Abstract
The paired box gene 6 (PAX6) is a powerful mediator of eye and brain organogenesis whose spatiotemporal expression is exquisitely controlled by multiple mechanisms, including post-transcriptional regulation by microRNAs (miRNAs). In the present study, we use bioinformatic predictions to identify three candidate microRNA-7 (miR-7) target sites in the human PAX6 3′ untranslated region (3′-UTR) and demonstrate that two of them are functionally active in a human cell line. Furthermore, transient transfection of cells with synthetic miR-7 inhibits PAX6 protein expression but does not alter levels of PAX6 mRNA, suggesting that miR-7 induces translational repression of PAX6. Finally, a comparison of PAX6 3′-UTRs across species reveals that one of the functional miR-7 target sites is conserved, whereas the second functional target site is found only in primates. Thus, the interaction between PAX6 and miR-7 appears to be highly conserved; however, the precise number of sites through which this interaction occurs may have expanded throughout evolution.
Keywords: miR-7, PAX6, miRNA, 3′-UTR
Introduction
MicroRNA (miRNA) regulation of protein expression adds a subtle layer of complex fine-tuning on top of numerous modes of protein production and destruction. miRNAs typically bind to specific sites within the 3′ untranslated regions (3′-UTRs) of their target mRNAs, to impart post-transcriptional silencing, either through translational repression and/or mRNA decay.1,2 In animals, miRNAs typically exhibit partial complementary base pairing to their mRNA targets, with the “seed region” of six to eight nucleotides toward the 5′end of the miRNA being important for target specificity.3 miRNA target efficacy is predicted to increase with the number of Watson-Crick matches to the seed region and is also dependent on base composition at nucleotide position 1.2,4 For example, a perfect match to nucleotides 2–8, referred to as a “7Mer-m8” target site, has a higher hierarchy rating compared to a “7Mer-A1” target site, which only confers Watson-Crick base pairing to nucleotides 2–7.3 Noticeably, the latter has an A at nucleotide position 1, which again rates it higher than a “6Mer” with the same 2–7 seed match, but a different number 1 nucleotide (C, G, or T).4 However, other factors such as AU-rich neighbor nucleotides and accessibility may also influence site-efficacy.5,6 Hence, miRNAs target specific genes for post-transcriptional regulation, fine-tuning protein expression levels during critical processes such as cell proliferation, differentiation, and maturation.7
The functional roles of microRNA-7 (miR-7) have been extensively studied, as it is highly conserved, being detected in 81 species (October 2013: http://www.mirbase.org),8–10 and controls multiple cell signaling networks, including epidermal growth factor receptor,11 insulin-like growth factor,12 Hedgehog,13 and the mammalian target of rapamycin (mTOR) signaling pathways.14 miR-7 is also a critical regulator of multiple regulatory genes, including paired box gene 6 (PAX6) in mice.8,9 PAX6 encodes a neurogenic transcription factor, which is highly conserved, both structurally and functionally, and is critical for neural tube polarization, brain regionalization, and eye formation.15–17 PAX6 is widely co-expressed with miR-7 in several tissues including the forebrain, pancreas, and retina where it is involved in regulating cellular differentiation.8–10
Given the functional importance of the miR-7/PAX6 regulatory apparatus in controlling cell fate in mouse, we sought to evaluate whether this mechanism was conserved in humans. Herein we use bioinformatics to predict three miR-7 target sites in the human PAX6 3′-UTR and reporter gene assays to confirm the functional capacity of two of these predicted target sites. Further, we compare the human PAX6 3′-UTR region across species and identify a conserved miR-7 3′-UTR target site present in many species, as well as a second functional miR-7 target site in the human PAX6 3′-UTR that is specific to primates.
Materials and Methods
Prediction of miR-7 target sites
3′-UTRs were identified using the “Spidey” freeware, which is part of the NCBI toolkit (http://www.ncbi.nlm.nih.gov/spidey/). Target sites for hsa-miR-7 within human PAX6 3′-UTR (GenBank accession number: NM_001127612) were predicted using TargetScan software (release 6.2)4 with the following search criteria: species: human, human Entrez gene symbol: PAX6, and miRNA name: hsa-miR-7. Target sites for miR-7 in 3′-UTRs of other species (GenBank accession numbers given in Supplementary Table 1) were predicted using a “Find in This Sequence” function available at GenBank to search for the miR-7 seed region (5′-GUCUUCC-3′). Notably, only species that encode miR-7 (mirbase.org) are included in the analysis. Nucleotide position numbers for 3′-UTRs are listed in Supplementary Table 1.
