium cells simplifies the use of localized OA to control membrane recruitment of CRY2 constructs.
Directionally responsive spatial gradients of PIP3 are believed to be one of the mediators of chemotaxis (Cai and Devreotes, 2011; Weiger and Parent, 2012). We examined whether local inhibition of G protein subunit activity could be used to direct the formation of PIP3 gradients in RAW cells exposed to a uniform extracellular stimulus. We examined the PIP3 response in RAW cells transfected with CRY2-mCh-RGS4Δ, CIBN-CaaX, PH(Akt)-Venus, and CXCR4. PIP3 dynamics in a live cell can be measured by imaging the translocation of a PH(Akt)-Venus sensor from the cytosol to the plasma membrane (James et al., 1996; Meili et al., 1999). We used the chemokine receptor CXCR4 to activate G proteins globally, since activation of this receptor by a gradient of the chemokine SDF-1α stimulates migration in many cell types (Bleul et al., 1996; Klein et al., 2001; Molyneaux et al., 2003).
First, we used localized OA to recruit CRY2-mCh-RGS4Δ to the plasma membrane at one side of a cell and followed this with global CXCR4 activation using 50 ng/ml SDF-1α (Figure 3). Before receptor activation, localized plasma membrane recruitment of CRY2-mCh-RGS4Δ did not produce any detectable PIP3 generation or cell shape changes. On receptor activation, cells responded by generating PIP3 gradients and initiating migration in the direction opposite to the CRY2-mCh-RGS4Δ gradient (Figure 3A and Supplemental Movie S1). Of 43 cells that provided a PIP3 response, all exhibited PIP3 gradients and directed lamellipodia. Of these, seven migrated at least 1 cell diameter in 15 min, 10 migrated between 1/2 and 1 cell diameter, and 26 migrated <1/2 cell diameter. Of those that migrated <1/2 cell diameter, five extended the front by at least 1/2 cell diameter but did not retract the back, and five initiated migration before snapping back to their initial positions, perhaps due to strong adhesion to the uncoated glass surface.
The directional responses were not due to unintended SDF-1α gradients, because two cells in close proximity could be made to respond in opposite directions (Supplemental Figure S2). The same directional control was observed when OA was applied after the uniform extracellular stimulus, with migration being initiated at the time of OA (Supplemental Movie S2). Furthermore, the direction of PIP3 accumulation and lamellipodia formation could be reversed by switching the location of OA to the opposite side of the cell (eight of eight cells) (Supplemental Figure S3).
Cells did not exhibit any of these directional responses to localized OA when a CRY2 construct (CRY2-mCh) without RGS4Δ was expressed in the cells or when a CRY2-mCh construct containing the cDNA for a glycolytic enzyme, PGK1, was expressed (CRY2-mCh-PGK1; Figure 3, B and D, and Supplemental Figure S4). These cells exhibited uniform PIP3 responses (29 of 36 cells) or polarized spontaneously in directions that did not depend on the side of OA with reference to the cell (7 of 36 cells). Compared to neutrophils, spontaneous polarization in response to a uniform stimulus appears to be much less common in RAW macrophage cells. This is consistent with their general lack of basal polarization and their greatly reduced basal motility compared with neutrophils. These controls show that the directional responses observed with CRY-mCh-RGS4Δ are due to localized inhibition of αi and βγ subunit activity by RGS4Δ rather than a nonspecific effect due to localized OA or accumulation of the CRY protein at the membrane.
We performed identical experiments using activation of endogenous C5 receptors to ensure that the migratory response induced by localized GAP activity was not peculiar to the CXCR4 receptor or due to overexpression of a GPCR. The anaphylatoxin C5a is known to stimulate chemotaxis of all myeloid cell lineages (Gerard and Gerard, 1994), and it has been shown to induce chemotaxis of RAW 264.7 cells (Wiege et al., 2012). We activated endogenous C5a receptors with 10 μM FKP-(D-Cha)-Cha-r, a peptide derived from the C-terminus of the full-length, 74–amino acid C5a. It has been reported to be a full agonist of the C5a receptor, eliciting responses comparable to those of full-length C5a in several assays, including chemotaxis (Konteatis et al., 1994). Localized OA of CRY2-mCh-RGS4Δ with uniform activation of endogenous C5a receptors generated directional responses similar to those seen with uniform activation of transfected CXCR4 (Supplemental Figure S5).
The ability to locally inhibit G protein signaling and generate a migratory response in immune cells showed that an internal gradient of αi and βγ activity is sufficient to direct cell migration in the absence of an external gradient. The results with endogenous C5a receptors showed that these internal gradients are sufficient to drive cell migration at levels of signaling activity normally achieved within a cell.
Gβγ signaling gradients generated by CRY2-mCh-GRK2ct direct PIP3 gradients and lamellipodia formation
To further dissect the roles of G protein subunits in cell migration, we sought to develop an optogenetic tool to specifically inhibit βγ signaling. We created a CRY2-mCh-GRK2ct construct that could be optically recruited to one side of a cell to produce a gradient of βγ activity (Figure 4A). The C-terminal domain of G protein–coupled receptor kinase 2 (GRK2ct) is capable of inhibiting responses downstream of βγ without inhibiting those generated by α subunit effectors (Koch et al., 1994). It has been widely used to sequester Gβγ and inhibit its activity, but this is the first time it was used asymmetrically within a single cell to study a polarized cell behavior.
