Abstract
A full-term pregnancy early in life reduces lifetime risk of developing breast cancer, and the effect can be mimicked in rodents by full-term pregnancy or short-term treatment with exogenous estrogen and progesterone. To gain insight into the protective mechanism, 15 3-mo-old postpubertal virgin Lewis rats were randomly assigned to three groups: control (C), pregnancy (P), or hormone (H). The P group animals underwent a full-term pregnancy, and H group animals were implanted subcutaneously with silastic capsules filled with ethynyl estradiol and megesterol acetate for 21 days. C and P animals were implanted with sham capsules. On day 21 capsules were removed, which was followed by a 49-day involution period, euthanasia, and mammary tissue collection. Global gene expression was measured using Rat Genome 230.2 Arrays. Histological analysis revealed that P and H treatments induced sustained morphological changes in the mammary gland with significantly increased percentages of mammary parenchyma and stromal tissues and higher ratio of stroma to parenchyma. Transcriptome analysis showed that P and H treatments induced sustained global changes in gene expression in the mammary gland. Analysis of commonly up- and downregulated genes in P and H relative to C treatment showed increased expression of three matrix metallopeptidases (Mmp3, 8, and 12), more differentiated mammary phenotype, enhanced innate and adaptive immunity, and reduced cell proliferation and angiogenic signatures. The sustained morphological and global gene expression changes in mammary tissue after pregnancy and hormone treatment may function together to provide the protective effect against breast cancer.
Keywords: breast cancer protection, hormone, pregnancy
breast cancer is one of the most common cancers in women and affects nearly 10% of all women in US. In the year 2009, 192,370 new cases of invasive and 62,280 new cases of in situ breast cancer were estimated to have occurred, with 40,170 estimated deaths (3). Various epidemiological studies have revealed that multiple factors including hormones, genetics, reproductive history, radiation, socio-economic status, place of residence, ethnicity, and the environment affect the incidence of breast cancer (9, 19, 28, 29, 50, 51, 60). It has been shown that full-term pregnancy early in life has a protective effect on women against the risk of breast cancer irrespective of genetic background, age, race, or ethnic background (2, 37, 39, 40, 64, 65, 72). For instance, Lambe et al. (1996) (39, 40) and Albrektsen et al. (2005) (2) reported that full-term gestation in a woman younger than 24 yr of age reduces her lifetime risk of developing breast cancer, and this parity-induced protection against breast cancer is significantly affected by the total number of pregnancies. The epidemiological data on breast-feeding and breast cancer risk in humans also show that prolonged breast-feeding confers additional protection against breast cancer (17). However, aborted pregnancies are not associated with decreased risk for breast cancer (8).
A similar type of pregnancy protection from breast cancer has also been observed in rodents (67, 74). In rats, pregnancy alone prior to carcinogen administration and after carcinogen challenge has significantly reduced the incidence and number of palpable carcinomas per rat (74). Sinha et al. (1988) (67) showed that interrupted pregnancies in rats at 5th, 10th, and 15th days resulted in lower protection against mammary tumor incidence (48, 50, and 45%, respectively) versus 70% in age-matched nonpregnant controls and 14% for full-term pregnancy. The protective effect of pregnancy was also observed to be persistent, indicating a long-lasting alteration in the sensitivity of the mammary gland against tumorigenesis in rat models similar to humans.
An endocrine milieu similar to that of pregnancy can be mimicked by exogenous estrogen and progesterone administration. Reproductive hormones, progesterone and estrogen, are required by the mammary gland for proliferation and secretory differentiation (59). Progesterone plays key role in alveolar proliferation, and estrogen is involved in ductal development (30). It has been consistently shown that treatment of rats with both estrogen and progesterone for a short period of time confers significant protection against mammary carcinogenesis, although the studies using either estrogen or progesterone alone yielded contrary results, depending on the dose and length of hormonal treatment (10, 22, 24, 52, 53, 63). For instance, Grubbs et al. (1985, 1988) (22, 23) reported 88–90% fewer cancers in rats pretreated with 20 μg of 17 β-estradiol and 4 mg of progesterone or 5 μg of estrogen and 4 mg of progesterone for 5 wk. Sivaraman et al. (1998) (68) and Guzman et al. (1999) (24) reported 82 and 96% reductions, respectively, in mammary cancers in rats treated with 20–30 μg of 17 β-estradiol and 20–30 mg of progesterone for 3 wk.
Although rodent experimental data and human epidemiological data consistently show the protective effect of pregnancy and/or exogenous hormones (estrogen plus progesterone) on mammary carcinogenesis, the mechanisms underlying this protection are still largely unclear. The persistent pregnancy- or exogenous hormone-induced mammary gland changes, cellularly and/or biochemically, remain to be delineated. The goal of this study was to identify morphological and transcriptional changes that may be linked to the protective effect of hormones and pregnancy against mammary tumorigenesis.
MATERIALS AND METHODS
Animals
All of the animal work and handling was carried out in accordance with institutional policies and federal guidelines and approved by the University of Vermont Animal Care and Use Committee. Fifteen 3 mo old postpubertal virgin Lewis rats (Charles River) were kept in the University of Vermont small animal care facility. Rats were housed in air- and temperature-controlled cage shelves on a 12 h light-dark cycle and were fed rat chow (RMH 3000, Lab Diet, Scott Distributing) and water ad libitum.
Hormone and Pregnancy Treatments
Rats were randomly assigned to three groups (five rats per group): control (C), pregnancy (P), and hormone treatment (H). The P group animals had a full-term pregnancy (21–23 days), and rats in the group H were implanted subcutaneously on the dorsal midline with two silastic capsules [(0.078 inch inner diameter, 0.125 inch outer diameter) × 2 cm long; Dow Corning] filled separately with 100 μg ethynyl estradiol (Sigma) packed in a cellulose matrix (Sigma) and 30 mg of megesterol acetate (Sigma) for 21 days (63). The control animals had neither the hormone treatment nor pregnancy. The animals in C and P groups were also implanted with sham capsules filled with cellulose matrix only. The capsules were surgically implanted at the beginning of the experiment and removed from all animals after 21 days except that the capsules were removed from the P group following parturition (21–23 days). The delivered pups in the P group were euthanized within 4–6 h of delivery to avoid suckling. After the removal of capsules all groups were rested a total of ∼49 days before euthanasia.
Tissue Collection and RNA Isolation
All animals were euthanized during metestrus stage, determined by vaginal cytology as per Nelson et al. (57) and Marcondes et al. (46). Whole inguinal (4th pair) mammary glands were excised. For the histological analysis of the mammary gland, three or four small pieces (∼1 mm3) of mammary tissue were taken from the right inguinal mammary gland and fixed in 10% neutral buffered formalin solution overnight. The fixed mammary tissues from individual rats were paraffin-embedded, sectioned, and stained with hematoxylin and eosin (H&E). Remaining mammary tissue was immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA isolation. Total RNA was extracted from the mammary tissues with TRIzol reagent (Invitrogen), DNase-treated, and column-purified using Qiagen RNeasy Mini Kit (Qiagen), as per manufacturer's instructions.
Histological Photomicrograph Analysis
H&E-stained mammary sections were examined under an Olympus light microscope (Olympus, Tokyo, Japan) at ×100 magnification, and five images of 1,024 × 1,024 pixels with 32-bit per pixel depth were captured per two sections from each rat (for a total of 25 images per treatment). The images were saved in tagged image file format (TIFF) in Image Pro Plus 5.1 (Media Cybernetics, Bethesda, MD), and each image represented a tissue area of 0.0237 mm2. Histological measurements were conducted using the free-hand area-of-interest tool to measure area in Image Pro Plus. Total area and areas of fat-pad (adipose tissue), connective tissue stroma (extracellular matrix, connective tissue, and blood vessels), and parenchymal (alveolar and ductal epithelia) tissue in the image were measured. Results are presented as a percentage of total area in the images on average of five photomicrographs per animal. Mean area measured was statistically analyzed among the groups by one-way ANOVA at P < 0.05 for the significant effect of the treatment.
Measurement of Plasma 17-β Estradiol and Progesterone Hormone Levels
Blood samples were collected from tail vein from all the rats on day 0, 2, 7, 14, and 21 of the silastic capsule implantation/pregnancy. Blood was also collected at the time of tissue harvest from the rat hearts in all groups. Plasma was separated, frozen, and assayed for 17-β estradiol and progesterone using I125-labeled RIA kits (MP Biomedicals).