Luciferase reporter constructs
Luciferase reporter plasmids were generated by ligating annealed DNA oligo nucleotides containing each of the three predicted PAX6 3′-UTR hsa-miR-7 7mer targets sites (NM_001127612.1; m8#1: nt. 2389–2441, m8#2: nt. 3613–3664, and A1#3: nt. 4170–4216) to the pMiR-REPORT luciferase plasmid (Ambion) via HindIII and SpeI restriction sites. Mutated (Mut) miR-7 seed target sites with three nucleotide substitutions were generated following the same procedure (for sequences refer to Supplementary Table 2). A perfect hsa-miR-7 target site (forward: 5′-CAACAAAATCACTAGTCTTCCA-3′ and reverse: 5′-TGGAAGACTAGTGATTTTGTTG-3′) inserted in the pGL3-control (Promega) firefly luciferase reporter vector was used as a control as previously described.11,18 Sequences of all plasmids were confirmed by Sanger DNA sequencing.
Cell culture
HeLa and HEK293 cells were obtained from the American Type Culture Collection (ATCC) and maintained at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM)/F-12-GlutaMAX (Life Technologies) supplemented with 5% fetal bovine serum (Serana) and 100 units/mL of Penicillin/Streptomycin (Life Technologies).
Luciferase assays
HEK293 cells in 24-well plates were co-transfected with 1) firefly luciferase reporter plasmid DNA (100 ng) containing either PAX6 3′-UTR WT or Mut miR-7 target sites (7mer-m8#1, 7mer-m8#2, and 7mer-A1#3), 2) a control pRL-CMV Renilla luciferase reporter plasmid (5 ng), and 3) precursor miRNAs (30 nM) purchased from Ambion, corresponding to human miR-7 (ID: PM10047) or a miRNA negative control (miR-NC) (ID: AM17110) using Lipofectamine 2000 (Invitrogen). Cells were harvested 24 hours post-transfection and assayed using a Dual-Luciferase Reporter Assay System (Promega) and an EnSpire Multimode Plate Reader (PerkinElmer). For data analysis, firefly luciferase activity was firstly normalized to Renilla (transfection control) and then hsa-miR-7 was normalized to miR-NC transfected cells.
Transfection
HeLa cells were seeded 24 hours before transfection with miR-7 or miR-NC miRNA precursor molecules (30 nM) using RNAi/MAX transfection reagent (Invitrogen). Cells were harvested at 24 hours (for RNA) or 72 hours (for protein) post-transfection.
Western blotting
HeLa cells were lysed by sonication (4 × 5 seconds) in ice-cold lysis buffer (0.1% SDS, 10 mM Tris–HCl pH 7.5) containing 1× complete inhibitor cocktail tablet (Roche) and protein concentrations determined by Bradford assay (Bio-Rad). Samples (70 μg) were electrophoretically separated on a 12.5% polyacrylamide gel, transferred to a Hybond-P membrane (Amersham Biosciences), and probed with anti-PAX6 (1:1,000; Abcam Clone AD2.38) or anti-β-actin (1:10,000; Sigma-Aldrich Clone AC-15) mouse monoclonal antibodies before detection with the commercially available Qdot 625 system (Molecular Probes) and UV light using Universal Hood II (Bio-Rad) for visualization of a representative image. A colored ladder was used to determine protein size (Marker Precision Plus Protein Kaleidoscope standard; Bio-Rad).