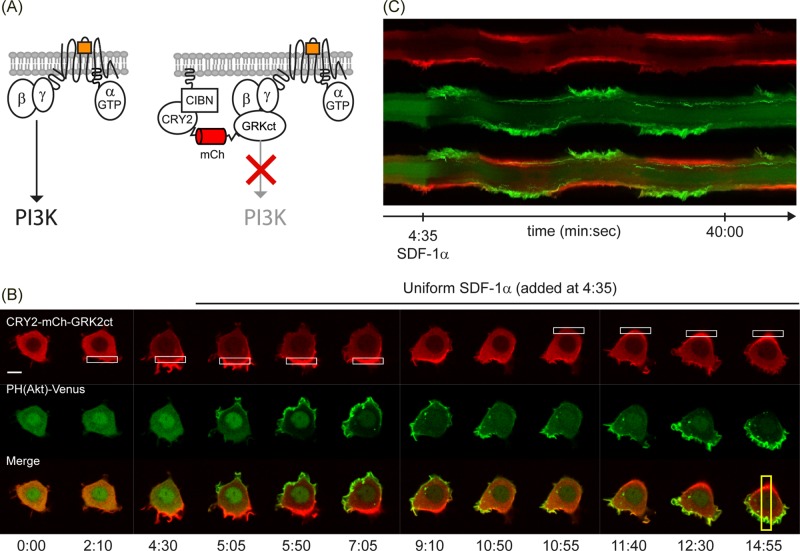
FIGURE 4.
Localized Gβγ inhibition directs PIP3 gradients and lamellipodia. (A) Light-triggered recruitment of CRY2-mCh-GRK2ct to the plasma membrane allows for spatially confined inhibition of βγ signaling. (B) Live-cell imaging of a RAW cell expressing CRY2-mCh-GRK2ct, CIBN-CaaX, PH(Akt)-Venus, and CXCR4. (C) The t-stack corresponding to the data in B. Localization of the GRKct construct generates reversible lamellipodia and PIP3 responses.
In RAW cells transiently transfected with CRY2-mCh-GRK2ct, CIBN-CaaX, PH(Akt)-Venus, and CXCR4, spatially confined OA resulted in localized recruitment of CRY2-mCh-GRK2ct from the cytosol to the plasma membrane. Subsequent activation of CXCR4 receptors with 50 ng/ml SDF-1α resulted in generation of a PIP3 gradient and lamellipodia toward the side of the cell that was opposite to the location of the OA (40 of 50 cells; Figure 4B). The direction of the lamellipodia and the PIP3 gradient could be reversed by switching the location of OA with reference to the cell (6 of 12 cells; Figure 4, B and C, and Supplemental Movie S3).
To ensure that these responses occurred due to localized sequestration of βγ and not some peculiar effect of GRK2ct, we performed identical experiments with a homologue, GRK3ct. GRK3ct binds to βγ subunits in biochemical (Daaka et al., 1997) and live-cell imaging assays (Hollins et al., 2009). GRK2 and GRK3 sequences are 85% identical, but their βγ-binding regions are only 52% identical (Daaka et al., 1997). The CRY2-mCh-GRK3ct construct was capable of producing similar directional (15 of 19 cells) and reversible (9 of 13 cells) responses (Supplemental Figure S6). The ability of both CRY2-GRKct constructs to elicit these directional responses, but not CRY2-mCh or CRY2-mCh-PGK1, confirmed that the directional responses occurred due to sequestration of Gβγ. Similar CRY2-mCh-GRKct directed responses were also observed when endogenous C5a receptors were activated with 10 μM FKP-(D-Cha)-Cha-r (Supplemental Figure S7).
Whereas the CRY2-mCh-GRKct constructs were capable of generating PIP3 gradients and directional lamellipodia similar to those generated by the CRY2-mCh-RGS4Δ construct, none of these cells exhibited appreciable cell migration. This difference could potentially be due to different magnitudes of βγ inhibition achieved by the GRKct versus RGS constructs, or it could it be that a gradient of αi activity is additionally required for migration. We suspect that the latter explanation is more likely, given that recent studies using a variety of cell types reported roles in chemotaxis for αi subunit interactions with proteins such as GIV (Ghosh et al., 2008), ELMO1/Dock180 (Li et al., 2013), and AGS3/mInsc (Kamakura et al., 2013). Overall, these results suggest that a gradient of activated Gβγ subunits stimulated by endogenous receptors is sufficient to elicit directional PIP3 responses and cell protrusions in the absence of an external gradient.
Generating light-triggered gradients in cells that have adapted to a uniform stimulus: evidence of global inhibition mediated by G protein subunits
Directional sensing in migratory cells is believed to be intimately related to their ability to adapt to a spatially uniform stimulus (Parent and Devreotes, 1999; Van Haastert and Devreotes, 2004; Levchenko and Iglesias, 2002). In this context, adaptation refers to a cell's ability to generate transient responses that return to near-basal levels after a uniform increase in chemoattractant concentration. This occurs through a mechanism other than desensitization, and it allows a cell to sense gradients over a wide range of background chemoattractant concentrations. The mechanisms that control adaptation in migratory cells are not fully understood.
