Abstract
During fetal development and early-infancy, environmental signals can induce epigenetic changes that alter neurobehavioral development and later-life mental health. Several neurodevelopmental genetic diseases influence epigenetic regulatory genes and genomic imprinting. Recently, brain epigenetic marks have been involved in idiopathic neurodevelopmental disorders including autism spectrum disorders (ASD). The placenta is an important regulator of the intrauterine environment that links maternal and fetal nervous systems. Placental epigenetic signatures have been associated with neurodevelopment of healthy newborns quantified through the NICU Network Neurobehavioral Scales (NNNS). Associations have been observed for DNA methylation of genes involved in cortisol (NR3C1, HSD11B), serotonin (HTR2A), and metabolic (LEP) pathways. Dysregulation of imprinted genes and microRNAs has also been associated with neurobehavior assessed by NNNS. Further analysis is needed to characterize the mechanisms by which the epigenome influences neurodevelopment, and the connection between this dysregulation and mental health disorders. In the future, epigenetic marks could serve as functional biomarkers of mental health and cognitive function.
Keywords: Neurobehavior, epigenetics, NNNS, Placenta, Autism, DNA Methylation
Introduction
The developmental origins of health and disease hypothesis (DOHaD) proposes that environmental cues during fetal development and early-infancy induce adaptive responses that can influence later-life disease susceptibility [1]. Populations exposed to prenatal famine show an increased risk of later-life mental outcomes, specifically schizophrenia, depression, addiction and dysregulation of stress response suggesting that intrauterine conditions program later-life mental health [2]. This early-life programming requires plasticity, thus epigenetic mechanisms have been proposed as molecular mediators because these integrate genetic and environmental signals with the control of gene expression [3].
Epigenetics is the study of heritable but feasibly environmentally modifiable control of gene expression potential without DNA sequence changes[4]. The major systems of epigenetic regulation include DNA methylation, genomic imprinting, non-coding RNAs (ncRNA) and histone modifications. DNA methylation is the best characterized epigenetic mark and involves the addition of a methyl group to cytosines usually within CpG dinucleotides that in promoters frequently results in gene silencing[4]. Epigenetic regulation is essential during development when somatic and germ cells experience a global epigenetic remodeling that regulates cell and tissue differentiation [5, 6]. The quality of the environment during this and other sensitive periods could alter this epigenetic reprogramming. Rodent studies have shown that maternal behavior in early-life influences offspring behavior during adulthood through epigenetic deregulation of NR3C1 and other loci [7-9] . This suggests that during intrauterine and early postnatal-life, epigenetic programming occurs that has long-term influences on mental health (Figure 1).
Figure 1.
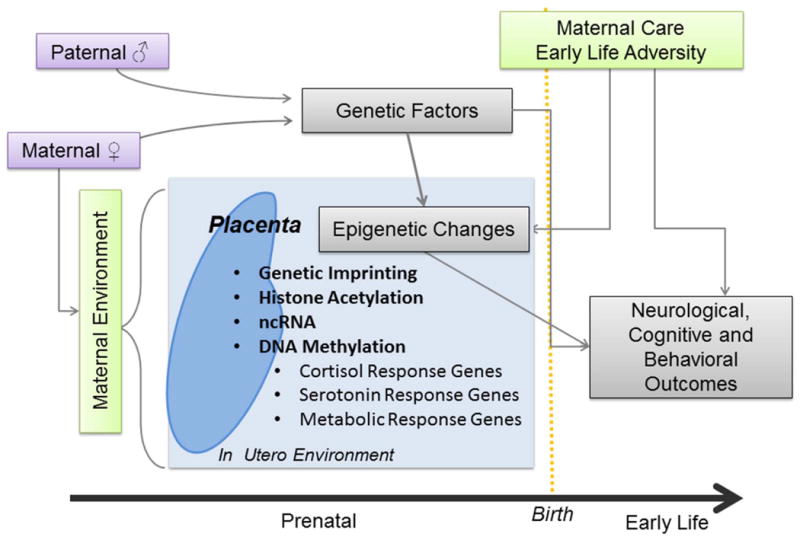
Diagram of principal factors influencing infant neurobehavior. Maternal and paternal genetics influence neurological, cognitive and behavioral outcomes. The in-utero and early-life environment can also influence these outcomes through epigenetic mechanisms. The placenta regulates the in-utero environment, and its epigenetic profiles can also contribute to infant neurobehavior.