RT-qPCR
Total RNA was extracted (SV Total RNA Isolation System; Promega), quality assessed, and quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific) with threshold cutoff for DNA contamination (260/280) at 2.0. cDNA was synthesized (SuperScript III; Invitrogen) using 250 ng RNA and random hexamers according to the manufacturer’s instructions. qPCR assays were performed using a Rotor-Gene Q (Qiagen) with SYBR-Green chemistry (GoTaq qPCR Master Mix; Promega) and previously published gene-specific primers: PAX619–21 forward: 5′-TCTTTGCTTGGGAAATCCG-3′ and reverse: 5′-CTGCCCGTTCAACATCCTTAG-3′; glyceraldehyde 3-phosphate dehydrogenase (GAPDH)22–25 forward: 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse: 5′-GAAGATGGTGATGGGATTTC-3′. Primer-specificity was tested using gel electrophoresis (to verify single band and amplicon size), DNA sequencing, and melting profile. A no-reverse transcription control was included in each run for detection of potential PCR contamination. Normalized PAX6 expression relative to GAPDH was calculated using the 2−ΔΔCt method.26
Statistical analysis
Data are presented as mean of three independent experiments (n = 3) + standard deviation (SD). Significance (P < 0.05) was assessed using a paired t-test (RT-qPCR) and ANOVA Tukey’s post-hoc analysis (luciferase assay) in R (version 3.0.2).27
Results and Discussion
The human PAX6 3′-UTR contains two functional miR-7 target sites
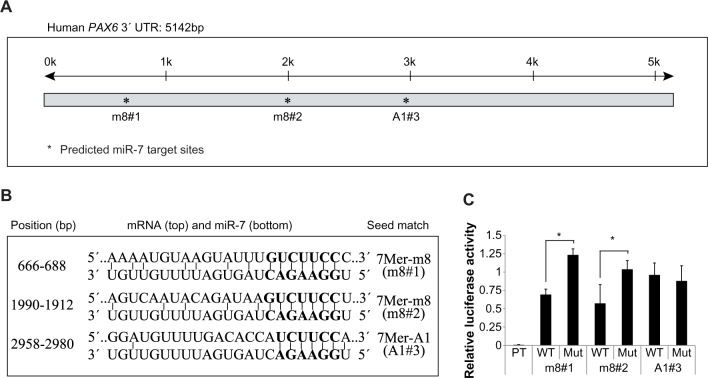
There are seven verified human PAX6 transcripts in the NCBI database, which result from alternative splicing and alternative initiation of transcription. However, it is noteworthy that they all share identical 3′-UTRs accounting for approximately 75% of the total mRNA sequence, which allows for the suggestion that PAX6 expression is possibly regulated by miRNAs. Using TargetScan software,4 we identified three miR-7 target sites: one of which (m8#1) has previously been shown to regulate PAX6 expression in mouse9 and two additional miR-7 target sites (m8#2 and A1#3) were predicted when poorly conserved target sites were considered (Fig. 1A). Two of the three predicted miR-7 target sites within the PAX6 3′-UTR contained 7mer-m8 seed matches (m8#1 and m8#2), whereas the third target site contained a lower hierarchy 7mer-A1 seed match (A1#3)4 (Fig. 1B). Noticeably, the A1#3 target site has a mismatch at position 8 in the miR-7 sequence (Fig. 1B), which has been shown to be essential for functional miR-7-target binding.28
Figure 1.
The human PAX6 3′-UTR contains two specific, functional miR-7 target sites. (A) Prediction of miRNA-7 target sites within the human PAX6 3′-UTR. Black line (top) illustrates the length of the PAX6 3′-UTR (k = kilobase), and asterisks illustrate predicted miR-7 target sites (m8#1, m8#2, and A1#3). (B) Alignment of predicted human PAX6 miR-7 target sites (top) to miR-7 (bottom). The seed target region (GUCUUCC) is in bold, and the seed matches, target sites, and positions are given. (C) Luciferase assays using firefly reporter constructs with predicted (WT: wild type and Mut) miR-7 7mer target sites (m8#1, m8#2, and A1#3) and a perfect miR-7 target site (PT) as a positive control. The firefly luciferase reporters were co-transfected into HEK293 cells with a Renilla luciferase reporter as a transfection control and either miR-7 or miR-NC precursors. Relative luciferase expression (firefly normalized to Renilla) values of hsa-miR-7 were normalized to miR-NC transfected controls, and is given as mean + SD. The relative luciferase activity (firefly normalized to Renilla) of the positive control was calculated as miR-7 relative to miR-NC and is given as mean + SD, *P < 0.05.
To examine the functional significance of the three predicted human miR-7 target sites within the PAX6 3′-UTR, we generated firefly luciferase reporter constructs containing each of these individual sites, as well as Mut constructs of each. We then co-transfected HEK293 cells with the firefly and Renilla (transfection control) luciferase constructs and either hsa-miR-7 or miR-NC (a negative control) precursor miRNAs to assess target specificity. To confirm the reliability of our assay, we first measured luciferase activity of a construct containing a perfect miR-7 target site (5′-CAACAAAATCACTAGTCTTCCA-3′), which resulted in a 99% reduction in luciferase activity (Renilla normalized) compared to cells transfected with miR-NC precursors (Fig. 1C). Assessment of cells transfected with PAX6 3′-UTR target site firefly luciferase reporter plasmids revealed that two of the three miR-7 target sites (m8#1 and m8#2) were functionally active, each yielding a significant reduction in luciferase activity of approximately 45% with miR-7 relative to their Mut target site reporters (Fig. 1C). Both of these active sites contained 7mer-m8-predicted miR-7 target sites, whereas the third site (7mer-A1#3) did not show any regulation by miR-7. Our study therefore confirms previous findings, showing that Watson-Crick base pairing to nucleotide position 8 is essential for functional miR-7-target binding.28
Taken together, these data indicate that the PAX6 3′-UTR contains two 7 mer-m8 miR-7 seed sites. A third predicted 7 mer-A1 miR-7 target site that did not comply with Watson-Crick pairing at nucleotide 8 failed to show functional activity in luciferase reporter assays.