An incoherent feedforward loop (IFFL) has been identified as a signaling motif capable of generating adaptive responses (Ma et al., 2009). In the IFFL, the input signal generates an activator with fast kinetics and an inhibitor with slower kinetics that converge on a downstream response such as PIP3. At short times after application of the stimulus, the activator generates an increase in PIP3 levels, but over time, the rising level of the inhibitor causes the PIP3 to decay back to its prestimulus level. Recent studies show that an IFFL can explain adaptation of PIP3 and Ras responses in Dictyostelium (Takeda et al., 2012; Wang et al., 2012)
A local-excitation global-inhibition (LEGI) mechanism that incorporates the IFFL motif has been proposed that can account for both adaptation and directional sensing (Parent and Devreotes, 1999; Levine et al., 2006). In the LEGI model, the activator signals locally, while the inhibitor diffuses throughout the cell to signal globally. As a result, downstream responses adapt to a uniform stimulus but exhibit sustained intracellular gradients in response to a gradient stimulus. Several models of chemotaxis incorporate the LEGI motif to account for directional sensing and adaptation, combining it with motifs that account for additional features of chemotaxis, such as basal motility, cell shape changes, or amplification of the external gradient (Xiong et al., 2011; Wang et al., 2012; Shi et al., 2013). However, a specific global inhibitor has not yet been identified. It is not known whether an inhibitor is generated by Gi signaling or by an independent pathway triggered by the GPCR.
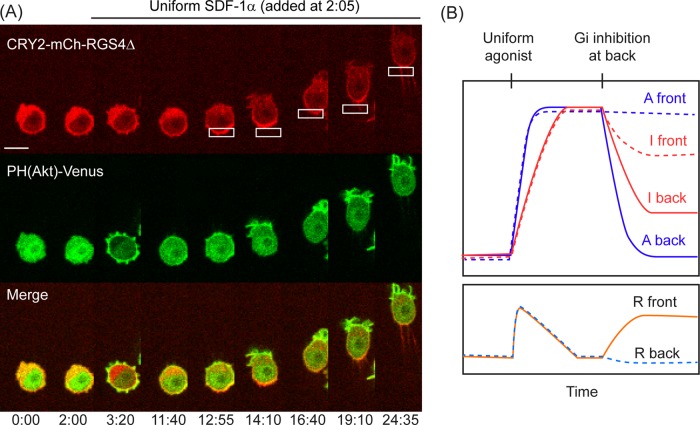
We designed an experiment to determine whether Gi signaling by itself leads to global inhibition (Figure 5, Supplemental Figure S5, and Supplemental Movie S4). First we exposed RAW cells to a uniform chemoattractant, either 50 ng/ml SDF-1α to activate transfected CXCR4 or 10 μM FKP-(D-Cha)-Cha-r to activate endogenous C5a receptors. This resulted in translocation of PH(Akt) to the plasma membrane and generation of cell protrusions. After the cells had adapted, as indicated by PH(Akt) returning to the cytosol and the cell protrusions subsiding, CRY2-mCh-RGS4Δ was optically recruited to one side of the cell to induce localized inhibition of αi and βγ activities. This resulted in the formation of a PIP3 gradient and initiation of cell migration in a direction that was opposite to the location of the OA (seven of eight cells).
FIGURE 5.
Local optical inhibition of Gi activity after adaptation to uniform stimulus. (A) Image sequence showing a RAW cell transfected with CRY2-mCh-RGS4Δ, CIBN-CaaX, PH(Akt)-Venus, and CXCR4. Addition of uniform SDF-1α (2:05) resulted in PIP3 accumulation and generation of cell protrusions (3:20). After several minutes, the PIP3 level and cell shape resembled those seen before the uniform stimulus (11:40). Localized OA of CRY2-mCh-RGS4Δ (12:55) was applied in this adapted state to inhibit Gi activity at one end of the cell. This resulted in the generation of a PIP3 gradient and cell migration directed toward the far side of the cell. Scale bar, 10 μm. (B) An illustration of the expected time dependence of an activator (A), inhibitor (I), and downstream response (R) in a LEGI model (Xiong et al., 2010) in which Gi signaling generates both A and I. Uniform activation of Gi signaling produces a transient downstream response that returns to the basal level due to the delayed increase in I. Subsequent optical inhibition of Gi signaling at the back causes a reduction in the level of Ifront but not Afront due to the differential movement of I and A throughout the cell. This leads to an increase in R at the front of the cell, resulting in directional migration.