Target Preparation and Microarray Analysis
Purified mammary gland total RNA was quantified using the Nanodrop spectrophotometer (Thermo Fisher Scientific) and analyzed for the integrity using the Agilent Bioanalyzer 2100 (Agilent Technologies). RNA amplification, fragmentation, and labeling were performed using the Ovation V2 and Encore reagents from NuGEN technologies. The labeled probes were hybridized to the Affymetrix Gene Chip, Rat Genome 230.2 Array for 16 h at 45°C at 60 rpm as per manufacturer's recommendations (n = 5). The arrays were double-stained with streptavidin-phycoerythrin using the Affymetrix 450 Fluidics station and scanned with the Affymetrix 3000-G7 scanner at the Vermont Genetics Network Microarray Core Facility. The microarray data were submitted to Gene Expression Omnibus (GSE32125).
Microarray Data Analysis
Analysis of GeneChip data included: 1) calculation of expression statistics for each probe set in each sample, 2) test of the genome-wide null hypothesis that no treatment effects are observed, 3) ordering of probe sets based on evidence against each of three null hypotheses (see below), and 4) identification of biological functions associated with sets of differentially expressed genes.
Calculation of probe set statistics.
Probe statistics were background corrected, normalized, and summarized using the R (62)/Bioconductor (21) aroma.affymetrix package (7), which implements the Robust Multichip Average statistic of Speed and coworkers (11, 33). Affymetrix MAS5 presence/absence calls were made using the Bioconductor affy package (20).
Multivariate analysis.
We tested the null hypothesis, that there was no effect of pregnancy or hormone treatment on mammary gene expression, using the Multi-Response Permutation Procedure (MRPP) of Mielke and Berry (54) using 1,000 permutations of sample labels. Principal component analysis was carried out using the R stats package.
Univariate analysis.
Genes were ordered based on differential expression statistics associated with three null hypotheses:
where Eg is the expression statistic of a probe set in sample group g ∈{C,P,H}. Genes were ordered using the P value obtained from the moderated t-statistic of Smyth (70), calculated using the Bioconductor limma package provided by Smyth (69).
Identification of biological functions associated with sets of differentially expressed genes.
The Functional Annotation Tool available through the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (31) was used to analyze gene sets significantly differentially expressed in the P and H groups relative to C group for ontological terms [Gene Ontology (GO) terms] (classification stringency = medium).
Quantitative Real-time PCR
DNase-treated total RNA (180 ng) was used to generate cDNA using the GeneAMP kit of Applied Biosystems. qRT-PCR analysis was performed on ABI PRISM 7900 using TaqMan Assays On Demand Gene Expression Kit (Applied Biosystems) specific for rat Tenascin-C (Tnc; catalog #Rn01454948_m1), B-cell leukemia/lymphoma 11B (Bcl11b; Rn01102259_m1), and housekeeping gene HPRT (Rn01527840_m1). Reactions were performed in duplicate in a 20 μl volume containing 10 μl Quanta PerfeCTa qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD), 1.0 μl Taqman assay, and 1 μl diluted cDNA (corresponding to 9 ng of reverse-transcribed total RNA). The relative expressions of Tnc and Bcl11b were normalized by HPRT and calculated by the 2−ΔΔCT method (44). The statistical significance of relative differences was analyzed using ANOVA with SAS software (SAS Institute, Cary, NC). The difference in the means of mRNA expression between different treatment groups was compared by least significant difference comparison analysis and Bonferroni's adjustments for the multiple comparisons.
Immunohistochemistry
Deparaffinization of the mammary tissue sections was carried out three times with xylene for 15 min at room temperature. The sections were then washed with 100, 95, 70, and 50% ethanol and distilled water, respectively. Antigen retrieval was done using 1× DAKO (Carpinteria, CA) Antigen Retrieval at 96°C for 20 min followed by 20 min cooling down at room temperature in 1× DAKO AR Buffer. Finally the sections were washed three times for 5 min in phosphate-buffered saline (PBS, pH 7.4).
Endogenous peroxidase was blocked by incubating the sections in the Dual Endogenous Block (DAKO #S2003) for 10 min. After being washed three times for 5 min with PBS, sections were exposed to monoclonal primary antibodies (anti-ED1: Hycult biotech, #HM3029; anti-MMP8: Abcam #ab81286) diluted 1:50 (100 μg/ml) in PBS with 1.0% bovine serum albumin (BSA, Sigma) at room temperature for 30 min followed by three times 5 min washes in PBS. Sections were exposed separately to DAKO LSAB2 link and LSAB2 streptavidin for 20 min followed by three times 5 min wash in PBS and incubated with 3′-3′ diaminobenzidine tetrahydrochloride (DAKO #S2003) solution for 4 min at 25°C followed by three times 5 min washes in distilled water. Sections were then stained with Mayer's hematoxylin for 30 s followed by 2 min rinse in running water and dipped two times in 0.5% lithium carbonate followed by 2 min rinse in running water. Finally, the sections were dehydrated two times for 2 min in 50, 70, 95, and 100% ethanol and xylene, respectively. Stained mammary gland tissue sections were photographed using the BX50 microscope (Olympus America, Center Valley, PA) with an attached QImaging Retiga 2000R Digital Camera (Quantitative Imaging, Surrey, BC, Canada).
Computer-assisted image analysis was performed to quantitate the proportion of ED1 positively stained areas per unit of mammary gland parenchymal and stromal tissue in all experimental animals. A total of 25 images (five per rat) were analyzed per treatment group using MetaMorph image analysis program (Universal Imaging, West Chester, PA). The same Metamorph color thresholding was applied to all the samples. The results are presented as a percentage of ED1 positively stained areas per unit of the measured parenchymal and stromal areas per treatment group. The statistical differences between treatment groups were analyzed by one-way ANOVA.
RESULTS
Blood Levels of Estradiol and Progesterone in Experimental Animals
Blood estradiol and progesterone levels were monitored in experimental period in all animals. Mean plasma estrogen and progesterone levels of control rats did not vary significantly across days of the experimental period and averaged 36.2 ± 18.2 pg/ml and 18.4 ± 13.78 ng/ml, respectively (Fig. 1). Plasma estradiol and progesterone levels in the exogenous hormone-treated animals increased to 74.8 ± 9.6 pg/ml and 47.3 ± 13.8 ng/ml, respectively, by day 2 of the silastic capsule implantation and reached the highest levels at day 14 (114.1 ± 43.3 pg/ml and 127.2 ± 26.1 ng/ml, respectively). The hormonal profiles during hormone treatment period in the exogenous hormone treatment group were similar to those of the pregnancy animals, except for a sharp estrogen hike in the P group at day 21 (Fig. 1).
Fig. 1.
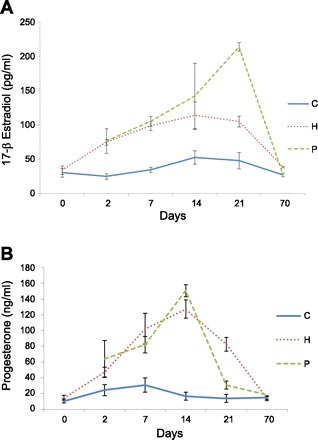
Profiles of serum 17-β estradiol (A) and progesterone (B) concentrations in the experimental rats of the control (C), pregnancy (P), and exogenous hormone treatment (H) groups.
Pregnancy and Hormone Treatments Induced Sustained Morphological Changes in the Mammary Gland
After 49 days of involution following the termination of pregnancy or exogenous hormone treatment, the mammary gland morphology of rats in the H and P groups appeared distinctly different from that of the control animals (Fig. 2A). Total area and total areas of fat-pad (adipose tissue), connective tissue stroma, and parenchymal (epithelial) tissue were measured to determine if there was a difference in distribution of tissue types among the treatments (Fig. 2B, Table 1). The percentages of connective tissue stroma and parenchymal tissue were significantly greater in P (21.35% ± 0.088 and 4.27% ± 0.03, respectively) and H (16.91% ± 0.43 and 3.79% ± 0.027) groups compared with C group (11.22% ± 0.027 and 3.12% ± 0.034) (P < 0.05). Control rats had significantly higher percent area of adipose tissue in the mammary gland than the other two treatments (P < 0.05). The ratio of the parenchyma to connective tissue stroma was significantly lower in animals that underwent with pregnancy or hormone treatment (0.2 and 0.224 vs. 0.278) (P < 0.05). These changes were significantly greater in P group animals compared with the H group animals (P < 0.05). Thus, our data showed that pregnancy and hormone treatment induced sustained and similar morphological changes in the rat mammary gland.