PAX6 protein expression is regulated by miR-7 in human cells
To assess whether miR-7 regulates PAX6 protein expression in human cells, we transiently transfected HeLa, a human cervical cancer cell line, with miR-7 precursor molecules and measured PAX6 levels by Western blotting. Compared with a negative miRNA precursor molecule (miR-NC), miR-7 reduced expression of PAX6 in HeLa cells (Fig. 2A). To determine whether miR-7 reduced PAX6 protein levels through interfering with the translational pathway or mediating mRNA decay, we assessed endogenous PAX6 mRNA levels by RT-qPCR; however, we saw no significant difference in PAX6 mRNA expression between HeLa cells transfected with hsa-miR-7 or miR-NC miRNA precursor molecules (Fig. 2B).
Figure 2.
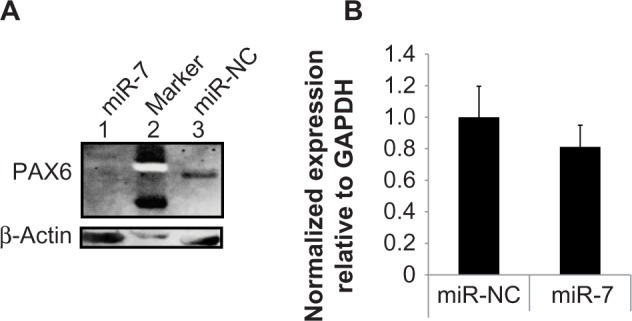
miR-7 reduces PAX6 protein expression in human cells without inducing PAX6 mRNA decay. (A) Western blot analysis of HeLa cells transfected with hsa-miR-7 (lane 1) or control non-coding miR-NC miRNA (lane 3) using antibodies against PAX6 (top) and β-actin (bottom). A protein marker (lane 2) confirmed the correct size of PAX6 (48 kDa) and β-actin (42 kDa) proteins. (B) RT-qPCR analysis of PAX6 mRNA isolated from HeLa cells transfected with miRNA precursors corresponding to hsa-miR-7 (miR-7) and a negative control (miR-NC). PAX6 message levels were normalized relative to GAPDH and are presented as mean + SD.
Taken together, these data indicate that miR-7 regulates PAX6 protein levels in human cells, but has no significant effect on mRNA levels, which suggests that miR-7 interferes with the translational pathway rather than mediating PAX6 mRNA decay.
Diversification of PAX6 3′-UTR miR-7 target sites across species
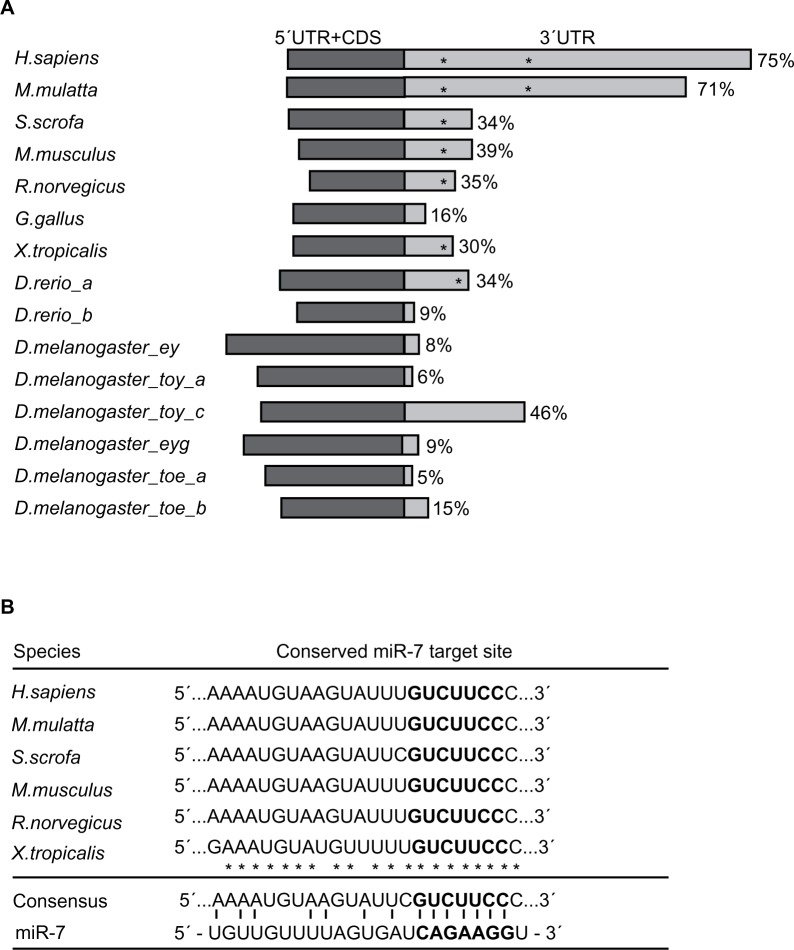
Recent experiments in mice have demonstrated that miR-7 directly represses the expression of PAX6 through a single miR-7 target site in the mouse PAX6 3′-UTR.8,9 Given our identification of a second functional miR-7 target site (7mer-m8) in human PAX6 3′-UTR compared to mouse, we examined the evolutionary conservation of PAX6 3′-UTRs and their miR-7 target sites across species, and found that the length of PAX6 3′-UTRs varied considerably and that this length seemed to be associated with the number of predicted miR-7 target sites (Fig. 3A). For example, in primates, human (Homo sapiens) and rhesus monkey (Macaca mulatta), PAX6 has relatively long 3′-UTRs (accounting for more than 70% of the total mRNA sequences), and we demonstrated the existence of two functional 7mer-m8 miR-7 target sites within these regions. In comparison, the 3′-UTRs of pig (Sus scrofa), rodents (Mus musculus and Rattus norvegicus), frog (Xenopus tropicalis) and fish (Danio rerio) account for between 30 and 39% of total PAX6 mRNA length and contain only one predicted miR-7 target site (Fig. 3A). In other species such as chicken (Gallus gallus) and fruit fly (Drosophila melanogaster), where the 3′-UTR accounts for less than 20% of the total PAX6 mRNA sequence, no miR-7 target sites were predicted (Fig. 3A). Notably, in zebrafish, there are two duplicated PAX6 gene variants, PAX6a and PAX6b, with distinct 3′-UTR lengths, accounting for 34%–39% of the total mRNA lengths, respectively (Fig. 3A). PAX6a contains a single predicted miR-7 target site, whereas no site was predicted in the shorter 3′-UTR of the PAX6b variant, suggesting that perhaps only PAX6a is post-transcription-ally