The ability to generate responses at the front of a cell simply by inhibiting G protein activity at the back provides direct evidence that Gi signaling can act at a distance to inhibit “frontness” signaling pathways. This result is consistent with a LEGI model in which both the local activator and the global inhibitor are generated by Gi signaling. Figure 5B shows schematic plots that illustrate the time dependence of the activator, the inhibitor, and the downstream response. Application of a uniform input initially leads to rapid generation of the activator and the downstream response. The delayed accumulation of the inhibitor causes the response to return to its prestimulus level. The levels of both activator and inhibitor remain high throughout the cell in the adapted state. When the cell is in this state, applying localized OA to inhibit Gi signaling causes the levels of activator and inhibitor to decrease on one side of the cell. Because the inhibitor acts globally, the cell encounters the reduced level of inhibitor over its entire space. In contrast, the level of activator is only reduced on one side. As a result, the level of activator overwhelms that of the inhibitor on one side of the cell, leading to the generation of downstream signaling gradients that drive cell migration.
DISCUSSION
Optical control of cell signaling by inhibition of endogenous proteins
Most of the current information about signaling molecules involved in cell migration comes from genetic manipulations that establish whether a given protein is required for migration and biochemical studies that identify its relevant interactions. Imaging methods have provided additional information about the localization of several signaling molecules to the front or back of a migrating cell. This information is valuable, but new kinds of information are required in order to understand how a network of dynamic interactions shapes the cellular response. Obtaining this kind of information has been limited due to a lack of methods to exert spatial and temporal control over the activity of intracellular signaling molecules.
Here we developed optogenetic tools that provide such control by locally inhibiting the activity of specific G protein subunits. We showed that light-triggered membrane recruitment of a truncated RGS4 can be used to spatially localize G protein subunit activity within a cell. We also showed that similar optical recruitment of GRK2ct to a spatially confined region of the plasma membrane can locally inhibit Gβγ-signaling activity. We combined the capabilities of these optogenetic tools with spatially uniform activation of GPCRs to generate intracellular gradients of G protein subunit activity.
An advantage of the optical inhibition approach used here is that it enables spatial and temporal control over the activity of endogenous untagged proteins. Inhibition is achieved by expression of a CRY2-tagged protein, but the cellular response is elicited by a pathway that is entirely in its native state at the distal end of the cell with reference to the site of OA. This ensures that the targeted protein retains all of its native signaling properties. It also provides control over intracellular signaling at levels that reflect those driving native cell behavior because all of the signaling is done by endogenous proteins.
A gradient of G protein αi and βγ activity is sufficient to drive cell migration
Inhibition by pertussis toxin showed that Gi signaling is required for cell migration toward many different chemoattractants (Spangrude et al., 1985; Hartmann et al., 1997). However, the dynamic roles for the αi and βγ subunits are not known, and it has not been possible to test whether a gradient in the activity of αi and βγ subunits is sufficient to generate cell migration. There have been suggestions that other G protein subunit types may also be required. For example, in N-formyl-methionyl-leucyl-phenylalanine (fMLP)–stimulated neutrophil chemotaxis, it has been reported that Gi signaling regulates “frontness,” whereas G12/13 regulates “backness” pathways (Xu et al., 2003). Cell migration could additionally require gradients of GPCR-triggered but G protein–independent signaling (Ge et al., 2003). It could also potentially require interactions between ligand-bound GPCRs and accessory proteins that modulate G protein–mediated signaling, for example by bringing specific effector molecules closer to the activated G protein (Ritter and Hall, 2009).
Here we activated receptors that couple to Gi heterotrimers. By breaking spatial symmetry downstream of the receptor, directly at the level of the Gi protein, we were able to identify molecular and cellular responses generated by a gradient of αi and βγ activity. The ability of optically localized CRY2-mCh-RGS4Δ to generate directional cell migration shows that a gradient of αi and βγ activity is sufficient to elicit the entire gamut of migratory responses, including generation of lamellipodia at the front of a cell, retraction of the back, directional changes, and ability to respond directionally after adapting to a uniform stimulus.
Directional sensing by a Gβγ signaling gradient
It has been shown that βγ inhibition by sequestering proteins (Arai et al., 1997; Neptune and Bourne, 1997) or small molecules (Lehmann et al., 2008; Kang et al., 2014) suppresses chemotaxis in many cell types. It was unknown, however, which features of cell migration are controlled by βγ signaling. Some reports implicated βγ signaling in directional sensing, whereas others proposed that it is primarily involved in controlling cell motility (Kamakura et al., 2013). Our results with CRY2-mCh-GRK2ct and CRY2-mCh-GRK3ct show that a gradient of activated βγ is sufficient to generate a PIP3 gradient and lamellipodia formation directed toward the side with a higher level of βγ activity. This directly demonstrates a role for βγ signaling in directional sensing.
Overall these results with CRY2-RGS and CRY2-GRKct suggest that in immune cells sensing a chemoattractant gradient, the occurrence of a gradient of activated G protein subunits is sufficient to initiate directionally sensitive migration.
Adaptation of cell migratory responses involves Gi-mediated global inhibition
There is limited understanding of the molecular interactions that allow eukaryotic migratory cells to adapt to uniform stimulation. Dynamic control over receptor activation using microfluidics provides evidence that these cells use an IFFL network motif for adaptation (Takeda et al., 2012; Wang et al., 2012). Examining whether and how specific signaling molecules map onto the IFFL motif can be aided by methods that provide acute control over their activities within a living cell.