In this review, we outline the evidence relating epigenetic variation and neurodevelopmental diseases, then discuss epigenetic marks in the placenta, a crucial organ for intrauterine development, and their role in infant neurodevelopmental outcomes.
Role of Epigenetics in Neurodevelopmental Disease
The significance of epigenetics in neurodevelopment is illustrated in genetic conditions that influence epigenetic regulatory genes and affect cognitive functions [10]. Rett syndrome (RTT) is a neurodevelopmental condition associated with autism spectrum disorder (ASD), and is caused by genetic mutations in the x-linked MECP2 [11]. MeCP2 is a chromatin-associated protein that binds to methylated DNA, is highly expressed in the brain, and is required for neuronal maturation. Loss or aberrant MeCP2 function leads to epigenetic deregulation and impaired synaptic function [10, 12]. Similarly, genomic imprinting disorders of 15q11-13 lead to Angelman syndrome (AS) and Prader-Willi syndrome (PWS), neurodevelopmental pathologies with structural and functional brain changes [13-15]. Imprinted genes are expressed in parent-of-origin-specific manner because DNA methylation silences the other allele [16]. A large proportion of imprinted genes are expressed in the brain, and imprinting disorders frequently exhibit neurodevelopmental delay [13]. Although, most AS and PWS cases are caused by genetic changes, in some cases loss of gene function is attributable to an imprinting defect or epimutation [17]. Moreover, 15q11-13 duplications are frequent cytogenetic abnormalities in ASD [18].
The majority of neurodevelopmental disorders, including ASD, cannot be directly associated with specific genetic changes, but have complex genetic and environmental influences contributing to disease [18]. Since epigenetic mechanisms integrate these signals, a number of studies suggest that idiopathic neurodevelopmental disorders may result from epigenetic dysregulation of neurological pathways. Most human studies of neurobehavioral disease and epigenetics (Table 1)[19-27] compare epigenetic profiles between ASD cases and controls in post-mortem brain samples, a highly relevant tissue, but not readily available. This limitation imposes cross-sectional study designs and reduces sample sizes. Thus, when selecting tissues for epigenetic studies of human neurobehavior, it is important to consider the high tissue-specificity of epigenetic marks, relevance to neural development and accessibility for prospective studies.
Table 1. Human Studies of Epigenetics and Neurobehavior.
| Gene(s) Epigenetic change | Major Findings | 1st author & year of publication | ||
|---|---|---|---|---|
| Epigenetics and Neurobehavioral Disease | ||||
| MECP2 DNA methylation | MECP2 promoter hypermethylation in prefrontal cortex of male ASD cases. Lower brain MECP2 expression in ASD cases. | Nagarajan et al 2006 | ||
| MECP2 DNA methylation | Increased methylation of a transition area (upstream of MECP2) in frontal cerebral cortex of male ASD cases. Aberrant MECP2 promoter methylation in ASD female brain. MECP2 methylation is locus-specific rather than global X chromosome changes. | Nagarajan et al 2008 | ||
| OXTR DNA methylation |
OXTR hypermethylation in blood and temporal cortex of ASD cases. Decreased OXTR expression in temporal cortex of ASD cases. |
Gregory et al 2009 | ||
| RORA, BCL-2 DNA methylation | Differentially methylated genes in blood of ASD cases enriched for transcription, nervous system development and cell death/ survival. RORA and BCL-2 exhibited decreased protein expression in tissue arrays (cerebellum and frontal cortex) in ASD cases. 8.1K CpG-island Array (HCGI8.1K) |
Nguyen et al 2010 | ||
| Genome-wide-scan H3K4me3 Histone methylation |