Fig. 2.
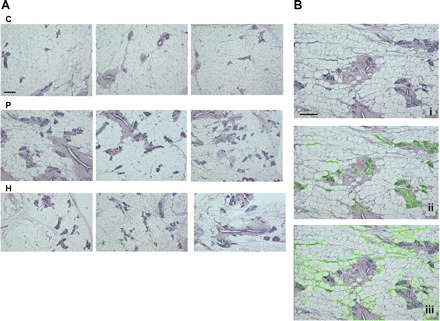
A: histological changes of the mammary gland of rats at day 49 after a full-term pregnancy (P) or exogenous hormone treatment (H) compared with the control (C) rats (scale bars = 100 μm). Each photomicrograph was randomly selected from an individual animal within the group. B: histological analyses of a mammary gland section using the free-hand area-of-interest (AOI) tool of Image Pro Plus for the areas of parenchyma (ii) or the areas of both parenchymal and stromal tissues (iii). i, The section of ii and iii without using the AOI tool.
Table 1.
Mean percent area of tissue types in rat mammary glands 49 days after full-term pregnancy or completion of 21 days of exogenous hormone treatment compared with control group
| Pregnancy | Hormone | Control | |
|---|---|---|---|
| Tissue type | % Area of Total | ||
| Parenchyma | 4.27 ± 0.03a* | 3.79 ± 0.027b | 3.12 ± 0.034c |
| Stroma (ECM/connective tissue) | 21.35 ± 0.088a | 16.91 ± 0.43b | 11.22 ± 0.027c |
| Adipose | 74.38 ± 0.12a | 79.3 ± 0.45b | 85.66 ± 0.032c |
| Ratio of parenchyma to stroma | 0.2 ± 0.0014 | 0.224 ± 0.0055 | 0.278 ± 0.0034 |
| (1: 5)a | (1:4.5)b | (1:3.6)c | |
*Data are from photo-micrographic image analysis of total 25 randomized images per group (5 images per rat). Values with different superscripts in the same row are significantly different (P < 0.05). ECM, extracellular matrix.
Pregnancy and Hormone Treatments Induced Sustained Gene Expression Changes in the Mammary Gland
Microarray analysis was used to examine global gene expression changes in rat mammary glands after 49 days of involution following a full term of pregnancy or exogenous hormone treatment. Our analysis indicates that pregnancy or hormone treatment resulted in a statistically significant changes in mammary gland gene expression genome wide (P = 0.03, MRPP test). The principal component plot analysis showed that the gene expression distances between control animals (C) and treated animals (P, H) were large compared with that between pregnant and hormone-treated animals (mean and range) (Fig. 3) and suggested that transcriptional changes due to exposure to pregnancy or hormone treatment may be common between these treatments.
Fig. 3.
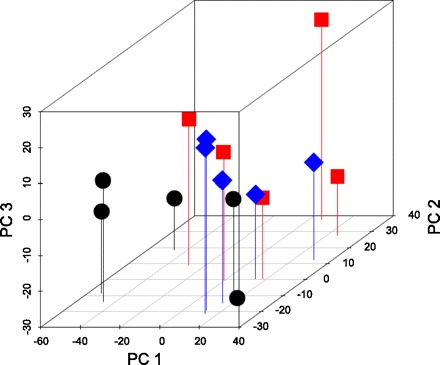
Three-dimensional principle component analysis plot (PCA) of genome-wide gene expression patterns in the mammary gland of rats at day 49 after a full-term pregnancy (square) or exogenous hormone treatment (diamond) compared with the control (circle) rats.
Common Genes that were Sustainably Up- or Downregulated in the Mammary Gland After Either Pregnancy or Hormone Treatment and Their Ontological Classifications
Microarray analysis revealed a total of 812 (476 up and 336 down) and 1,024 (245 up and 779 down) differentially expressed genes (fold change ≥ 1.4 and P < 0.05, the latter associated with a false discovery rate of 0.21), respectively, in the mammary gland at 49 days after either a full-term pregnancy or 21 days of hormone treatment compared with nonpregnancy and nonhormone treatment control animals (Fig. 4). Of these differentially changed genes, 243 genes were either up- (87) or down (156)-regulated in both P and H groups, representing 30 and 24% differentially expressed gene in P and H group, respectively (Fig. 4).
Fig. 4.
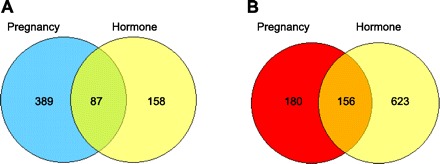
Venn diagram analysis of the differentially up (A)- and down (B)-regulated genes in the mammary gland at day 49 after a full-term pregnancy or 21-day exogenous hormone treatment. The cut-off criteria were set at fold change ≥ 1.4 and P < 0.05.
The top upregulated genes in the mammary gland common to both treatment animals were immunoglobulin heavy chain (alpha polypeptide) (Igha), fatty acid binding protein 3 (Fabp3), WAP four-disulfide core domain 3 (Wfdcd3), two milk protein genes (β-casein and whey acidic protein), and three matrix metallopeptidases (MMP12, 8, and 3). Other notable commonly upregulated genes included the transcription factor CCAAT/enhancer binding protein, alpha (C/EBPα), leptin, growth hormone receptor, metallothionein 1E&F (Mt1E&F), and G0/G1 switch gene 2 (G0s2). The top downregulated genes in both treatment animals were Bcl11B, brain and acute leukemia, cytoplasmic (Baalc), glutamate receptor, ionotropic, AMPA3 (Bria3), plasticity-related gene 1 (Prg1), and Tnc. Other noticeable common downregulated genes include cyclin D2; growth arrest and DNA-damage-inducible 45 beta (GADD45B); phosphoinositide-3-kinase, class 2, beta polypeptide (C2-PI3K); and ret proto-oncogene (RET).
The sets of common up- and downregulated genes were sorted by ontology using DAVID. The ontological analysis revealed distinct differences in the molecular signatures for the up- or downregulated genes common amongst treatment groups (Tables 2 and 3). Top ontology terms (GO) assigned to common upregulated genes were extracellular region, peptidase/MMP, response to organic substance, plasma membrane part and signal (Table 2). Approximately 29% (46 of 157) commonly downregulated genes were not annotated. Top ontologies assigned to the annotated commonly downregulated genes were ATP/adenyl ribonucleotide binding, chromosome organization, and protein localization (Table 3).
Table 2.