Previously it had not been possible to test directly whether Gi activity generates the inhibitory signaling that is required for adaptation in a migratory cell. It was only known that an inhibitory pathway should be present downstream of the receptor. Our results show that Gi signaling is capable of triggering a delayed inhibitory pathway that acts throughout the entire space of a cell. The ability to generate a postadaptation PIP3 gradient by local suppression of αi and βγ activity shows that Gi stimulates a signaling pathway capable of inhibiting PIP3 globally. The response is reflected at the cellular level, because the cells demonstrate directional migration. Many downstream responses have been observed to adapt, and there is evidence that pathways acting in parallel to PIP3 signaling are involved in controlling cell migration. The ability of local Gi inhibition in an adapted cell to elicit a directional migratory response suggests that all of the relevant pathways are under control of G protein αi and βγ subunit activity.
G proteins remain active when downstream responses adapt
A fluorescence resonance energy transfer–based G protein sensor in Dictyostelium indicated that G protein heterotrimers remain dissociated after transient downstream responses such as PIP3 have adapted (Janetopoulos et al., 2001). This suggested that adaptation does not require deactivation of G protein subunits. There are examples, however, such as the response to mating pheromone in yeast, in which adaptation occurs at the level of the G protein (Cole and Reed, 1991). In the case of mammalian migratory cells, it has not been clear whether G protein subunit deactivation plays a role in adaptation. It has not been directly tested whether G protein subunit activity continues after an immune cell adapts to a uniform signal (Iglesias, 2012). Here, in immune cells that have adapted to a uniform stimulus, asymmetric G protein deactivation triggered a directional migratory response. This showed that in a fully adapted cell, G protein subunits continue to be in the activated state.
Optical control over G protein subunits to dissect their dynamic signaling roles
GPCRs have been implicated in other polarized cell behaviors, such as yeast budding (Bi and Park, 2012), neurite outgrowth (Fricker et al., 2005; Georganta et al., 2013), and orientation of asymmetric cell divisions (Yoshiura et al., 2012). The ability of our optogenetic tools to locally inhibit G protein subunits can be used to help determine their dynamic roles in these polarized responses.
G protein subunits were classically believed to carry out all of their signaling functions at the plasma membrane, but mounting evidence suggests that they can have signaling activities at other locations within a cell (Hewavitharana and Wedegaertner, 2012). There is a lack of methods to determine the functions of G protein subunit signaling at intracellular locations. Existing methods to interfere with G protein signaling act over an entire cell. Optical recruitment of the CRY2-RGS to specific intracellular locations could be achieved through the use of appropriately targeted CIBN constructs. This could provide a means to acutely perturb G protein subunit activity at different locations within a cell. This could help dissect the functions of GPCR stimulated signaling at different locations of a cell. For example, signaling can be perturbed at a growth cone or a synapse. It can also be used to examine the temporal role of GPCR signaling in cell differentiation or development by inhibiting it at specific time points.
MATERIALS AND METHODS
Reagents
SDF-1α/CXCL12 (S190; Sigma-Aldrich, St. Louis, MO) was dissolved to 10 μg/ml in 1× phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin and stored as aliquots at −20°C. The C5a receptor agonist FKP-(D-Cha)-Cha-r (65121; Anaspec, Freemont, CA) was dissolved to 2.5 mM in 1× PBS containing 0.1% albumin and stored as aliquots at −20°C.
DNA constructs
CRY2PHR-mCh was obtained from AddGene (Cambridge, MA) (plasmid #26866). CIBN-CaaX was a kind gift from the lab of P. Di Camilli (Boyer Center for Molecular Medicine, Yale School of Medicine, New Haven, CT) (Idevall-Hagren et al., 2012). CXCR4 was a kind gift from the lab of I. Schraufstatter (Torrey Pines Institute for Molecular Studies, San Diego, CA) (Zhao et al., 2006). YFP-γ9 has been described before (Saini et al., 2007). A PCR product of PGK1 (38071; Addgene) was inserted into the KpnI and XbaI sites of CRY2PHR-mCh to create CRY2-mCh-PGK1. A PCR product of GRK2ct (Irannejad and Wedegaertner, 2010) was inserted into the KpnI and XbaI sites of CRY2PHR-mCh to make CRY2-mCh-GRK2ct. A PCR product of GRK3ct (Hollins et al., 2009) was inserted into the KpnI and XbaI sites of CRY2PHR-mCh to make CRY2-mCh-GRK3ct. A PCR product of RGS4 lacking residues 1–33 was inserted into the KpnI and XbaI sites of CRY2PHR-mCh to make CRY2-mCh-RGS4Δ. PH(Akt)–green fluorescent protein (GFP; 18836; Addgene) was cut with BamHI and XbaI to release GFP, and a PCR product of Venus was inserted in its place to make PH(Akt)-Venus.
Tissue culture
HeLa cells were obtained from ATCC and cultured in MEM (CellGro 10-010-CM) supplemented with 10% dialyzed fetal bovine serum (FBS; Atlanta Biologicals) and 1× antibiotic-antimycotic solution (CellGro) at 37°C and 5% CO2. RAW 264.7 cells were obtained from the Washington University Tissue Culture Support Center and cultured in DMEM supplemented with 10% dialyzed FBS and 1× antibiotic-antimycotic solution at 37°C and 5% CO2. RAW cells ranging from passage 3 to passage 12 were used for experiments.