Subset of ASD cases exhibited H3K4me3 spreading into nucleosomes in prefrontal cortex. Identification of 711 loci with altered H3K4me3 signal in brain of ASD cases compared to controls.H3K4me3 peaks enriched in genes implicated in neurodevelopmental disease. Aberrant H3K4 methylation at a specific TSS is a predictor of transcriptional dysregulation. |
Shulha et al 2012 | ||
| EN-2 DNA and histone methylation |
EN-2 promoter hypermethylation in cerebellar cortex associated with ASD, methylation positively correlated with EN-2 expression - Decreased histone H3K27 in the EN-2 promoter in ASD cases. |
James et al 2013 | ||
| PRRT1 TSPAN32/C11orf21 Near ZFP57 SDHAP3 DNA methylation | 3 DMRs Temporal cortex: PRRT1 (3′ UTR) hypomethylation in ASD cases TSPAN32/ C11orf21 hypomethylation in cases Near ZFP57 hypermethylated in cases 1 DMR Cerebellum SDHAP3 hypermethylated in ASD cases |
Ladd-Acosta et al 2013 | ||
| SHANK3 DNA methylation | SHANK3 hypermethylation in cerebellum and cerebral cortex of ASD cases compared to controls. Altered expression an alternative splicing of SHANK3 isoforms in brain tissue. | Zhu et al 2013 | ||
| DRD4 and 5-HTT DNA methylation | Cord blood DNA methylation of DRD4 and 5-HTT regions negatively associated with ADHD symptom score at age 6. | van Mil el al 2014 | ||
| Placental Epigenetics and Newborn Neurobehavior | ||||
| HSD11B2 DNA methylation | Inverse association between placental HSD11B2 methylation and quality of movement scores in RICHS newborns. Pregnancy anxiety and placental HS11B2 methylation (CpG4) interaction influences hypotonicity in RICHS infants |
Marsit et al 2012 Conradt et al 2013 | ||
| NR3C1 DNA methylation | Higher NR3C1 placental promoter methylation associated with higher quality of movement scores and lower infant attention scores in RICHS newborns. Potential interaction between methylation and genotype on infant attention score Pregnancy depression and placental NR3C1 methylation (CpG2) interaction influences self-regulation, hypotonicity, and lethargy in RICHS infants. |
Bromer et al 2012 Conradt et al 2013 |
||
| HTR2A DNA methylation | Higher HTR2A placental methylation associated with lower quality of movement and higher infant attention scores in RICHS newborns | Paquette at al 2013 | ||
| LEP DNA methylation | Higher LEP promoter placental methylation associated with membership in a NNNS neurobehavioral profile marked by increased lethargy and hypotonicity and reduced risk of membership in a profile with opposite characteristics in RICHS newborns. | Lesseur et al 2014 | ||
| Expression of 22 imprinted genes | Placental imprinted gene expression classes associated with quality of movement and handling in RICHS newborns. | Marsit et al 2012 | ||
| Expression of 6 placental miRNAs | Increased miR-16 placental expression associated with reduced attention, Increased miR-146a and miR-182 placental expression associated with increased quality of movement in RICHS newborns. | Maccani et al 2013 | ||
ASD, autism spectrum disorder; MECP2, methyl CpG binding protein 2; OXTR, oxytocin receptor; RORA, RAR-related orphan receptor A; BCL-2, B-cell CLL/lymphoma 2; H3K4me3, trimethylation of lysine 4 of histone 3; EN-2, engrailed homeobox 2; DMR, differentially methylated region; PRRT1, proline-rich transmembrane protein 1; TSPAN32, tetraspanin 32; C11orf21, chromosome 11 open reading frame 21; ZFP57, zinc finger protein; SDHAP3, succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3; SHANK3, SH3 and multiple ankyrin repeat domains 3; DRD4, dopamine receptor D4; SLC6A4 solute carrier family 6; HSD11B2, hydroxysteroid (11-beta) dehydrogenase 2; NR3C1, glucocorticoid receptor; HTR2A 5-hydroxytryptamine (serotonin) receptor 2A; LEP, leptin.
