SUMMARY
Dengue virus (DENV) infection induces the expansion of plasmablasts, which produce antibodies that can neutralize DENV but also enhance disease upon secondary infection with another DENV serotype. To understand how these immune responses are generated, we used a systems biological approach to analyse immune responses to dengue in humans. Transcriptomic analysis of whole blood revealed that genes encoding pro-inflammatory mediators and type I IFN-related proteins were associated with high DENV levels during initial symptomatic disease. Additionally, CD14+CD16+ monocytes increased in the blood. Similarly, in a non-human primate model, DENV infection boosted CD14+CD16+ monocyte numbers in the blood and lymph nodes. Upon DENV infection in vitro, monocytes up-regulated CD16 and mediated differentiation of resting B cells to plasmablasts as well as IgG and IgM secretion. These findings provide a detailed picture of innate responses to dengue and highlight a role for CD14+CD16+ monocytes in promoting plasmablast differentiation and anti-DENV antibody responses.
INTRODUCTION
Dengue is an emerging, mosquito-borne infectious disease which causes clinical disease in nearly 100 million people annually (Bhatt et al., 2013). Infection with one of the four serotypes DENV can result in dengue fever (DF), dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS), which is a life threatening illness (Simmons et al., 2012). The initial targets of DENV infection in vivo are poorly understood, although DENV can infect skin resident Langerhans cells (LC) (Wu et al., 2000), monocytes, macrophages (MQ), dendritic cells (DC) (Durbin et al., 2008; Ho et al., 2001; Wu et al., 2000), and endothelial cells in vitro (Bosch et al., 2002). Consistent with this, recent studies implicate molecules commonly expressed on myeloid cells such as DC-SIGN (Tassaneetrithep et al., 2003), mannose receptor (MMR) (Miller et al., 2008) and TIM and TAM proteins (Meertens et al., 2012) as receptors for DENV entry, and CLEC5A was shown to directly interact with DENV to promote inflammatory response (Chen et al., 2008). DENV infection can be also mediated by interactions of the virus Ab complexes with Fc-γ receptors during secondary infection with a heterologous serotype (Boonnak et al., 2008; Halstead and O’Rourke, 1977).
Another characteristic feature of dengue infection is the massive expansion of antibody-producing plasmablasts in the blood, which occurs within a few days of infection (Balakrishnan et al., 2011; Garcia-Bates et al., 2013; Wrammert et al., 2012). However, although infection with a given serotype can induce antibodies that are cross reactive to the other serotypes, generally long term immunity is generated only against the original serotype (Green and Rothman, 2006). In fact, in many cases, immunity against a heterologous serotype is not protective, but may augment the severity of disease (Burke et al., 1988; Guzman et al., 2000; Sangkawibha et al., 1984), possibly through a mechanism termed “antibody-dependent enhancement” (ADE) (Halstead et al., 2010). Although it is clear that both the virus strain and the immune response play a role in disease outcome, the specific mechanisms that lead to protective versus non-protective immune responses or mild versus severe disease are poorly understood.
Monocytes, the most abundant blood mononuclear phagocytes and one of the main cell targets of DENV (Durbin et al., 2008), originate from myeloid precursors in bone marrow and differentiate into tissue MQ and DCs (Auffray et al., 2009). In fact, human blood monocytes represent a diverse group of cells that can be distinguished by their phenotype and function in to at least 3 populations (Saha and Geissmann, 2011; Ziegler-Heitbrock and Hofer, 2013). The “classical” CD14+CD16- monocytes or “intermediate” CD14+CD16+ monocytes demonstrate similarity to the mouse Gr1+Ly6Chi monocytes and respond to CCL2 (MCP-1) that signals via CCR2 (Ingersoll et al., 2011). CD14+CD16- monocytes produce IL-10 as well as IL-6, IL-8, CCL2 (MCP-1) and RANTES upon LPS stimulation (Cros et al., 2010; Serbina et al., 2009; Wong et al., 2011); in contrast the “intermediate” CD14+CD16+ monocytes can sense ligands for TLR2, TLR4 as well as TLR8 and secrete IL-6, IL-8, CCL2 (MCP-1), CXCL10 (IP-10), IL-1β and TNF-α (Cros et al., 2010; Wong et al., 2011). The “non-classical” CD14dimCD16++ resemble murine Gr1- Ly6Clo cells, express CX3CR1, can detect viral RNA via TLR7 and 8 and are predominant producers of IL-1β, TNF-α and CXCL10 (IP-10) (Cros et al., 2010; Wong et al., 2011).