GO terms enriched with commonly upregulated genes (n = 87) in rat mammary gland 49 days after either pregnancy or hormone treatment
| P |
H |
||||||
|---|---|---|---|---|---|---|---|
| GO Term | % | P Value | Affy ID | Gene Symbol | Gene Name | Log FC | Log FC |
| Extracellular region | 20.8 | 3.8E-4 | 1388605_at | Wfdc3 | WAP four-disulfide core domain 3 (predicted) | 3.02 | 1.49 |
| 1368924_at | Ghr | growth hormone receptor | 0.52 | 0.56 | |||
| 1387748_at | Lep | leptin | 1.06 | 0.72 | |||
| 1379677_at | Tnfsf13 | tumor necrosis factor (ligand) superfamily, member 13 | 0.56 | 0.5 | |||
| 1371099_at | polymeric immunoglobulin receptor | 3.14 | 2.21 | ||||
| 1387735_at | Mmp8 | matrix metallopeptidase 8 | 1.56 | 0.84 | |||
| 1392172_at | Ccl9 | chemokine (C-C motif) ligand 9 | 1.53 | 0.6 | |||
| 1369539_at | St3 gal3 | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 | 0.64 | 0.49 | |||
| 1368913_at | Csn2 | casein beta | 2.96 | 2.36 | |||
| 1368657_at | Mmp3 | matrix metallopeptidase 3 | 1.16 | 0.73 | |||
| 1387751_at | Wap | whey acidic protein | 2.12 | 1.25 | |||
| 1391323_at | Tf | signal recognition particle receptor, B subunit; transferrin | 1.07 | 0.66 | |||
| 1368008_at | Prom1 | prominin 1 | 0.71 | 0.72 | |||
| 1371130_at | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 0.49 | 0.58 | |||
| Peptidase M10A, matrix metallopeptidase | 4.2 | 1.4E-3 | 1387735_at | Mmp8 | matrix metallopeptidase 8 | 1.56 | 0.84 |
| 1368657_at | Mmp3 | matrix metallopeptidase 3 | 1.16 | 0.73 | |||
| 1368530_at | Mmp12 | matrix metallopeptidase 12 | 2.22 | 1.56 | |||
| Response to organic substance | 13.9 | 5.5E-3 | 1367660_at | Fabp3 | fatty acid binding protein 3 | 3.04 | 2.09 |
| 1394028_at | Dusp10 | dual specificity phosphatase 10 (predicted) | 0.75 | 0.74 | |||
| 1368924_at | Ghr | growth hormone receptor | 0.52 | 0.56 | |||
| 1387748_at | Lep | leptin | 1.06 | 0.72 | |||
| 1369337_at | Adcy10 | adenylate cyclase 10 (soluble) | 0.85 | 0.52 | |||
| 1369658_at | Cebpa | CCAAT/enhancer binding protein (C/EBP), alpha | 1.14 | 0.65 | |||
| 1368657_at | Mmp3 | matrix metallopeptidase 3 | 1.16 | 0.73 | |||
| 1391323_at | Tf | signal recognition particle receptor, B subunit; transferrin | 1.07 | 0.66 | |||
| 1382692_at | RGD15 | similar to Clecsf12 protein (predicted) | 1.15 | 0.63 | |||
| 65140 | |||||||
| 1370026_at | Cryab | crystallin, alpha B | 0.54 | 0.71 | |||
| Plasma membrane part | 18.1 | 6.8E-3 | 1388240_a_at | Itga7 | integrin alpha 7 | 0.64 | 0.52 |
| 1368317_at | Aqp7 | aquaporin 7 | 0.55 | 0.54 | |||
| 1371130_at | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 0.49 | 0.58 | |||
| 1368924_at | Ghr | growth hormone receptor | 0.52 | 0.56 | |||
| 1368187_at | Gpnmb | glycoprotein (transmembrane) nmb | 1.32 | 0.87 | |||
| 1369337_at | Adcy10 | adenylate cyclase 10 (soluble) | 0.85 | 0.52 | |||
| 1379677_at | Tnfsf13 | tumor necrosis factor (ligand) superfamily, member 13 | 0.56 | 0.5 | |||
| 1382692_at | RGD15 | similar to Clecsf12 protein (predicted) | 1.15 | 0.63 | |||
| 65140 | |||||||
| 1370614_s_at | Stk39 | serine/threonine kinase 39, STE20/SPS1 homolog (yeast) | 0.65 | 0.5 | |||
| 1375756_at | RGD13 | similar to ionized calcium binding adapter molecule 2 (Iba2) (predicted) | 0.54 | 0.51 | |||
| 05081 | |||||||
| 1391323_at | Tf | signal recognition particle receptor, B subunit; transferrin | 1.07 | 0.66 | |||
| 1368008_at | Prom1 | prominin 1 | 0.71 | 0.72 | |||
| 1387889_at | Folr1 | folate receptor 1 (adult) | 1.14 | 1.03 | |||
| Signal | 20.8 | 8.9E-3 | 1388240_a_at | Itga7 | integrin alpha 7 | 0.64 | 0.52 |
| 1371099_at | polymeric immunoglobulin receptor | 3.14 | 2.21 | ||||
| 1368924_at | Ghr | growth hormone receptor | 0.52 | 0.56 | |||
| 1368187_at | Gpnmb | glycoprotein (transmembrane) nmb | 1.32 | 0.87 | |||
| 1387748_at | Lep | leptin | 1.06 | 0.72 | |||
| 1398241_a_at | Spt1 | salivary protein 1 | 1.76 | 2.16 | |||
| 1374933_at | Mcam | melanoma cell adhesion molecule | 0.49 | 0.54 | |||
| 1369468_at | Fzd4 | frizzled homolog 4 (Drosophila) | 0.62 | 0.54 | |||
| 1387735_at | Mmp8 | matrix metallopeptidase 8 | 1.56 | 0.84 | |||
| 1368913_at | Csn2 | casein beta | 2.96 | 2.36 | |||
| 1396947_at | Lgr4 | leucine-rich repeat-containing G protein-coupled receptor 4 | 0.57 | 0.51 | |||
| 1368657_at | Mmp3 | matrix metallopeptidase 3 | 1.16 | 0.73 | |||
| 1387751_at | Wap | whey acidic protein | 2.12 | 1.25 | |||
| 1391323_at | Tf | signal recognition particle receptor, B subunit; transferrin | 1.07 | 0.66 | |||
| 1368530_at | Mmp12 | matrix metallopeptidase 12 | 2.22 | 1.56 |
GO, Gene Ontology; P, full-term pregnancy; H, 21 days of exogenous hormone treatment; FC, fold change.
Table 3.
GO terms enriched with commonly downregulated genes (n = 156) in rat mammary gland 49 days after either pregnancy or hormone treatment
| P |
H |
||||||
|---|---|---|---|---|---|---|---|
| GO Term | % | P Value | Affy ID | Gene Symbol | Gene Name | Log FC | Log FC |
| ATP binding | 13.4 | 7.7E-3 | 1372995_at | Prkd2 | protein kinase D2 | −0.8 | −0.9 |
| 1379218_at | RGD15 | similar to nemo like kinase (predicted) | −0.63 | −0.58 | |||
| 61602 | |||||||
| 1388821_at | RGD15 | similar to Tribbles homolog 2 (predicted) | −0.65 | −0.78 | |||
| 64451 | |||||||
| 1383229_at | Abca7 | ATP-binding cassette, sub-family A (ABC1), member 7 | −0.54 | −0.61 | |||
| 1396839_at | Nsf | N-ethylmaleimide sensitive fusion protein | −0.58 | −0.74 | |||
| 1372756_at | Map4k2 | mitogen activated protein kinase kinase kinase kinase 2 (predicted) | −0.55 | −0.66 | |||
| 1368477_at | Atp2a3 | ATPase, Ca2+ transporting, ubiquitous | −0.6 | −0.57 | |||
| 1370989_at | Ret | ret proto-oncogene | −0.68 | −0.62 | |||
| 1379830_at | Chd2 | chromodomain helicase DNA binding protein 2 (predicted) | −0.68 | −0.62 | |||
| 1384200_at | RGD13 | similar to RIKEN cDNA 4931400A14 (predicted) | −0.65 | −0.54 | |||
| 07234 | |||||||
| 1389359_at | Smc4l1 | SMC4 structural maintenance of chromosomes 4-like 1 (yeast) | −0.63 | −0.76 | |||
| 1383008_at | −0.67 | −0.79 | |||||
| 1385266_at | LOC36 | similar to nemo like kinase | −0.53 | −0.57 | |||
| 5949 | |||||||
| 1373256_at | Chd3 | chromodomain helicase DNA binding protein 3 | −0.66 | −0.82 | |||
| 1368247_at | Hspa1a | heat shock 70kD protein 1A | −0.73 | −0.53 | |||
| Chromosome organization | 5.8 | 0.027 | 1379830_at | Chd2 | chromodomain helicase DNA binding protein 2 | −0.68 | −0.62 |
| 1373256_at | Chd3 | chromodomain helicase DNA binding protein 3 | −0.66 | −0.82 | |||
| 1368247_at | Hspa1a | heat shock 70kD protein 1B (mapped); heat shock 70kD protein 1A | −0.73 | −0.53 | |||
| 1384200_at | RGD13 | similar to RIKEN cDNA 4931400A14 | −0.65 | 0.54 | |||
| 07234 | |||||||
| 1392512_at | Hist3 h2ba | similar to histone 3, H2ba; histone cluster 3, H2ba | −0.68 | −0.69 | |||
| 1383008_at, 1389359_at | Smc4l1 | structural maintenance of chromosomes 4 | −0.67 | −0.79 | |||
| −0.63 | −0.76 | ||||||
| Protein localization | 7.7 | 0.047 | 1372830_at | Arl6ip1 | ADP-ribosylation factor-like 6 interacting protein 1 | −0.48 | −0.50 |
| 1396839_at | Nsf | N-ethylmaleimide-sensitive factor | −0.59 | −0.73 | |||
| 1377062_at | RGD15 | Similar to centrosomal protein 250 kDa | −0.49 | −0.51 | |||
| 62262 | |||||||
| 1394805_at | Xpo6 | exportin 6 | −0.55 | −0.74 | |||
| 1379934_at | Lyst | lysosomal trafficking regulator | −0.59 | −0.68 | |||
| 1395436_at | Ppp3ca | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform | −0.53 | −0.65 | |||
| 1397565_at | Stx7 | syntaxin 7 | −0.58 | −0.65 | |||
| 1373510_at | Vamp1 | vesicle-associated membrane protein 1 | −0.62 | −0.77 |
Abbreviations are defined in Table 1 footnote.