Placental Epigenetics and Infant Neurobehavior
During intrauterine life, the placenta is the essential regulator of the fetal environment [28], and has been described as a third brain linking the mother and infant[29]. Recent evidence suggests similarities between neuronal and placental DNA methylation profiles in areas associated with neuronal development genes [30]. In order to study epigenetic changes that occur during prenatal development and their relationship with infant neurobehavioral outcomes, we have explored placental epigenetic marks as functional biomarkers of the in-utero environment in a large population-based cohort of healthy term infants: the Rhode Island Child Health Study (RICHS). We assessed newborn neurobehavior using the Neonatal Intensive Care Unit Network Neurobehavioral Scales (NNNS), a comprehensive evaluation of neurobehavioral performance, including neurologic and behavioral measures and signs of stress [31]. Profiles of neurobehavior derived through NNNS have previously shown to predict neurodevelopmental and cognitive performance in childhood [32].
Maternal cortisol influences the development of the fetal HPA axis and is metabolized through the placenta [33]. Thus, changes in placental cortisol metabolism may alter infant neurobehavioral outcomes. We have analyzed epigenetic changes in cortisol response genes HSD11B2 and NR3C1 within the RICHS cohort. HSD11B2 inactivates cortisol by metabolizing it to cortisone, protecting the infant from excess glucocorticoids [34]. HSD11B2 promoter methylation was associated with decreased quality of movement [35]. In an expanded study, we observed an interaction between maternal anxiety and HSD11B2 methylation that contributed to infant hypotonia [36]. NR3C1 encodes the glucocorticoid receptor, is expressed in the placenta and is involved in metabolism of maternal cortisol. NR3C1 placental methylation is positively associated with infant attention and quality of movement NNNS scores, and negatively associated with stress abstinence scores [37]. In a larger study, we observed an interaction between maternal depression and NR3C1 methylation on infant hypotonicity, lethargy and self-regulation[36]. Both HSD11B2 and NR3C1 promoter methylation are negatively associated with expression [35, 37], suggesting that infants with higher methylation of these genes are exposed to increased cortisol. The cortisol response pathway influences infant cognitive development and physical maturation in humans [38, 39]. Altered placental cortisol response may alter infant neuromuscular and stress responses, as reflected in the infant attention, stress-abstinence and quality of movement scores. Further analysis of other genes involved in cortisol response, such as FKBP5, is needed to fully understand the contribution of these epigenetic changes to infant neurobehavior.
Cortisol response and serotonergic tone are intimately linked, and serotonin can stimulate the HPA axis[34]. During fetal development, serotonin is important for the development of brain circuits[40], and the placenta acts as a transient source of serotonin during early development [41]. Infants that experienced maternal depression in-utero had decreased promoter methylation of the serotonin receptor SLC6A4 in blood[42], but we did not find associations between placental promoter methylation of SLC6A4 and infant neurobehavioral outcomes within the RICHS cohort (unpublished data). Methylation of the serotonin receptor HTR2A was positively associated with NNNS attention scores and negatively associated with quality of movement [43]. This study provided evidence for epigenetics as a potential regulator of components of the placental serotonin response pathway, which influence behavioral outcomes. More research is needed to determine if other genes in this pathway are epigenetically regulated.
Rodent studies have linked the adipokine leptin (LEP) with neurodevelopment; leptin deficient mice (ob/ob) display brain abnormalities and decreased locomotor activity [44]. Leptin is epigenetically regulated and produced by the placenta [45, 46]. Recently, we detected an associations between higher LEP promoter methylation and increased odds of membership in a neurobehavioral profile characterized by lethargy and hypotonicity and with reduced odds of membership in a profile with opposite characteristics [47]. These observations were significant only in males and are consistent with a marked negative correlation between methylation and LEP gene expression that was absent in placentas from females. These are the first results that link an energy-homeostasis gene with human neurobehavior and resemble the phenotype of ob/ob mice. Future research is needed to assess if epigenetic marks in other metabolic genes can influence neurobehavior.
MicroRNAs (miRNAs) post-transcriptionally target mRNAs and induce gene silencing, regulating a substantial amount of the mammalian genome[48]. miRNAs have been linked to placental functions and pathology and to neuronal survival and differentiation during development [49] [50]. We assessed placental expression of 6 miRNAs and their relationship to neurobehavior in the RICHS study [51]. Increases in miR-16 were associated with reduced attention scores, and increased miR-146a and miR-182 expression was associated with increased quality of movement scores. Some of the targets of these miRNAs are involved in regulation of the serotonin [52], NFκβ [53] and reward pathways [54], this could help explain our observation regarding infant neurobehavior.