Transfections
HeLa cells were transfected using Lipofectamine 2000. Cells were plated at 2 × 105 cells/dish in 29-mm glass-bottom dishes (In Vitro Scientific) 1 d before transfection. RAW cells were transfected by electroporation using Cell Line Nucleofection Kit V (Lonza) with a Nucleofector II device (Amaxa). For each sample, 2 × 106 cells were pelleted by spinning at 90 × g for 10 min, resuspended in 100 μl of Nucleofection solution containing between 0.2 and 2.5 μg of each plasmid DNA, depending on the specific construct (0.2 μg of PH(Akt)-Venus, 2 μg of CXCR4, and 2.5 μg of others), and electroporated using program D-032. Immediately after electroporation, 500 μl of prewarmed medium was added to the cuvette, and this was split among 29-mm glass-bottom dishes (8–10 dishes) containing 500 μl of prewarmed medium in the center well. After transfection, dishes were kept in a 37°C, 5% CO2 incubator until imaging.
Live-cell imaging and optical activation
All imaging was performed using a spinning-disk confocal imaging system consisting of a Leica DMI6000B microscope with adaptive focus control, a Yokogawa CSU-X1 spinning-disk unit, an Andor iXon electron-multiplying charge-coupled device camera, an Andor fluorescence recovery after photobleaching–photoactivation unit, and a laser combiner with 445-, 448-, 515-, and 594-nm solid-state lasers, all controlled using Andor iQ2 software. This system allows live-cell imaging to be combined with localized OA within a selected region of the sample that can be redefined in between images in a sequence. For OA of CRY2, the 445-nm laser was used at 5 μW and scanned across the selected region at a rate of 0.9 ms/μm2. This was performed once every 5 s. Solid-state lasers with wavelengths of 515 and 594 nm were used for excitation of Venus and mCherry, respectively. Emission filters were Venus 528/20 and mCherry 628/20 (Semrock). All images were acquired using a 63× oil immersion objective. A single confocal plane was imaged at a rate of 1 frame/5 s. All imaging was performed inside a temperature-controlled chamber held at 37°C. Imaging of HeLa cells was performed 1 d after transfection with Lipofectamine 2000. Imaging of RAW 264.7 cells was performed 2–10 h after electroporation. Before imaging, the culture medium was replaced with 500 μl of Hank's balanced salt solution supplemented with 1 g/l glucose (HBSSg). An equal volume of agonist in warm HBSSg was added at the time specified in the figures to achieve the final concentration given in the corresponding figure legends. Only cells that exhibited a detectable PIP3 response were included in the analysis. Approximately 10% of the cells did not exhibit any detectable PIP3 response to SDF-1α. This was true regardless of which CRY construct was expressed.
Supplementary Material
Acknowledgments
We thank W.K.A. Karunarathne and V. Kalyanaraman for useful discussions, Y. Ordabayev for assistance in making a construct, and R. Gereau for use of an electroporator. We thank P. De Camilli, P. Wedegaertner, N. Lambert, and I. Schraufstatter for DNA constructs. This work was supported by National Institutes of Health Grants GM069027 and GM080558 to N.G. and National Research Service Award Postdoctoral Fellowship GM099351 to P.R.O.
Abbreviations used:
- CRY2
cryptochrome 2
- GAP
GTPase-accelerating protein
- GPCR
G protein–coupled receptor
- GRK
G protein–coupled receptor kinase
- OA
optical activation
- RGS
regulator of G protein signaling
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-04-0870) on June 11, 2014.