Here, we used an integrated approach to obtain a detailed picture of the innate response during the acute dengue. Our transcriptional profiling and immunological analysis of clinical dengue patients, together with results from a non-human primate (NHP) model of DENV infection and in-vitro experiments suggest a distinctive role of CD14+CD16+ monocytes in mediating humoral immunity to DENV infection.
RESULTS
Transcriptional signatures correlate with DENV viral loads and duration of illness, but do not discriminate between DF and DHF
We analyzed whole blood samples from 28 secondary dengue patients (DF n=18, DHF=10) hospitalized at the Siriraj Hospital in Bangkok, Thailand during the 2009 season. A single blood collection was acquired between days 2 and 9 after onset of symptoms (acute illness) as reported by patients at admission, and for 19 patients (DF n=13, DHF=6) also at the convalescence at 4 weeks or later after discharge (Figure 1A). Additionally, blood was sampled from 9 local, healthy, young adults to provide controls for transcriptomic and immunological analysis (full characteristic of the cohorts in Supplemental Materials and Table S1 and S2). Among the patients there was a considerable variation in the viral load (VL, from <100 to 9.37 × 109 copies/ml) and concentration of DENV NS-1 antigen in plasma (Figure 1B). The VL and NS-1 antigen concentration were positively correlated with each other (Figure 1B) and correlated inversely with the duration of acute illness (Figure 1C), with the highest VL and NS-1 concentration at early illness between 2 and 4 days of symptomatic dengue for both DF and DHF patients (Figure 1C).
Figure 1. The transcriptional response induced by dengue infection is correlated with viral load.
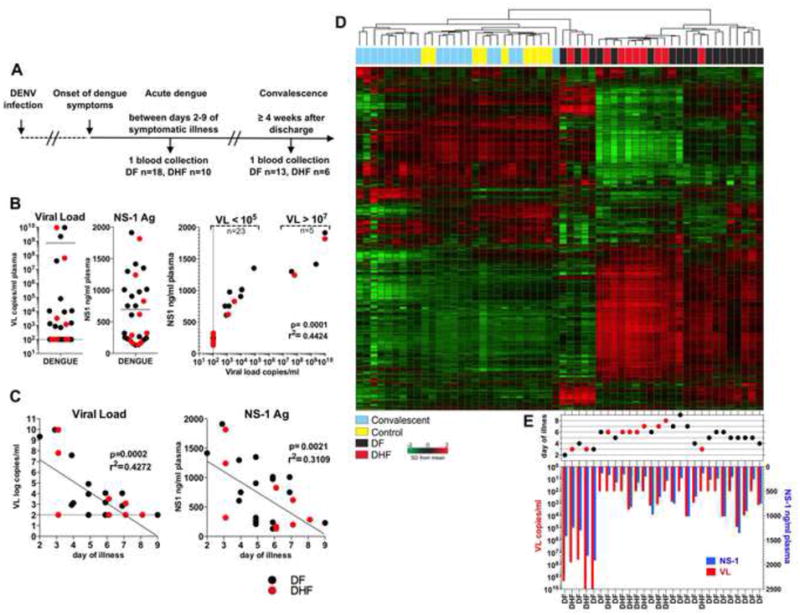
(A) Clinical study design. (B) VL and NS-1 antigen concentration in plasma from acute dengue patients. Correlation analysis shows formation of two distinct groups with high (VL>107, n=23) and low (VL<105, n=5) viral burden. (C) Correlation analysis of VL (left) and NS-1 Ag concentration (right) with days of dengue illness at the specimen collection. (D) Unsupervised clustering analysis. (E) Reported days of illness at collection and matching VL (red bars) and NS-1 Ag (blue bars) corresponding to the individual dengue patients on the heat map. DF n=18: black dots, DHF n=10: red dots. See also Figures S1 and Table S1 and S2.