Imprinted gene expression is abundant in human placenta and is involved in growth and neural development [13, 55]. We observed associations between expression profiles of 22 placental-imprinted genes and quality of movement and handling scores of RICHS infants [56]. Quality of movement was associated with decreased expression of imprinted genes involved in neurological and motor functions during development, including MEG3, HOXA11, and HOXD10. We also observed a high-degree of correlation in expression of adjacent imprinted genes, suggesting that in-utero exposures produce coordinated expression changes and/or disrupt imprinting within control regions. Further research is required to determine the role of epigenetic marks in imprinted genes and infant neurobehavior.
Future Directions
The field of neurobehavioral epigenetics is growing, with human studies complimenting animal models. The human environment is multifaceted, and the fetus is exposed to nonspecific stressors, which are difficult to capture in laboratory conditions. The laboratory environment may induce epigenetic alterations independently of experimental conditions, confounding analysis. However, there are limitations to the observations made from human population studies. Epigenetic changes are tissue-specific[57]; the placenta is a relevant and accessible tissue for infant neurobehavioral studies [30], but we cannot definitely assess if these epigenetic patterns are conserved in brain tissue. These studies are also limited by their observational nature; we cannot establish mechanisms based on observed associations, and we cannot presently assess the prognostic value of the neurobehavioral outcomes observed at birth. Most studies have used candidate-gene approaches of targets known to be important in the developing brain, and we encourage validation of findings from candidate-gene studies in different populations. However, this has a limited scope in complex neurobehavioral phenotypes, highlighting the need for epigenome-wide, agnostic analyses to identify novel genes that contribute to infant neurobehavior.
A number of neurobehavioral diseases exhibit sex-differences in their prevalence and onset including autism, ADHD, and affective disorders [58]. Placental epigenetic marks also exhibit sexual-dimorphism [59-61] [47] which could influence these neurobehavioral differences. More research is needed to define sexually-dimorphic epigenetic patterning in autosomal loci and their potential role in infant neurobehavioral outcomes.
DNA sequence variation also exerts effects on epigenetic signatures across the genome [62]. Thus, it is important to consider possible contributions of single nucleotide polymorphisms (SNPs) to epigenetic regulation of neurobehavior. It has been suggested that individuals may be able to adapt to deleterious polymorphisms through epigenetic changes, which may explain the inability of these polymorphisms alone to predict disease[63]. In particular, monozygotic twins represent a desirable population to study because of reduced genetic confounding. Differential epigenetic patterning in combination with genetic factors may help explain differences in behavioral responses.
As our understanding of epigenetic changes and their role in newborn behavior increases, they could serve as biomarkers of neurobehavioral risk, facilitating early screening. In neurobehavioral diseases that manifest in early-childhood, such as autism and ADHD, prompt interventions are important to improve long-term mental health [64, 65]. Future advancements may move this field beyond risk-assessment to identification of prognostic biomarkers to evaluate response to therapy. The brain epigenome exhibits plasticity throughout life [66], and response to cognitive therapies alters gene expression [67, 68], which may be driven by epigenetic changes. Tracking responses to cognitive interventions through epigenetic markers could provide a quantitative assessment of therapeutic response. Pharmacologic agents that alter gene expression through epigenetic changes are established treatment of some psychiatric and neurologic conditions; this is the case of valproic acid, and it has been proposed that this could be used to correct epigenetic changes in cognitive disorders [69, 70]. Maternal cognitive intervention may induce epigenetic effects in offspring, as epigenetic changes have been observed in children born to mothers who underwent bariatric surgery [71]. More groundwork is needed to understand the normal epigenome, the consequences of its deregulation and the connection with mental health disorders before these tools can be used as functional biomarkers.