regulated by miR-7. Also, the fruit fly has multiple PAX6 gene loci, from which six different PAX6 3′-UTRs are transcribed; none have a predicted miR-7 target site in the 3′-UTR despite one transcript having a 3′-UTR that accounts for more than 45% of the total mRNA sequence (Fig. 3A), suggesting that miR-7 might not regulate PAX6 expression levels in fruit flies. These results are in agreement with previous work that suggested the expansion of 3′-UTR sequences increases with organism complexity throughout evolution.29
Figure 3.
Diversification of the PAX6 3′-UTR and prediction of 7mer-m8 target sites. (A) Schematic representation of validated PAX6 mRNAs from the following species: H. sapiens (human), M. mulatta (rhesus monkey), S. scrofa (pig), M. musculus (mouse), R. norvegicus (rat), G. gallus (chicken), X. tropicalis (frog), D. rerio (zebrafish): PAX6a and PAX6b, and D. melanogaster (fruit fly), and eyeless (Ey), twin of eyeless (Toy) isoforms a and c, eyegone (Eyg), and twin of eyegone (Toe) variants a and b. The 5′-UTRs and coding DNA sequences (CDSs) are represented in dark gray, and 3′-UTRs in light gray. Percentages illustrate the size of individual 3′-UTRs compared to full-length transcripts, and asterisks illustrate predicted miR-7 target sites. (B) Alignment of the conserved predicted 7mer-m8 miR-7 target sites of H. sapiens, M. mulatta, S. scrofa, M. musculus, R. norvegicus, and X. tropicalis. The seed region is in bold, and identical nucleotides are illustrated with an asterisk. The consensus sequence is given and base pairing to miR-7 is illustrated.
In species with a predicted miR-7 target site in the PAX6 3′-UTR, sequence alignment revealed that 20 of 23 nucleotides were conserved and that all predicted miR-7 target sites shared identical 7mer-m8 seed regions (Fig. 3B). Thus, the first functional human 7mer-m8 PAX6 miR-7 target site (more 5′-located; m8#1 in Figure 1) appears to be conserved, whereas the second functional human 7mer-m8 miR-7 target site (more 3′-located; m8#2 in Figure 1) does not. The identification of a miR-7 target site positioned outside the conserved region suggests that this site may have resulted from the expansion of the PAX6 3′-UTR. The appearance of multiple functional miR-7 binding sites within the human PAX6 3′-UTR suggests an added level of complexity by allowing the combinatorial regulation of PAX6 expression by miR-7.7
In summary, given our finding that miR-7 can regulate PAX6 expression via two miR-7 target sites in a human cell line, future experiments are needed to examine the physiological importance of this regulatory system, in particular, examining whether there is a combinatory effect between the two functional human miR-7 target sites (m8#1 and m8#2) or if a dysfunctional miR-7/PAX6 regulatory system promotes tumorigenesis, as both factors are highly associated with cancer.30–34
Conclusions
In conclusion, we provide the first demonstration that miR-7 regulates the expression of PAX6 protein in human cells, and show that this regulation occurs via two target sites within the PAX6 3′-UTR: the first site is highly conserved across species, whereas the second functional miR-7 site is conserved only in primates. This finding leads to the intriguing possibility that the expanded 3′-UTR found in human PAX6 may allow for a tighter control of PAX6 expression by miR-7 than that already documented in mouse.