Gene microarrays were used to assess the whole blood transcriptome of 28 acute patients, 19 convalescents and 9 healthy controls. Our initial unsupervised analysis using the 7,528 genes with highest variance demonstrated that samples from the acutely infected patients, including both DF and DHF patients, clustered differently from the convalescent and control individuals. However, within the acutely infected patients we observed large heterogeneity of gene expression patterns as classified by either hierarchical clustering (Figure 1D) or principal component analysis (PCA; Figure S1A). Furthermore, no defined clusters between acute DF and DHF patients could be detected (Figure 1D). Instead, we observed two distinct clusters that were related to VL and NS-1 concentration at the early stage of symptomatic illness (Figure 1E). A group of five patients with the highest VL (VL>107) and NS-1 concentration formed a distinct cluster from those patients with lower viremia (VL<105) (Figure 1D and E). Thus, transcriptional profiling could clearly discriminate between patients with acute dengue and convalescent or control subjects, as well as those with high versus low VL, but not between DF and DHF.
Distinct molecular pathways discriminate between subjects with high versus low viral loads, and early versus late disease
The blood transcriptome in dengue patients can be dependent on both the disease duration and VL (Popper et al., 2012; Sun et al., 2013). To determine molecular pathways associated with VL or disease duration, we first performed Pearson’s correlation analysis between all the 18,146 genes and the VL (Figure 2A) or days of symptomatic illness (Figure S1B) in the 28 acute dengue patients. Pathway enrichment analyses were performed using the 1,000 genes with highest positive (high VL) or negative (low VL) correlation (Figure 2B-F). Genes associated with the innate sensing of viruses and inflammatory responses to viral infection were positively correlated with high VL (107-109 copies/ml; Figure 2B, Table S3), and inversely correlated with duration of illness (Figure S2C). Similar results were established by analyzing the VL-scored specimens by GSEA using the Reactome database (Table S4). Genes with the highest correlation to VL and early symptomatic disease were associated with the upstream regulation of type-I IFN signaling (Figure 2C). The top 1000 genes whose expression was negatively correlated with VL, i.e., highly expressed in patients with low or undetectable VL (Figure 2D) at the late stage of illness (Figure S1D) were associated with pathways involved in cell cycle, proliferation, cell metabolism and translational control (Figure 2D, S1D, Table S3 and S4).
Figure 2. Transcriptomic analysis identifies molecular networks that correlate with DENV viral load.