Ontological Classifications of Genes that were Sustainably Up- and Downregulated in the Mammary Gland After Either a Full-term Pregnancy or Exogenous Hormone Treatment
Surprisingly, except the GO terms shared by commonly regulated genes in both treatment animals, ontological classifications of the up- or downregulated genes in P or H group revealed very different GO terms enriched in the mammary gland of these two groups of animals. There were more upregulated genes in the P animals (476 in P group vs. 245 in H group), while there were more downregulated genes in the H animals (336 in P group vs. 779 in H group). The functional annotation clustering of upregulated genes in the P group showed major GO terms in lipid biosynthesis process, responses to nutrient, hormonal stimulus, hypoxia and wounding, glucose metabolic process, cell fraction, protein dimerization activity, regulation of phosphorylation, regulation of lipid metabolic process, extracellular region, skeletal system development, lung development, membrane organization, and ion homeostasis, while the functional annotation clustering of upregulated genes in the H group showed only a few clusters with significant enrichment scores in transmembrane, transmembrane protein, O-acyltransferase activity, and extracellular region.
The functional annotation clustering of downregulated genes in the P group showed only three GO term clusters in extracellular region/matrix (mainly collagens) and cell adhesion with enrichment scores of >2. However, analysis of downregulated genes in the H group showed significant enriched GO term clusters in lymphocyte differentiation, regulation of leukocyte activation, positive regulation of immune response, plasma membrane part, GTPase regulator activity, Src homology-3 domain, Bromodomain, cytokine, regulation of cytokine production, phosphorylation, regulation of programmed cell death, cell morphogenesis, and cytoskeleton organization.
Quantitative Real-time PCR Analysis of Tnc and Bcl11b
Real-time qRT-PCR analysis of Bcl11b and Tnc in rat mammary glands was consistent with the results of our microarray analysis. The mRNA expression of Bcl11b in the involuted mammary gland of pregnancy and hormone-treated rats was six (P = 0.07)- and eight (P < 0.01)-fold lower than that of the control rats in qRT-PCR analysis, respectively (Fig. 5A), and the mRNA expression of Tnc in the pregnancy- and hormone-treated rats was 12- and 11-fold lower than the control rats, respectively (P < 0.05) (Fig. 5B). There were no statistical differences in Bcl11b and Tnc mRNA expression between pregnancy and hormone treatment animals.
Fig. 5.
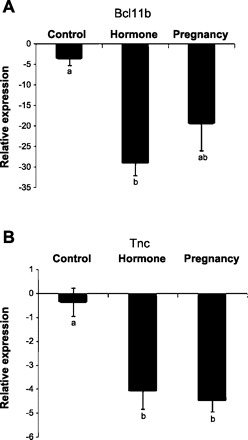
Real-time qRT-PCR analysis of B-cell leukemia/lymphoma 11B (Bcl11b; A) and tenascin-C (Tnc; B) mRNA expression in the mammary gland of rats at day 49 after a full-term pregnancy or exogenous hormone treatment compared with the control rats. Relative expression values (means ± SE) were normalized against housekeeping gene HPRT. Different lowercase letters represent P < 0.05.
Immunological Staining of MMP8 and the Macrophage Marker ED1 in Rat Mammary Gland
Consistent with our microarray analysis, immunohistochemical staining showed stronger MMP8 staining in rat mammary gland after 49 days of involution following either a full-term pregnancy or exogenous hormone treatment compared with the control animals (Fig. 6A, right panels). The MMP8 was mainly expressed in the epithelial cells of the mammary gland. In addition, immunohistochemical staining of ED1, a cellular marker specific for activated rat microglia, monocytes, and macrophages, also showed that the involuted mammary gland had more microglia, monocytes, and macrophages in the parenchyma and connective tissue stroma compared with the control rats (Fig. 6A, left panels), as the mean percentages of ED1-positive areas per unit of parenchymal and connective tissue stroma in the pregnancy and hormone treatment animals were six- to eightfold higher than the control animals by a semiobjective image analysis (P < 0.01) (Fig. 6B).
Fig. 6.
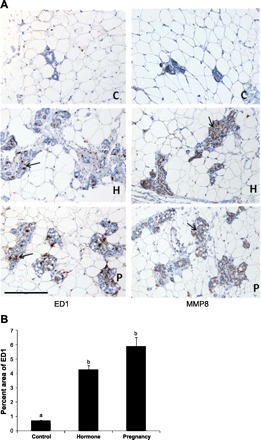
A: immunohistochemical staining (brown color as indicated by arrows) of the macrophage marker ED1 (left) and the matrix metallopeptidase MMP8 (right) in the mammary gland of rats at day 49 after a full-term pregnancy (P) or exogenous hormone treatment (H) compared with the control (C) rats. One representative staining is shown for each group. Scale bar = 200 μm. B: graphs representing the percentages of ED1-stained areas per unit of parenchyma and stroma. Different lowercase letters represent P < 0.01.
DISCUSSION
It is well established that a full-term pregnancy in women and rodent animals or treatment of rodents with exogenous progesterone and estrogen for 21 days can induce significant protection of the mammary gland from tumorigenesis (52). In this study, our objectives were to identify the sustained pregnancy- or hormone-induced mammary gland changes that may contribute to this protective effect. We examined the morphological and gene expression changes in whole mammary gland of rats 49 days after a full-term pregnancy or exogenous progesterone and estrogen treatment. The levels of blood estrogen and progesterone in experimental animals during the exogenous hormone treatment matched well with the hormonal profiles of pregnancy animals and were in the range of other exogenous hormone treatment studies in which significant protections of the mammary gland from tumorigenesis were observed (24, 63).
There were distinct morphological differences between control mammary glands and after the 49-day involution period following a full-term pregnancy or exogenous hormone treatment. Glands from rats that were treated with exogenous hormones or that had undergone full-term pregnancy had significantly more percent areas of connective tissue stroma and parenchyma tissue, while the adipose tissue area was decreased. Stroma area of extracellular matrix and connective tissue increased 90 and 51% in the pregnancy and hormone treatment rats, respectively. Increased connective tissue stroma and parenchyma tissue percent areas in the mammary gland seem contradictory to the decreased rate of mammary tumorigenesis in animals exposed to these treatments, as high breast density, which is determined by the amount of parenchyma and connective tissue stroma, is a risk factor for developing breast cancer (13). However, our study showed that the pregnancy and hormone treatment not only increased the total percent areas of parenchyma and connective tissue stroma but also significantly reduced the ratios of parenchyma to connective tissue stroma in the mammary gland. While breast cancer is an epithelial cell phenomenon, more and more evidence has shown that the microenvironment and surrounding connective tissue stroma provide important role in directing epithelial phenotype in mammary development and cancer (14, 61). Pregnancy and exogenous hormone treatment may remodel the extracellular matrix to create a microenvironment that is inhibitory to cancer cell progression.
In addition to the morphological changes, the involuted mammary gland also showed sustained genome-wide gene expression changes after pregnancy and exogenous hormone treatment. Some of these expression changes were very likely linked to the morphological differences among the treatments, in particular the distribution in amounts of epithelial, stromal, and adipose tissues in mammary glands. However, >70% of genes with significantly altered expression from controls were unique to each of the treatment groups, and GO term analysis of these genes revealed few shared GO terms between the two groups. Thus, it appears that the majority of the gene expression changes in the two groups mainly resulted from the sustained expression changes in the same cell types, rather than from the differences in distribution of tissue types, as the two treatments induced similar types of morphological changes although the degrees of the changes were different.