REFERENCES
- Akgoz M, Kalyanaraman V, Gautam N. Receptor-mediated reversible translocation of the G protein beta gamma complex from the plasma membrane to the Golgi complex. J Biol Chem. 2004;279:51541–51544. doi: 10.1074/jbc.M410639200. [DOI] [PubMed] [Google Scholar]
- Arai H, Tsou CL, Charo IF. Chemotaxis in a lymphocyte cell line transfected with C-C chemokine receptor 2B: evidence that directed migration is mediated by betagamma dimers released by activation of Galphai-coupled receptors. Proc Natl Acad Sci USA. 1997;94:14495–14499. doi: 10.1073/pnas.94.26.14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu I, Akgoz M, Kalyanaraman V, Gautam N. G protein betagamma11 complex translocation is induced by Gi, Gq and Gs coupling receptors and is regulated by the alpha subunit type. Cell Signal. 2006;18:1190–1200. doi: 10.1016/j.cellsig.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagorda A, Parent CA. Eukaryotic chemotaxis at a glance. J Cell Sci. 2008;121:2621–2624. doi: 10.1242/jcs.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar A, Lazar J. Dissociated GalphaGTP and Gbetagamma protein subunits are the major activated form of heterotrimeric Gi/o proteins. J Biol Chem. 2014;289:1271–1281. doi: 10.1074/jbc.M113.493643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Devreotes PN. Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Semin Cell Dev Biol. 2011;22:834–841. doi: 10.1016/j.semcdb.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole GM, Reed SI. Pheromone-induced phosphorylation of a G protein beta subunit in S. cerevisiae is associated with an adaptive response to mating pheromone. Cell. 1991;64:703–716. doi: 10.1016/0092-8674(91)90500-x. [DOI] [PubMed] [Google Scholar]
- Daaka Y, Pitcher JA, Richardson M, Stoffel RH, Robishaw JD, Lefkowitz RJ. Receptor and G beta gamma isoform-specific interactions with G protein-coupled receptor kinases. Proc Natl Acad Sci USA. 1997;94:2180–2185. doi: 10.1073/pnas.94.6.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker AD, Rios C, Devi LA, Gomes I. Serotonin receptor activation leads to neurite outgrowth and neuronal survival. Brain Res Mol Brain Res. 2005;138:228–235. doi: 10.1016/j.molbrainres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Ge L, Ly Y, Hollenberg M, DeFea K. A beta-arrestin-dependent scaffold is associated with prolonged MAPK activation in pseudopodia during protease-activated receptor-2-induced chemotaxis. J Biol Chem. 2003;278:34418–34426. doi: 10.1074/jbc.M300573200. [DOI] [PubMed] [Google Scholar]
- Georganta EM, Tsoutsi L, Gaitanou M, Georgoussi Z. δ-Opioid receptor activation leads to neurite outgrowth and neuronal differentiation via a STAT5B-G αi/o pathway. J Neurochem. 2013;127:329–341. doi: 10.1111/jnc.12386. [DOI] [PubMed] [Google Scholar]
- Gerard C, Gerard NP. C5A anaphylatoxin and its seven transmembrane-segment receptor. Annu Rev Immunol. 1994;12:775–808. doi: 10.1146/annurev.iy.12.040194.004015. [DOI] [PubMed] [Google Scholar]
- Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann K, Henz BM, Kruger-Krasagakes S, Kohl J, Burger R, Guhl S, Haase I, Lippert U, Zuberbier T. C3a and C5a stimulate chemotaxis of human mast cells. Blood. 1997;89:2863–2870. [PubMed] [Google Scholar]
- Hepler JR, Berman DM, Gilman AG, Kozasa T. RGS4 and GAIP are GTPase-activating proteins for Gq alpha and block activation of phospholipase C beta by gamma-thio-GTP-Gq alpha. Proc Natl Acad Sci USA. 1997;94:428–432. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewavitharana T, Wedegaertner PB. Non-canonical signaling and localizations of heterotrimeric G proteins. Cell Signal. 2012;24:25–34. doi: 10.1016/j.cellsig.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollins B, Kuravi S, Digby GJ, Lambert NA. The C-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signal. 2009;21:1015–1021. doi: 10.1016/j.cellsig.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA. 2012;109:E2316–2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias PA. Chemoattractant signaling in Dictyostelium: adaptation and amplification. Sci Signal. 2012;5:pe8. doi: 10.1126/scisignal.2002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Wedegaertner PB. Regulation of constitutive cargo transport from the trans-Golgi network to plasma membrane by Golgi-localized G protein beta gamma subunits. J Biol Chem. 2010;285:32393–32404. doi: 10.1074/jbc.M110.154963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SR, Downes CP, Gigg R, Grove SJ, Holmes AB, Alessi DR. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem J. 1996;315:709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- Kamakura S, Nomura M, Hayase J, Iwakiri Y, Nishikimi A, Takayanagi R, Fukui Y, Sumimoto H. The cell polarity protein mInsc regulates neutrophil chemotaxis via a noncanonical G protein signaling pathway. Dev Cell. 2013;26:292–302. doi: 10.1016/j.devcel.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Kang BH, Shim YJ, Tae YK, Song JA, Choi BK, Park IS, Min BH. Clusterin stimulates the chemotactic migration of macrophages through a pertussis toxin sensitive G-protein-coupled receptor and Gβγ-dependent pathways. Biochem Biophys Res Commun 445, 645–650. 2014 doi: 10.1016/j.bbrc.2014.02.071. [DOI] [PubMed] [Google Scholar]
- Karunarathne WK, Giri L, Kalyanaraman V, Gautam N. Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc Natl Acad Sci USA. 2013a;110:E1565–E1574. doi: 10.1073/pnas.1220697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, Giri L, Patel AK, Venkatesh KV, Gautam N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc Natl Acad Sci USA. 2013b;110:E1575–1583. doi: 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, O'Neill PR, Martinez-Espinosa PL, Kalyanaraman V, Gautam N. All G protein betagamma complexes are capable of translocation on receptor activation. Biochem Biophys Res Commun. 2012 doi: 10.1016/j.bbrc.2012.04.054. 421, 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973-U948. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, Luster AD. SDF-1 alpha induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128:1971–1981. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- Koch WJ, Hawes BE, Inglese J, Luttrell LM, Lefkowitz RJ. Cellular expression of the carboxyl terminus of a G protein-coupled receptor kinase attenuates G beta gamma-mediated signaling. J Biol Chem. 1994;269:6193–6197. [PubMed] [Google Scholar]