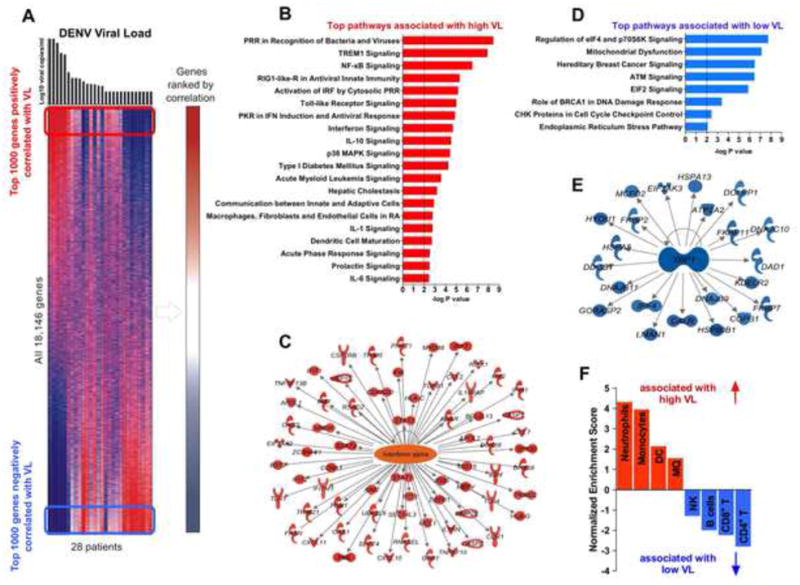
(A) Heat map of probe sets (rows) and subjects (columns) ranked by VL (top), colors in map indicate relative gene expression. Right margin: probe sets with negative (blue) or positive correlation (red) to VL. P < 0.05 (Pearson). Frames: approximate range of top 1000 genes positively (red) or negatively (blue) correlated to VL used for analysis in B-E. (B and D) Top Ingenuity pathways that positively (B) or negatively (D) correlate with VL. Dotted line represents a cut-off value (−log p=2). (E and C) IPA analysis of positively (C) or negatively (E) VL correlated genes regulated by IFN-α (C) or XBP-1 (E). P < 0.05 (Pearson). (F) GSEA analysis of the whole blood transcriptome of acute dengue patients using cell-specific data sets. Normalized enrichment score (NES) indicates positive (NES>0, red bars) or negative (NES<0, blue bars) correlation of distinct cell-specific gene sets to VL. See also Figures S1 and S2, and Tables S3 and S4.
The fact that molecular pathways that directly correlate with VL are inversely correlated with the duration of illness is consistent with the reciprocal relationship between VL and disease duration (Figure 1C). However to determine the impact of VL on transcriptional profiles, at a given time during the disease, we compared gene signatures between patients with high VL versus low VL enrolled during first days 2-4 of symptoms (Figure S1F). Distinct innate and inflammatory immune pathways were enriched in high versus low VL patients (Figure S1G and H) even at early illness (Figure 2B and D), suggesting that the VL can impact the transcriptional profiles independently of disease duration.
Additional analysis revealed a significant enrichment of XBP-1 target genes among the genes highly expressed in patients with low VL (Figure 2E). XBP1 is a transcription factor that mediates the unfolded protein response and is involved in the differentiation of plasma cells (Reimold et al., 2001). The signature of XBP-1 activation was consistent with increased numbers of plasmablast cells (Figure S2A), which correlated with the duration of illness (Figure S2B). Enhanced numbers of plasmablasts has been previously observed with dengue infections (Wrammert et al., 2012), and in humans vaccinated with the influenza vaccine (Nakaya et al., 2011).
Finally, we used gene set enrichment analysis (GSEA) to identify immune cell types in blood of dengue patients. We detected significant enrichment in genes specific for neutrophils, monocytes, MQ and DCs in the specimens with high VL (Figure 2F) at early stage of acute dengue (Figure S1E). In contrast, patients at the late stage of symptomatic dengue with low VL were associated with genes expressed specifically in NK cells, CD4+ and CD8+ T cells as well as B cells (Figure 2F and S1E). Thus, these data reveal enrichment in the innate immune pathways in patients with high viral burden at the early phase of infection. Gene signatures specific for innate cells including monocytes, DCs and MQ were associated with high VL and early illness.
DENV infection induces a cellular, transcriptional and cytokine signature of CD14+CD16+ monocytes in blood
Alterations in blood cells during acute dengue were dependent on the VL and duration of the symptomatic disease (Figure S2C and Table S2). Hence, we evaluated the innate cell populations by flow cytometry (Figure S3A). DENV infection increased the frequency of blood monocytes, especially in high VL patients (Figure 3A), however we did not detect a significant increase in absolute numbers of monocytes (Figure S3B). In contrast, there was a striking reduction in the proportion and absolute numbers of the BDCA-1+ mDC-1 population in individuals with high VL at the early stage of illness (Figure 3A, S3B and C). Low numbers of the mDC-1 correlated with the duration of disease (Figure S3C). Similar trends were also evident with basophils, but not with the cross presenting BDCA-1neg mDC-2 subset or pDCs and NK cells (Figure 3A and S3B and C).