Acknowledgments
This work is supported by NIH-NIMH R01MH094609, NIH-NIEHS R01ES022223 and NIH-NIEHS P01 ES022832/EPA RD83544201
References
- 1.Gluckman PD, Hanson MA, Spencer HG, Bateson P. Environmental influences during development and their later consequences for health and disease: implications for the interpretation of empirical studies. Proceedings of the Royal Society B: Biological Sciences. 2005;272:671–677. doi: 10.1098/rspb.2004.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Räikkönen K, Pesonen AK, Roseboom TJ, Eriksson JG. Early determinants of mental health. Best Practice & Research Clinical Endocrinology & Metabolism. 2012;26:599–611. doi: 10.1016/j.beem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Gluckman PD, Hanson MA, Pinal C. The developmental origins of adult disease. Maternal & child nutrition. 2005;1:130–141. doi: 10.1111/j.1740-8709.2005.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 5.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 6.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nature structural & molecular biology. 2013;20:282–9. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 7.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki A, Vega WCd, McGowan PO. Biological embedding in mental health: An epigenomic perspective 1. Biochemistry and Cell Biology. 2013;91:14–21. doi: 10.1139/bcb-2012-0070. [DOI] [PubMed] [Google Scholar]
- 9.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting Epigenetic Influence of Early-Life Adversity on the BDNF Gene. Biological Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature reviews Neuroscience. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 11.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 12.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annual review of cell and developmental biology. 2011;27:631–52. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nature reviews Neuroscience. 2007;8:832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 14.Yamada K, Matsuzawa H, Uchiyama M, Kwee IL, Nakada T. Brain developmental abnormalities in Prader-Willi syndrome detected by diffusion tensor imaging. Pediatrics. 2006;118:e442–8. doi: 10.1542/peds.2006-0637. [DOI] [PubMed] [Google Scholar]
- 15.Williams CA. Neurological aspects of the Angelman syndrome. Brain and Development. 2005;27:88–94. doi: 10.1016/j.braindev.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Temple IK, Clayton-Smith J, Mackay DJ. Epigenetic Epidemiology. Springer; 2012. Imprinting Disorders of Early Childhood; pp. 137–160. [Google Scholar]
- 17.Buiting K, Groβ S, Lich C, Gillessen-Kaesbach G, El-Maarri O, Horsthemke B. Epimutations in Prader-Willi and Angelman Syndromes: A Molecular Study of 136 Patients with an Imprinting Defect. The American Journal of Human Genetics. 2003;72:571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schanen NC. Epigenetics of autism spectrum disorders. Human molecular genetics. 2006;15:R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 19.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagarajan RP, Patzel KA, Martin M, Yasui DH, Swanberg SE, Hertz-Picciotto I, Hansen RL, Van de Water J, Pessah IN, Jiang R, Robinson WP, LaSalle JM. MECP2 promoter methylation and X chromosome inactivation in autism. Autism research : official journal of the International Society for Autism Research. 2008;1:169–78. doi: 10.1002/aur.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, Lintas C, Abramson RK, Wright HH, Ellis P, Langford CF, Worley G, Delong GR, Murphy SK, Cuccaro ML, Persico A, Pericak-Vance MA. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC medicine. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:3036–51. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, Guo Y, Lessard A, Akbarian S, Weng Z. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Archives of general psychiatry. 2012;69:314–24. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- 24.James SJ, Shpyleva S, Melnyk S, Pavliv O, Pogribny IP. Complex epigenetic regulation of engrailed-2 (EN-2) homeobox gene in the autism cerebellum. Translational psychiatry. 2013;3:e232. doi: 10.1038/tp.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, Bowman R, Yang H, Goldstein J, Li YJ, Jiang YH. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Human molecular genetics. 2014;23:1563–1578. doi: 10.1093/hmg/ddt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Mil NH, Steegers-Theunissen RP, Bouwland-Both MI, Verbiest MM, Rijlaarsdam J, Hofman A, Steegers EA, Heijmans BT, Jaddoe VW, Verhulst FC, Stolk L, Eilers PH, Uitterlinden AG, Tiemeier H. DNA methylation profiles at birth and child ADHD symptoms. Journal of psychiatric research. 2014;49:51–9. doi: 10.1016/j.jpsychires.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Maccani MA, Marsit CJ. Epigenetics in the placenta. American journal of reproductive immunology (New York, NY: 1989) 2009;62:78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yen SS. The placenta as the third brain. The Journal of reproductive medicine. 1994;39:277–80. [PubMed] [Google Scholar]