There were 30 and 24% of genes that were significantly up- or downregulated in both P and H groups, respectively. Transcriptional changes in these genes likely contribute to the protective effect against mammary tumorigenesis that occurs in both groups of animals (52). GO enrichment analysis revealed that genes commonly upregulated in both groups were mainly related to extracellular region, MMP, and plasma membrane part, consistent with the morphological changes of increased stromal tissues observed in two groups. Among the upregulated genes were three MMPs (MMP3, 8, and 12). Increased expression of MMP8 protein in the mammary epithelial cells after pregnancy and hormone treatment was confirmed by our immunohistochemical staining. MMPs are a group of zinc-dependent endopeptidases that are able to degrade the main protein components of extracellular matrix and basement membranes. Because of this role, the MMPs have long been considered to help tumor cells to migrate, invade, and spread (12). However, most clinical trials with MMP inhibitors have shown negative results in cancer treatment, and there is emerging evidence to support a protective role of some MMPs in tumor progression (16, 45). Transgenic mice deficient in MMP3 have an increased sensitivity to the development of squamous cell carcinoma, and keratinocyte expression of MMP3 prevents tumor establishment (48, 49). Similarly, MMP8-null mice have an increased incidence of skin tumors, and neutrophil-derived MMP8 can sufficiently rescue the increased tumorigenesis in MMP8-deficient mice (4). In addition, experimental manipulation of MMP8 expression has shown an inverse correlation of MMP8 expression levels with metastatic potential of breast cancer cells (55). The presence of MMP8 correlates with a lower incidence of metastasis, and a high-expression allele of the MMP8 gene is associated with lower susceptibility to metastasis in human breast carcinomas (18). A protective role of the MMP12 was also seen in lung tumor growth (1). These findings strongly support the notion that increased expression of MMP3, 8, and 12 in pregnancy and hormone treatment rats observed in our study may play an important role in the protective effect against mammary tumorigenesis in these animals. Consistent with this hypothesis, Casey et al. (2008) (15) showed higher expression of MMP3 in the normal breast fibroblasts compared with fibroblasts isolated from breast cancers.
Two milk protein genes, β-casein and whey acidic protein, are highly upregulated in both pregnancy and hormone treatment rats, consistent with more alveolar structures appeared in these animals in our histology staining and with the observation by Uehara et al. (2006) (71). This indicates that even after 49 days of involution following pregnancy or exogenous hormone treatment, the mammary glands still retain significant amounts of differentiated epithelial cells. Russo et al. (65, 66) proposed that the mammary differentiation status is a key inhibitor of cancer initiation. They postulated that breasts of nulliparous women contain undifferentiated cells that are susceptible to neoplastic transformation, while the epithelial cell population present in breasts of parous women acquires “genomic signature” changes and becomes more differentiated during early pregnancy, making it refractory to transformation. Our results in this study support this hypothesis.
The top commonly upregulated gene in both treatment rats was immunoglobulin heavy chain (alpha polypeptide). This may indicate an enhanced immunity in these animals. Supporting this notion is also the common upregulation of MMP12, triggering receptor expressed on myeloid cells 2 (TREM2), glycoprotein 49b (GP49B), chemokine ligand 9 (CCL9), CCAAT/enhancer binding protein alpha (C/EBPα), macrophage scavenger receptor 2 (MSR2), tumor necrosis factor (ligand) superfamily member 13 (TNFSF13), immunoreceptor Ly49si3, and metallothionein 1a (MT1a). MMP12 is a macrophage metalloelastase. TREM2 is a key regulator in chronic inflammation by stimulating the production of constitutive inflammatory cytokines (73). GP49B is a member of the immunoglobulin superfamily and has a regulatory role in T-cell priming (36). CCL9 is an interferon-γ-inducible β-chemokine and has shown potent antitumor activity through attraction of cytotoxic T lymphocytes and inhibition of angiogenesis (32, 56). The C/EBPα is required for the maintenance of CCL9 in transformation of hematopoietic cells by the BCR/ABL oncogene (32). TNFSF13 plays an important role in B cell development (58). Metallothionein is implicated in heavy metal detoxification and oxidative stress (35). Thus, increased expression of these genes in pregnancy and hormone treatment animals may suggest enhanced innate and adaptive immunities in these animals. In fact, our immunochemical staining of the ED1, a common cellular marker specific for activated rat microglia, monocytes, and macrophages, showed that the involuted mammary glands had increased these immune cells compared with the control glands, consistent with the discoveries of Zhao et al. (2010) (75).
About 29% of the commonly downregulated genes were not annotated, indicating that the functions of a big portion of genes in this group are unknown in contrast to the commonly upregulated genes. GO enrichment analysis of the annotated genes revealed significant downregulation of genes involved in ATP binding, chromosome organization, and protein localization in postinvolution mammary gland's transcriptome of pregnancy and hormone treatment rats. Genes enriching the ATP binding GO term included several kinases: protein kinase D2 (PKD2), phosphoinositide-3-kinase, class 2, beta polypeptide (PI3Kbeta), RET, nemo-like kinase (NLK), and mitogen activated protein kinase kinase kinase kinase 2 (MAP4K2). Both protein kinase D (PKD) and PI3Kbeta regulate DNA synthesis and replication (47), and PKD2 has been found to be a pivotal regulator of endothelial cell proliferation, migration, and angiogenesis (27). RET encodes a receptor tyrosine kinase that is implicated in the development of endocrine tumors (38). As MAP4K2, NLK is also a mitogen-activated protein kinase (MAPK)-like kinase and suppresses Notch signaling (34). The Notch and MAPK signaling have crucial functions in determining cell fates and in regulation of cell growth, differentiation, and stress responses. Thus, downregulation of PKD2, PI3Kbeta, RET, NLK, and MAP4K2 may imply a decreased cell growth and proliferation and angiogenesis in the involuted mammary gland. Consistently, cyclin D2 and polymerase (DNA directed), μ were also downregulated in these animals.
Consistent with this transcriptional signature changes was the downregulated expression of genes in chromosome organization in the involuted mammary gland. These genes include chromodomain helicase DNA binding proteins 2 and 3 (CHD2 and CHD3). The CHD proteins are known to modulate gene transcription by remodeling chromatin and changing histone deacetylation (25).
The top downregulated genes in both treatment rats were Bcl11b (verified by qRT-PCR analysis) and brain and acute leukemia, cytoplasmic (Baalc). Both genes have shown critical roles in lymphocyte development. Bcl11b is T-cell specific and is essential for T-cell development and for maintenance of T-cell integrity (41–43). It has been shown that when Bcl11b is deleted, T cells from all developmental stages acquire properties of NK cells and are able to kill tumor cells in vitro and effectively prevent tumor metastasis in vivo (42). Baalc is a marker of early hematopoietic progenitor cells, and overexpression of Baalc is an adverse prognostic factor in adults with acute myeloid leukemia and T cell-acute lymphoblastic leukemia (5, 6). Thus, downregulation of Bcl11b and Baalc is consistent with enhanced immunity in the involuted mammary gland discussed above. In addition, another highly downregulated gene is Tnc (verified by qRT-PCR analysis). Tnc is an extracellular matrix glycoprotein and has been implicated in the modulation of cell migration, proliferation, invasion, and angiogenesis. Tnc has been shown to promote breast cancer cell invasion and growth (26). A downregulation of Tnc is consistent with remodeling of extracellular matrix in the involuted mammary gland observed in our study. In addition, it is worth mentioning that several oncogenes were downregulated in the involuted mammary gland, including v-raf murine sarcoma 3611 viral oncogene homolog (Araf), Vav2 oncogene, RET, and Wnt2.
In summary, the results from our study showed that the involuted mammary gland following a full-term pregnancy or exogenous estrogen and progesterone treatment retains a high ratio of connective tissue stroma to parenchyma tissue and acquires genome-wide gene expression changes. The gene expression profiling indicated that the involuted mammary gland has remodeled extracellular matrix with increased expression of MMPs and decreased expression of Tnc, more differentiated mammary epithelial cells, enhanced innate and adaptive immunity, reduced cell proliferation and angiogenesis by modulating kinases and phosphatases, and reduced expression of pro-oncogenes (Fig. 7). These changes may all contribute to the parity/hormone-induced protection of breast cancer.
Fig. 7.