Supplementary Data
Table 1.
PAX6 3′untranslated regions.
| SPECIES | VARIANTS | ACCESSION NUMBERS | 3′UTR (START-STOP) | |
|---|---|---|---|---|
| Homo Sapiens | N/A | NM_001127612.1 | 1739–6883 | |
| Macaca mulatta | N/A | NM_001266257.1 | 1711–5908 | |
| Sus scrofa | N/A | NM_001244172.1 | 1728–2643 | |
| Mus musculus | N/A | NM_001244198.1 | 1597–2619 | |
| Rattus norvegicus | N/A | NM_013001.2 | 1430–2191 | |
| Gallus gallus | N/A | NM_205066.1 | 1641–1961 | |
| Xenopus tropicalis | N/A | NM_001006762.1 | 1651–2361 | |
| Danio rerio | PAX6a | NM_131304.1 | 1845–2809 | |
| PAX6b | NM_131641.1 | 1575–1733 | ||
| Drosophila Melanogaster | Eyeless (Ey) | NM_166789.2 | 2621–2851 | |
| Twin of eyeless (Toy) | Variant a | NM_079899.4 | 2160–2287 | |
| Variant c | NM_00127215.1 | 2109–3870 | ||
| Eyegone (Eyg) | NM_001014582.1 | 2350–2586 | ||
| Twin of eyegone (Toe) | Variant a | NM_07 9 317. 3 | 2279–2400 | |
| Variant b | NM_001274833.1 | 2030–2303 |
Notes: List of PAX6 GenBank accession numbers, N/A = “not applicable.” Nucleotide position numbers for the 5′end (start) and 3′end (stop) of the 3′-UTRs are listed.
Table 2.
Oligonucleotides used for generating firefly luciferase constructs.
| TARGET SITES | OLIGONUCLEOTIDES (5′→3′) | ||
|---|---|---|---|
| m8#1 | WT | SE | AGCTTTCTGAGGATTTCTAGGGAAGACAAATACTTACATTTTGACATAAAACAAATT |
| AS | CTAGAATTTGTTTTATGTCAAAATGTAAGTATTTGTCTTCCCTAGAAATCCTCAGAA | ||
| Mut | SE | AGCTTTCTGAGGATTTCTAGGAAGGGCAAATACTTACATTTTGACATAAAACAAATT | |
| AS | CTAGAATTTGTTTTATGTCAAAATGTAAGTATTTGCCCTTCCTAGAAATCCTCAGAA | ||
| m8#2 | WT | SE | AGCTAGTGAAGTTTGCAGAGGAAGACTTATCTGTATTGACTTATATGTTGCACAGA |
| AS | CTAGTCTGTGCAACATATAAGTCAATACAGATAAGTCTTCCTCTGCAAACTTCACT | ||
| Mut | SE | AGCTAGTGAAGTTTGCAGAGAAGGGCTTATCTGTATTGACTTATATGTTGCACAGA | |
| AS | CTAGTCTGTGCAACATATAAGTCAATACAGATAAGCCCTTCTCTGCAAACTTCACT | ||
| A1# 3 | WT | SE | AGCTCAAATAATCTCCATCCTGGAAGATGGTGTCAAAACATCCCTGCAGATACCCCA |
| AS | CTAGTGGGGTATCTGCAGGGATGTTTTGACACCATCTTCCAGGATGGAGATTATTTG | ||
| Mut | SE | AGCTCAAATAATCTCCATCCTGAAGGGTGGTGTCAAAACATCCCTGCAGATACCCCA | |
| AS | CTAGTGGGGTATCTGCAGGGATGTTTTGACACCACCCTTCAGGATGGAGATTATTTG | ||
Notes: List of sense (SE) and antisense (AS) oligonucleotides used for generating firefly luciferase constructs containing WT and Mut miR-7 7mer target sites (m8#1, m8#2, and A1#3). Italic, bold, and underlined nucleotides represent restriction recognition-, miR-7 target- and Mut sites, respectively.