- Konteatis ZD, Siciliano SJ, Van Riper G, Molineaux CJ, Pandya S, Fischer P, Rosen H, Mumford RA, Springer MS. Development of C5a receptor antagonists. Differential loss of functional responses. J Immunol. 1994;153:4200–4205. [PubMed] [Google Scholar]
- Lehmann DM, Seneviratne AM, Smrcka AV. Small molecule disruption of G protein beta gamma subunit signaling inhibits neutrophil chemotaxis and inflammation. Mol Pharmacol. 2008;73:410–418. doi: 10.1124/mol.107.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: a balanced inactivation model. Proc Natl Acad Sci USA. 2006;103:9761–9766. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yang L, Fu H, Yan J, Wang Y, Guo H, Hao X, Xu X, Jin T, Zhang N. Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat Commun. 2013;4:1706. doi: 10.1038/ncomms2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of cdc42. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- Lin Y, Smrcka AV. Understanding molecular recognition by G protein betagamma subunits on the path to pharmacological targeting. Mol Pharmacol. 2011;80:551–557. doi: 10.1124/mol.111.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TB, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux KA, et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- Neptune ER, Bourne HR. Receptors induce chemotaxis by releasing the betagamma subunit of Gi, not by activating Gq or Gs. Proc Natl Acad Sci USA. 1997;94:14489–14494. doi: 10.1073/pnas.94.26.14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neptune ER, Iiri T, Bourne HR. Galphai is not required for chemotaxis mediated by Gi-coupled receptors. J Biol Chem. 1999;274:2824–2828. doi: 10.1074/jbc.274.5.2824. [DOI] [PubMed] [Google Scholar]
- O'Neill PR, Karunarathne WKA, Kalyanaraman V, Silvius JR, Gautam N. G-protein signaling leverages subunit-dependent membrane affinity to differentially control beta gamma translocation to intracellular membranes. Proc Natl Acad Sci USA. 2012;109:E3568–E3577. doi: 10.1073/pnas.1205345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Runne C, Chen S. PLEKHG2 promotes heterotrimeric G protein betagamma-stimulated lymphocyte migration via Rac and Cdc42 activation and actin polymerization. Mol Cell Biol. 2013;33:4294–4307. doi: 10.1128/MCB.00879-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini DK, Kalyanaraman V, Chisari M, Gautam N. A family of G protein βγ subunits translocate reversibly from the plasma membrane to endomembranes on receptor activation. J Biol Chem. 2007;282:24099–24108. doi: 10.1074/jbc.M701191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Huang CH, Devreotes PN, Iglesias PA. Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol. 2013;9:e1003122. doi: 10.1371/journal.pcbi.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude GJ, Sacchi F, Hill HR, Van Epps DE, Daynes RA. Inhibition of lymphocyte and neutrophil chemotaxis by pertussis toxin. J Immunol. 1985;135:4135–4143. [PubMed] [Google Scholar]
- Srinivasa SP, Bernstein LS, Blumer KJ, Linder ME. Plasma membrane localization is required for RGS4 function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:5584–5589. doi: 10.1073/pnas.95.10.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Shao D, Adler M, Charest PG, Loomis WF, Levine H, Groisman A, Rappel WJ, Firtel RA. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci Signal. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Bergmann A, Lin B, Kim K, Levchenko A. Diverse sensitivity thresholds in dynamic signaling responses by social amoebae. Sci Signal. 2012;5, ra17 doi: 10.1126/scisignal.2002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedegaertner PB, Wilson PT, Bourne HR. Lipid modifications of trimeric G-proteins. J Biol Chem. 1995;270:503–506. doi: 10.1074/jbc.270.2.503. [DOI] [PubMed] [Google Scholar]
- Weiger MC, Parent CA. Phosphoinositides in chemotaxis. Subcell Biochem. 2012;59:217–254. doi: 10.1007/978-94-007-3015-1_7. [DOI] [PubMed] [Google Scholar]
- Wiege K, Le DD, Syed SN, Ali SR, Novakovic A, Beer-Hammer S, Piekorz RP, Schmidt RE, Nurnberg B, Gessner JE. Defective macrophage migration in G alpha(i2)- but not G alpha(i3)-deficient mice. J Immunol. 2012;189:980–987. doi: 10.4049/jimmunol.1200891. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Huang CH, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci USA. 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Huang CH, Iglesias PA, Devreotes PN. A LEGI-biased excitable network controls temporal and spatial responses to chemoattractants. Biophys J. 2011;100:165–165. [Google Scholar]
- Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, Bourne HR. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L, Veenstra TD, Parent CA, Jin T. A Gbetagamma effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Dev Cell. 2012;22:92–103. doi: 10.1016/j.devcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura S, Ohta N, Matsuzaki F. Tre1 GPCR signaling orients stem cell divisions in the Drosophila central nervous system. Dev Cell. 2012;22:79–91. doi: 10.1016/j.devcel.2011.10.027. [DOI] [PubMed] [Google Scholar]
- Zhao M, Discipio RG, Wimmer AG, Schraufstatter IU. Regulation of CXCR4-mediated nuclear translocation of extracellular signal-related kinases 1 and 2. Mol Pharmacol. 2006;69:66–75. doi: 10.1124/mol.105.016923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.