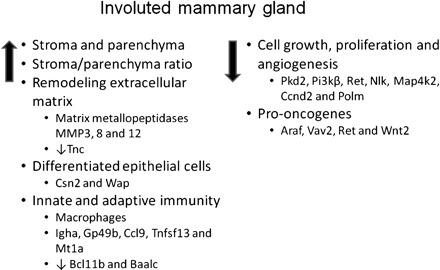
Schematic diagram of proposed physiological changes and associated expression changes of key genes in the involuted mammary gland following a full-term pregnancy and exogenous estrogen and progesterone treatment that may contribute to the protection of mammary tumorigenesis.
GRANTS
This work was supported by grants from the Vermont Cancer Center and Vermont Genetic Network.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We are grateful to Dr. Theresa Casey for her expertise in microarray data analysis and for reading the manuscript. We are thankful to Douglas Taatjes, Marilyn Wadsworth, Nicole Bishop, and Mary Lou Shane for technical assistance. We thank the staffs of the Small Animal Facility of the University of Vermont for the excellent animal care.
REFERENCES
- 1. Acuff HB, Sinnamon M, Fingleton B, Boone B, Levy SE, Chen X, Pozzi A, Carbone DP, Schwartz DR, Moin K, Sloane BF, Matrisian LM. Analysis of host- and tumor-derived proteinases using a custom dual species microarray reveals a protective role for stromal matrix metalloproteinase-12 in non-small cell lung cancer. Cancer Res 66: 7968– 7975, 2006 [DOI] [PubMed] [Google Scholar]
- 2. Albrektsen G, Heuch I, Hansen S, Kvale G. Breast cancer risk by age at birth, time since birth and time intervals between births: exploring interaction effects. Br J Cancer 92: 167– 175, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. American Cancer Society. Breast Cancer Facts & Figures 2009–2010, 2009 [Google Scholar]
- 4.Balbin M, Fueyo A, Tester AM, Pendas AM, Pitiot AS, Astudillo A, Overall CM, Shapiro SD, Lopez-Otin C. Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nat Genet 35: 252– 257, 2003 [DOI] [PubMed] [Google Scholar]
- 5. Baldus CD, Tanner SM, Kusewitt DF, Liyanarachchi S, Choi C, Caligiuri MA, Bloomfield CD, de la Chapelle A. BAALC, a novel marker of human hematopoietic progenitor cells. Exp Hematol 31: 1051– 1056, 2003 [PubMed] [Google Scholar]
- 6. Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, Caligiuri MA, Carroll AJ, Vardiman JW, Powell BL, Allen SL, Moore JO, Larson RA, Kolitz JE, de la Chapelle A, Bloomfield CD. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood 102: 1613– 1618, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Bengtsson H, Simpson K, Bullard J, Hansen K. aroma.affymetrix: a generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory, in Technical Report #745. Berkeley, CA: Department of Statistics, University of California, Berkeley, 2008 [Google Scholar]
- 8.Beral V, Bull D, Doll R, Peto R, Reeves G. Breast cancer and abortion: collaborative reanalysis of data from 53 epidemiological studies, including 83000 women with breast cancer from 16 countries. Lancet 363: 1007– 1016, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Bernstein L, Ross RK. Endogenous hormones and breast cancer risk. Epidemiol Rev 15: 48– 65, 1993 [DOI] [PubMed] [Google Scholar]
- 10. Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, D'Cruz CM, Chodosh LA. Hormone-induced protection against mammary tumorigenesis is conserved in multiple rat strains and identifies a core gene expression signature induced by pregnancy. Cancer Res 66: 6421– 6431, 2006 [DOI] [PubMed] [Google Scholar]
- 11. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185– 193, 2003 [DOI] [PubMed] [Google Scholar]
- 12. Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): positive and negative regulators in tumor cell adhesion. Semin Cancer Biol 20: 161– 168, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boyd NF, Martin LJ, Bronskill M, Yaffe MJ, Duric N, Minkin S. Breast tissue composition and susceptibility to breast cancer. J Natl Cancer Inst 102: 1224– 1237, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Weaver D, Muss H, Plaut K. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat 114: 47– 62, 2009 [DOI] [PubMed] [Google Scholar]
- 15. Casey TM, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Bunn JY, Weaver D, Muss H, Plaut K. Cancer associated fibroblasts stimulated by transforming growth factor beta1 (TGF-beta 1) increase invasion rate of tumor cells: a population study. Breast Cancer Res Treat 110: 39– 49, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Chen EX, Siu LL. Development of molecular targeted anticancer agents: successes, failures and future directions. Curr Pharm Des 11: 265– 272, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Collaborative Group on Hormonal Factors in Breast Cancer. Breast cancer and breastfeeding: collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet 360: 187– 195, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Decock J, Long JR, Laxton RC, Shu XO, Hodgkinson C, Hendrickx W, Pearce EG, Gao YT, Pereira AC, Paridaens R, Zheng W, Ye S. Association of matrix metalloproteinase-8 gene variation with breast cancer prognosis. Cancer Res 67: 10214– 10221, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Dupont WD, Page DL. Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med 312: 146– 151, 1985 [DOI] [PubMed] [Google Scholar]
- 20. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307– 315, 2004 [DOI] [PubMed] [Google Scholar]
- 21. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grubbs CJ, Farnell DR, Hill DL, McDonough KC. Chemoprevention of N-nitroso-N-methylurea-induced mammary cancers by pretreatment with 17 beta-estradiol and progesterone. J Natl Cancer Inst 74: 927– 931, 1985 [PubMed] [Google Scholar]
- 23. Grubbs CJ, Juliana MM, Whitaker LM. Short-term hormone treatment as a chemopreventive method against mammary cancer initiation in rats. Anticancer Res 8: 113– 117, 1988 [PubMed] [Google Scholar]
- 24. Guzman RC, Yang J, Rajkumar L, Thordarson G, Chen X, Nandi S. Hormonal prevention of breast cancer: mimicking the protective effect of pregnancy. Proc Natl Acad Sci USA 96: 2520– 2525, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85: 463– 476, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Hancox RA, Allen MD, Holliday DL, Edwards DR, Pennington CJ, Guttery DS, Shaw JA, Walker RA, Pringle JH, Jones JL. Tumour-associated tenascin-C isoforms promote breast cancer cell invasion and growth by matrix metalloproteinase-dependent and independent mechanisms. Breast Cancer Res 11: R24, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao Q, Wang L, Zhao ZJ, Tang H. Identification of protein kinase D2 as a pivotal regulator of endothelial cell proliferation, migration, and angiogenesis. J Biol Chem 284: 799– 806, 2009 [DOI] [PubMed] [Google Scholar]
- 28. Harris RE, Namboodiri KK, Wynder EL. Breast cancer risk: effects of estrogen replacement therapy and body mass. J Natl Cancer Inst 84: 1575– 1582, 1992 [DOI] [PubMed] [Google Scholar]
- 29. Henderson IC. Risk factors for breast cancer development. Cancer 71: 2127– 2140, 1993 [DOI] [PubMed] [Google Scholar]
- 30. Hovey RC, Trott JF, Vonderhaar BK. Establishing a framework for the functional mammary gland: from endocrinology to morphology. J Mammary Gland Biol Neoplasia 7: 17– 38, 2002 [DOI] [PubMed] [Google Scholar]
- 31. Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics Chapter 13: Unit 13 11, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Iotti G, Ferrari-Amorotti G, Rosafio C, Corradini F, Lidonnici MR, Ronchetti M, Bardini M, Zhang Y, Martinez R, Blasi F, Calabretta B. Expression of CCL9/MIP-1gamma is repressed by BCR/ABL and its restoration suppresses in vivo leukemogenesis of 32D-BCR/ABL cells. Oncogene 26: 3482– 3491, 2007 [DOI] [PubMed] [Google Scholar]
- 33. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Kitagawa M, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol 12: 278– 285, 2010 [DOI] [PubMed] [Google Scholar]
- 35. Kang YJ. Metallothionein redox cycle and function. Exp Biol Med (Maywood) 231: 1459– 1467, 2006 [DOI] [PubMed] [Google Scholar]