Footnotes
Author Contributions
MN, RBW, KMG, SAD, and MGT conceived and designed the experiments. MN, RBW, KMG, and MGT analyzed the data. MN wrote the first draft of the manuscript. MN, RBW, KMG, SAD, and MGT contributed to the writing of the manuscript. MN, RBW, KMG, SAD, and MGT agreed with manuscript results and conclusions. MN, RBW, KMG, SAD, and MGT jointly developed the structure and arguments for the paper. MN, RBW, KMG, SAD, and MGT made critical revisions and approved the final version. All authors reviewed and approved the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
ACADEMIC EDITOR: Jike Cui, Associate Editor
FUNDING: MGT was funded by Edith Cowan University’s Office of Research and Innovation, the Faculty of Computing, Health and Science, and the Vario Health Institution. RBW was funded by a Raine Medical Research Foundation priming grant and an Early Career grant from the Cancer Council of Western Australia. SAD was funded by an NHMRC Principal Research’s fellowship (1002347), MN received a postgraduate research scholarship from Edith Cowan University, and KG was the recipient of a Royal Perth Hospital Medical Research Foundation’s fellowship.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 7.Vo NK, Cambronne XA, Goodman RH. MicroRNA pathways in neural development and plasticity. Curr Opin Neurobiol. 2010;20(4):457–65. doi: 10.1016/j.conb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Kredo-Russo S, Mandelbaum AD, Ness A, et al. Pancreas-enriched miRNA refines endocrine cell differentiation. Development. 2012;139(16):3021–31. doi: 10.1242/dev.080127. [DOI] [PubMed] [Google Scholar]
- 9.de Chevigny A, Coré N, Follert P, et al. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci. 2012;15(8):1120–6. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- 10.Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48(9):3962–7. doi: 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- 11.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284(9):5731–41. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 12.Jiang L, Liu X, Chen Z, et al. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432(1):199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da R, Gutierrez-Perez I, Ferres-Marco D, Dominguez M. Dampening the signals transduced through Hedgehog via microRNA miR-7 facilitates notch-induced tumourigenesis. PLoS Biol. 2013;11(5):1–16. doi: 10.1371/journal.pbio.1001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic beta-cells. Diabetes. 2013;62(3):887–95. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericson J, Rashbass P, Schedl A, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90(1):169–80. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 16.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21(5):1031–44. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 17.Stoykova A, Gruss P. Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J Neurosci. 1994;14(3 pt 2):1395–412. doi: 10.1523/JNEUROSCI.14-03-01395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giles KM, Daly JM, Beveridge DJ, et al. The 3’-untranslated region of p21 WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J Biol Chem. 2003;278(5):2937–46. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Huang CT, Chen J, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7(1):90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136(23):4055–63. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belinsky GS, Sirois CL, Rich MT, et al. Dopamine receptors in human embryonic stem cell neurodifferentiation. Stem Cells Dev. 2013;22(10):1522–40. doi: 10.1089/scd.2012.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin MC, Liu YC, Tam MF, Lu YJ, Hsieh YT, Lin LY. PTEN interacts with metal-responsive transcription factor 1 and stimulates its transcriptional activity. Biochem J. 2012;441(1):367–77. doi: 10.1042/BJ20111257. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Wu X, Yu FS, Zhao J. Toll-like receptor 2 mediates the induction of IL-10 in corneal fibroblasts in response to. Fusarium solu Immunol Cell Biol. 2008;86(3):271–6. doi: 10.1038/sj.icb.7100158. [DOI] [PubMed] [Google Scholar]
- 24.Lukasiak S, Schiller C, Oehlschlaeger P, et al. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27(46):6068–74. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Chen Y, Qin X, et al. MiR-138 downregulates miRNA processing in HeLa cells by targeting RMND5 A and decreasing Exportin-5 stability. Nucleic Acids Res. 2014;42(1):458–74. doi: 10.1093/nar/gkt839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.R Foundation for Statistical Computing . R: A Language and Environment for Statistical Computing [Computer Program]. Version 3.0.2. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 28.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazumder B, Seshadri V, Fox PL. Translational control by the 3’-UTR: the ends specify the means. Trends Biochem Sci. 2003;28(2):91–8. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 30.Shyr CR, Tsai MY, Yeh S, et al. Tumor suppressor PAX6 functions as androgen receptor co-repressor to inhibit prostate cancer growth. Prostate. 2010;70(2):190–9. doi: 10.1002/pros.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mascarenhas JB, Young KP, Littlejohn EL, Yoo BK, Salgia R, Lang D. PAX6 is expressed in pancreatic cancer and actively participates in cancer progression through activation of the MET tyrosine kinase receptor gene. J Biol Chem. 2009;284(40):27524–32. doi: 10.1074/jbc.M109.047209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong X, Yang H, Yu Y, et al. Possible role of Pax-6 in promoting breast cancer cell proliferation and tumorigenesis. BMB Rep. 2011;44(9):595–600. doi: 10.5483/bmbrep.2011.44.9.595. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Yue W, Zhang L, et al. Downregulation of PAX6 by shRNA inhibits proliferation and cell cycle progression of human non-small cell lung cancer cell lines. PLoS ONE. 2014;9(1):e85738. doi: 10.1371/journal.pone.0085738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1.