- 30.Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, LaSalle JM. The human placenta methylome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6037–42. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lester BM, Tronick EZ. The neonatal intensive care unit network neurobehavioral scale procedures. Pediatrics. 2004;113:641–667. [PubMed] [Google Scholar]
- 32.Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, Bauer C, Shankaran S, Bada H. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics. 2010;125:e90–8. doi: 10.1542/peds.2009-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis EP, Sandman CA. The Timing of Prenatal Exposure to Maternal Cortisol and Psychosocial Stress Is Associated With Human Infant Cognitive Development. Child Development. 2010;81:131–148. doi: 10.1111/j.1467-8624.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lester BM, Conradt E, Marsit CJ. Epigenetic basis for the development of depression in children. Clinical obstetrics and gynecology. 2013;56:556–65. doi: 10.1097/GRF.0b013e318299d2a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsit CJ, Maccani MA, Padbury JF, Lester BM. Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PloS one. 2012;7:e33794. doi: 10.1371/journal.pone.0033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The role of DNA methylation of <em>NR3C1</em> and <em>11β-HSD2</em> and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8:0–1. doi: 10.4161/epi.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bromer C, Marsit CJ, Armstrong DA, Padbury JF, Lester B. Genetic and Epigenetic Variation of the Glucocorticoid Receptor (NR3C1) in Placenta and Infant Neurobehavior. Dev Psychobiol. 2013;55:673–683. doi: 10.1002/dev.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis EP, Sandman CA. The timing of prenatal exposure to maternal cortisol and psychosocial stress is associated with human infant cognitive development. Child development. 2010;81:131–48. doi: 10.1111/j.1467-8624.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellman LM, Schetter CD, Hobel CJ, Chicz-DeMet A, Glynn LM, Sandman CA. Timing of fetal exposure to stress hormones: Effects on newborn physical and neuromuscular maturation. Developmental psychobiology. 2008;50:232–241. doi: 10.1002/dev.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deneris ES, Wyler SC. Serotonergic transcriptional networks and potential importance to mental health. Nature neuroscience. 2012;15:519–27. doi: 10.1038/nn.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, Blakely RD, Deneris ES, Levitt P. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472:347–50. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devlin AM, Brain U, Austin J, Oberlander TF. Prenatal Exposure to Maternal Depressed Mood and the <italic>MTHFR</italic> C677T Variant Affect <italic>SLC6A4</italic> Methylation in Infants at Birth. PloS one. 2010;5:e12201. doi: 10.1371/journal.pone.0012201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paquette AG, Lesseur C, Armstrong DA, Koestler DC, Appleton AA, Lester BM, Marsit CJ. Placental HTR2A methylation is associated with infant neurobehavioral outcomes. Epigenetics-Us. 2013;8:796–801. doi: 10.4161/epi.25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouret SG. Neurodevelopmental actions of leptin. Brain research. 2010;1350:2–9. doi: 10.1016/j.brainres.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambino YP, Maymó JL, Pérez Pérez A, Calvo JC, Sánchez-Margalet V, Varone CL. Elsevier Trophoblast Research Award Lecture: Molecular mechanisms underlying estrogen functions in trophoblastic cells- Focus on leptin expression. Placenta. 2011 doi: 10.1016/j.placenta.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Bouchard L, Thibault S, Guay SP, Santure M, Monpetit A, St-Pierre J, Perron P, Brisson D. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes care. 2010;33:2436–2441. doi: 10.2337/dc10-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesseur C, Armstrong DA, Murphy MA, Appleton AA, Koestler DC, Paquette AG, Lester BM, Marsit CJ. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology. 2014;40:1–9. doi: 10.1016/j.psyneuen.