- 36. Kasai S, Inui M, Nakamura K, Kakizaki Y, Endo S, Nakamura A, Ito S, Takai T. A novel regulatory role of gp49B on dendritic cells in T-cell priming. Eur J Immunol 38: 2426– 2437, 2008 [DOI] [PubMed] [Google Scholar]
- 37. Kelsey JL, Gammon MD. The epidemiology of breast cancer. CA Cancer J Clin 41: 146– 165, 1991 [DOI] [PubMed] [Google Scholar]
- 38. Lai AZ, Gujral TS, Mulligan LM. RET signaling in endocrine tumors: delving deeper into molecular mechanisms. Endocr Pathol 18: 57– 67, 2007 [DOI] [PubMed] [Google Scholar]
- 39. Lambe M, Hsieh C, Tsaih S, Ekbom A, Adami HO, Trichopoulos D. Maternal risk of breast cancer following multiple births: a nationwide study in Sweden. Cancer Causes Control 7: 533– 538, 1996 [DOI] [PubMed] [Google Scholar]
- 40. Lambe M, Hsieh CC, Chan HW, Ekbom A, Trichopoulos D, Adami HO. Parity, age at first and last birth, and risk of breast cancer: a population-based study in Sweden. Breast Cancer Res Treat 38: 305– 311, 1996 [DOI] [PubMed] [Google Scholar]
- 41. Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329: 89– 93, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329: 85– 89, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu P, Li P, Burke S. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol Rev 238: 138– 149, 2010 [DOI] [PubMed] [Google Scholar]
- 44. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25: 402– 408, 2001 [DOI] [PubMed] [Google Scholar]
- 45. Lopez-Otin C, Palavalli LH, Samuels Y. Protective roles of matrix metalloproteinases: from mouse models to human cancer. Cell Cycle 8: 3657– 3662, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marcondes FK, Bianchi FJ, Tanno AP. Determination of the estrous cycle phases of rats: some helpful considerations. Braz J Biol 62: 609– 614, 2002 [DOI] [PubMed] [Google Scholar]
- 47. Marques M, Kumar A, Poveda AM, Zuluaga S, Hernandez C, Jackson S, Pasero P, Carrera AC. Specific function of phosphoinositide 3-kinase beta in the control of DNA replication. Proc Natl Acad Sci USA 106: 7525– 7530, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCawley LJ, Crawford HC, King LE, Jr, Mudgett J, Matrisian LM. A protective role for matrix metalloproteinase-3 in squamous cell carcinoma. Cancer Res 64: 6965– 6972, 2004 [DOI] [PubMed] [Google Scholar]
- 49. McCawley LJ, Wright J, LaFleur BJ, Crawford HC, Matrisian LM. Keratinocyte expression of MMP3 enhances differentiation and prevents tumor establishment. Am J Pathol 173: 1528– 1539, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McPherson K. Childbearing, oral contraceptive use, and breast cancer. Lancet 341: 1604– 1605, 1993 [DOI] [PubMed] [Google Scholar]
- 51. McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ 321: 624– 628, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Medina D. Breast cancer: the protective effect of pregnancy. Clin Cancer Res 10: 380S- 384S, 2004 [DOI] [PubMed] [Google Scholar]
- 53. Medina D, Peterson LE, Moraes R, Gay J. Short-term exposure to estrogen and progesterone induces partial protection against N-nitroso-N-methylurea-induced mammary tumorigenesis in Wistar-Furth rats. Cancer Lett 169: 1– 6, 2001 [DOI] [PubMed] [Google Scholar]
- 54. Mielke PW, Berry KJ. Permutation Methods: a Distance Function Approach. New York: Springer, 2007, p. xvii [Google Scholar]
- 55.Montel V, Kleeman J, Agarwal D, Spinella D, Kawai K, Tarin D. Altered metastatic behavior of human breast cancer cells after experimental manipulation of matrix metalloproteinase 8 gene expression. Cancer Res 64: 1687– 1694, 2004 [DOI] [PubMed] [Google Scholar]
- 56. Nardi V, Naveiras O, Azam M, Daley GQ. ICSBP-mediated immune protection against BCR-ABL-induced leukemia requires the CCL6 and CCL9 chemokines. Blood 113: 3813– 3820, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nelson JF, Felicio LS, Randall PK, Sims C, Finch CE. A longitudinal study of estrous cyclicity in aging C57BL/6J mice: I. Cycle frequency, length and vaginal cytology. Biol Reprod 27: 327– 339, 1982 [DOI] [PubMed] [Google Scholar]
- 58. Ng LG, Mackay CR, Mackay F. The BAFF/APRIL system: life beyond B lymphocytes. Mol Immunol 42: 763– 772, 2005 [DOI] [PubMed] [Google Scholar]
- 59. Pang WW, Hartmann PE. Initiation of human lactation: secretory differentiation and secretory activation. J Mammary Gland Biol Neoplasia 12: 211– 221, 2007 [DOI] [PubMed] [Google Scholar]
- 60. Pike MC, Spicer DV, Dahmoush L, Press MF. Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol Rev 15: 17– 35, 1993 [DOI] [PubMed] [Google Scholar]
- 61. Polyak K, Kalluri R. The Role of the Microenvironment in Mammary Gland Development and Cancer. Cold Spring Harb Perspect Biol 2: a003244, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. R Development Core Team. R: A Language and Environment for Statistical Computing. http://www.R-project.org, 2009
- 63.Rajkumar L, Guzman RC, Yang J, Thordarson G, Talamantes F, Nandi S. Short-term exposure to pregnancy levels of estrogen prevents mammary carcinogenesis. Proc Natl Acad Sci USA 98: 11755– 11759, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Russo J, Mailo D, Hu YF, Balogh G, Sheriff F, Russo IH. Breast differentiation and its implication in cancer prevention. Clin Cancer Res 11: 931s– 936s, 2005 [PubMed] [Google Scholar]
- 65. Russo J, Moral R, Balogh GA, Mailo D, Russo IH. The protective role of pregnancy in breast cancer. Breast Cancer Res 7: 131– 142, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Russo J, Russo IH. Differentiation and breast cancer. Medicina (B Aires) 57 Suppl 2: 81– 91, 1997 [PubMed] [Google Scholar]
- 67. Sinha DK, Pazik JE, Dao TL. Prevention of mammary carcinogenesis in rats by pregnancy: effect of full-term and interrupted pregnancy. Br J Cancer 57: 390– 394, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sivaraman L, Stephens LC, Markaverich BM, Clark JA, Krnacik S, Conneely OM, O'Malley BW, Medina D. Hormone-induced refractoriness to mammary carcinogenesis in Wistar-Furth rats. Carcinogenesis 19: 1573– 1581, 1998 [DOI] [PubMed] [Google Scholar]
- 69. Smyth GK. Limma: linear models for microarray data. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor, edited by Gentleman RC, Carey VJ, Dudoit S, Irizarry R, and H, uber W. New York: Springer, 2005, p. 397– 420 [Google Scholar]
- 70. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004 [DOI] [PubMed] [Google Scholar]
- 71.Uehara N, Unami A, Kiyozuka Y, Shikata N, Oishi Y, Tsubura A. Parous mammary glands exhibit distinct alterations in gene expression and proliferation responsiveness to carcinogenic stimuli in Lewis rats. Oncol Rep 15: 903– 911, 2006 [PubMed] [Google Scholar]
- 72. Ursin G, Bernstein L, Wang Y, Lord SJ, Deapen D, Liff JM, Norman SA, Weiss LK, Daling JR, Marchbanks PA, Malone KE, Folger SG, McDonald JA, Burkman RT, Simon MS, Strom BL, Spirtas R. Reproductive factors and risk of breast carcinoma in a study of white and African-American women. Cancer 101: 353– 362, 2004 [DOI] [PubMed] [Google Scholar]
- 73. Whittaker GC, Orr SJ, Quigley L, Hughes L, Francischetti IM, Zhang W, McVicar DW. The linker for activation of B cells (LAB)/non-T cell activation linker (NTAL) regulates triggering receptor expressed on myeloid cells (TREM)-2 signaling and macrophage inflammatory responses independently of the linker for activation of T cells. J Biol Chem 285: 2976– 2985, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang J, Yoshizawa K, Nandi S, Tsubura A. Protective effects of pregnancy and lactation against N-methyl-N-nitrosourea-induced mammary carcinomas in female Lewis rats. Carcinogenesis 20: 623– 628, 1999 [DOI] [PubMed] [Google Scholar]
- 75. Zhao W, Grubbs CJ, Myers RK, Nilsen-Hamilton M. Parity is associated with an expanded macrophage population in the mammary gland. Int J Oncol 37: 1195– 1202, 2010 [DOI] [PubMed] [Google Scholar]