PAX6 3′untranslated regions.
| SPECIES | VARIANTS | ACCESSION NUMBERS | 3′UTR (START-STOP) | |
|---|---|---|---|---|
| Homo Sapiens | N/A | NM_001127612.1 | 1739–6883 | |
| Macaca mulatta | N/A | NM_001266257.1 | 1711–5908 | |
| Sus scrofa | N/A | NM_001244172.1 | 1728–2643 | |
| Mus musculus | N/A | NM_001244198.1 | 1597–2619 | |
| Rattus norvegicus | N/A | NM_013001.2 | 1430–2191 | |
| Gallus gallus | N/A | NM_205066.1 | 1641–1961 | |
| Xenopus tropicalis | N/A | NM_001006762.1 | 1651–2361 | |
| Danio rerio | PAX6a | NM_131304.1 | 1845–2809 | |
| PAX6b | NM_131641.1 | 1575–1733 | ||
| Drosophila Melanogaster | Eyeless (Ey) | NM_166789.2 | 2621–2851 | |
| Twin of eyeless (Toy) | Variant a | NM_079899.4 | 2160–2287 | |
| Variant c | NM_00127215.1 | 2109–3870 | ||
| Eyegone (Eyg) | NM_001014582.1 | 2350–2586 | ||
| Twin of eyegone (Toe) | Variant a | NM_07 9 317. 3 | 2279–2400 | |
| Variant b | NM_001274833.1 | 2030–2303 |
Notes: List of PAX6 GenBank accession numbers, N/A = “not applicable.” Nucleotide position numbers for the 5′end (start) and 3′end (stop) of the 3′-UTRs are listed.
Table 2.
Oligonucleotides used for generating firefly luciferase constructs.
| TARGET SITES | OLIGONUCLEOTIDES (5′→3′) | ||
|---|---|---|---|
| m8#1 | WT | SE | AGCTTTCTGAGGATTTCTAGGGAAGACAAATACTTACATTTTGACATAAAACAAATT |
| AS | CTAGAATTTGTTTTATGTCAAAATGTAAGTATTTGTCTTCCCTAGAAATCCTCAGAA | ||
| Mut | SE | AGCTTTCTGAGGATTTCTAGGAAGGGCAAATACTTACATTTTGACATAAAACAAATT | |
| AS | CTAGAATTTGTTTTATGTCAAAATGTAAGTATTTGCCCTTCCTAGAAATCCTCAGAA | ||
| m8#2 | WT | SE | AGCTAGTGAAGTTTGCAGAGGAAGACTTATCTGTATTGACTTATATGTTGCACAGA |
| AS | CTAGTCTGTGCAACATATAAGTCAATACAGATAAGTCTTCCTCTGCAAACTTCACT | ||
| Mut | SE | AGCTAGTGAAGTTTGCAGAGAAGGGCTTATCTGTATTGACTTATATGTTGCACAGA | |
| AS | CTAGTCTGTGCAACATATAAGTCAATACAGATAAGCCCTTCTCTGCAAACTTCACT | ||
| A1# 3 | WT | SE | AGCTCAAATAATCTCCATCCTGGAAGATGGTGTCAAAACATCCCTGCAGATACCCCA |
| AS | CTAGTGGGGTATCTGCAGGGATGTTTTGACACCATCTTCCAGGATGGAGATTATTTG | ||
| Mut | SE | AGCTCAAATAATCTCCATCCTGAAGGGTGGTGTCAAAACATCCCTGCAGATACCCCA | |
| AS | CTAGTGGGGTATCTGCAGGGATGTTTTGACACCACCCTTCAGGATGGAGATTATTTG | ||
Notes: List of sense (SE) and antisense (AS) oligonucleotides used for generating firefly luciferase constructs containing WT and Mut miR-7 7mer target sites (m8#1, m8#2, and A1#3). Italic, bold, and underlined nucleotides represent restriction recognition-, miR-7 target- and Mut sites, respectively.