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales-Prieto DM, Ospina-Prieto S, Schmidt A, Chaiwangyen W, Markert UR. Elsevier Trophoblast Research Award Lecture: Origin, evolution and future of placenta miRNAs. Placenta. doi: 10.1016/j.placenta.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Maccani MA, Padbury JF, Lester BM, Knopik VS, Marsit CJ. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatric research. 2013;74:272–8. doi: 10.1038/pr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329:1537–41. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 53.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. Journal of immunology (Baltimore, Md : 1950) 2009;183:2150–8. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 54.Gerdjikov TV, Giles AC, Swain SN, Beninger RJ. Nucleus accumbens PKA inhibition blocks acquisition but enhances expression of amphetamine-produced conditioned activity in rats. Psychopharmacology. 2007;190:65–72. doi: 10.1007/s00213-006-0590-1. [DOI] [PubMed] [Google Scholar]
- 55.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marsit CJ, Lambertini L, Maccani MA, Koestler DC, Houseman EA, Padbury JF, Lester BM, Chen J. Placenta-imprinted gene expression association of infant neurobehavior. The Journal of pediatrics. 2012;160:854–860. e2. doi: 10.1016/j.jpeds.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones PA, Takai D. The Role of DNA Methylation in Mammalian Epigenetics. Science (New York, NY) 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 58.Viveros MP, Mendrek A, Paus T, Lopez Rodriguez AB, Marco EM, Yehuda R, Cohen H, Lehrner A, Wagner E. A comparative, developmental and clinical perspective of neurobehavioral sexual dimorphisms. Frontiers in Neuroscience. 2012;6 doi: 10.3389/fnins.2012.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C. Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics. Biology of sex differences. 2013;4:5. doi: 10.1186/2042-6410-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PloS one. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–14. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 62.Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. The American Journal of Human Genetics. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lester BM, Marsit CJ, Conradt E, Bromer C, Padbury JF. Behavioral epigenetics and the developmental origins of child mental health disorders. Journal of Developmental Origins of Health and Disease. 2012;3:395–408. doi: 10.1017/S2040174412000426. [DOI] [PubMed] [Google Scholar]
- 64.Chronis AM, Jones HA, Raggi VL. Evidence-based psychosocial treatments for children and adolescents with attention-deficit/hyperactivity disorder. Clinical Psychology Review. 2006;26:486–502. doi: 10.1016/j.cpr.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Moree BN, Davis TE., Iii Cognitive-behavioral therapy for anxiety in children diagnosed with autism spectrum disorders: Modification trends. Research in Autism Spectrum Disorders. 2010;4:346–354. [Google Scholar]
- 66.LaSalle JM, Powell WT, Yasui DH. Epigenetic layers and players underlying neurodevelopment. Trends in Neurosciences. 2013;36:460–470. doi: 10.1016/j.tins.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moustafa AA. Increased hippocampal volume and gene expression following cognitive behavioral therapy in PTSD. Frontiers in human neuroscience. 2013;7:747. doi: 10.3389/fnhum.2013.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atkinson D, Iannotti S, Cozzolino M, Castiglione S, Cicatelli A, Vyas B, Mortimer J, Hill R, Chovanec E, Chiamberlando A, Cuadros J, Virot C, Kerouac M, Kallfass T, Krippner S, Frederick C, Gregory B, Shaffran M, Bullock M, Soleimany E, Rossi AC, Rossi K, Rossi E. A new bioinformatics paradigm for the theory, research, and practice of therapeutic hypnosis. The American journal of clinical hypnosis. 2010;53:27–46. doi: 10.1080/00029157.2010.10401745. [DOI] [PubMed] [Google Scholar]
- 69.Narayan P, Dragunow M. Pharmacology of epigenetics in brain disorders. British journal of pharmacology. 2010;159:285–303. doi: 10.1111/j.1476-5381.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peedicayil J. Epigenetic Drugs in Cognitive Disorders. Curr Pharm Des. 2013 doi: 10.2174/13816128113199990526. [DOI] [PubMed] [Google Scholar]
- 71.Guénard F, Deshaies Y, Cianflone K, Kral JG, Marceau P, Vohl MC. Differential methylation in glucoregulatory genes of offspring born before vs. after maternal gastrointestinal bypass surgery. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1216959110. [DOI] [PMC free article] [PubMed] [Google Scholar]