Figure 3. DENV infection enhances the frequency of CD14+ CD16+ monocytes in blood.
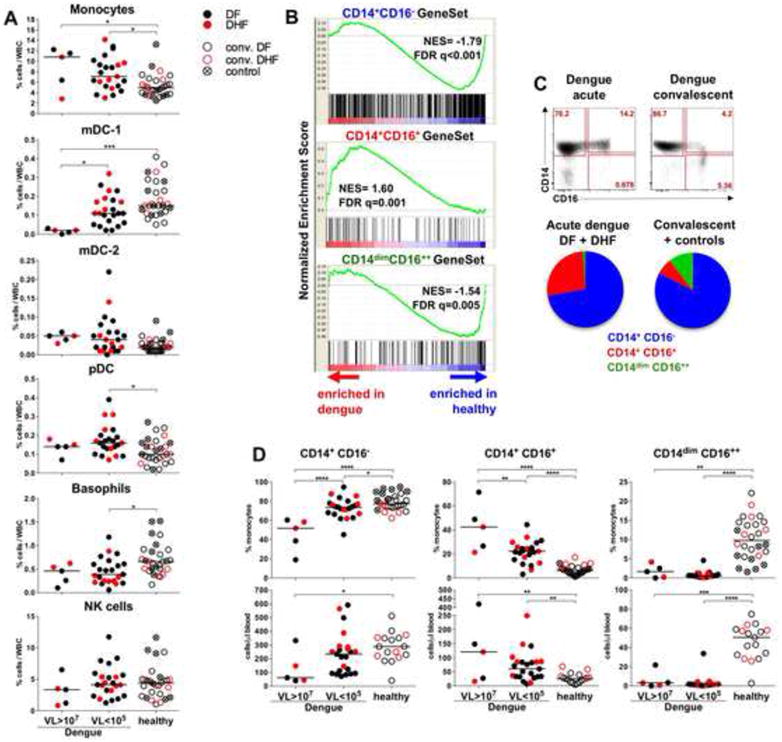
(A) Proportions of innate cell subsets within white blood cells (WBC). (B) GSEA analysis of the whole blood transcriptome of the acute dengue patients and healthy individuals (convalescent and controls) using CD14+CD16-, CD14+CD16+ and CD14dimCD16++ cell-specific gene sets. NES: overrepresentation of each of the monocyte subset in dengue or healthy cohorts. False discovery rate (FDR) is indicated as q. (C) Dot plots show blood monocytes in a representative patient during acute dengue and convalescence. Pie-charts represent mean proportions of the CD14+CD16- (blue), CD14+CD16+ (red) and CD14dimCD16++ (green) cells within all monocytes in acute dengue patients and healthy individuals. (D) Proportions (top) and absolute counts (bottom) of the monocyte subsets in blood. Absolute counts comprise only convalescent subjects in the healthy cohort. (A and D) Data compared between dengue patients with high VL (VL>107, n=5), low VL (VL<105, n=23), and healthy (convalescent, n=19 and controls, n=9). Symbols: acute dengue: DF black dots, DHF red dots; convalescent: DF black circles, DHF red circles; and controls: crossed circles. See also Figures S2 and S3 and Table S2.
The change in the frequency of blood monocytes was consistent with the enrichment of monocyte-associated genes in the transcriptional signature (Figure 2F and S1E); therefore, we applied a meta-analysis of the whole blood transcriptome using monocyte-specific datasets (Cros et al., 2010) to discriminate monocytes-specific signatures. GSEA analysis determined a significant enrichment in genes specific for the “intermediate” CD14+CD16+ monocytes in acute dengue (Figure 3B). This is consistent with the enhanced frequency of CD14+CD16+ subset as measured by flow cytometry (Figure 3C, D). Consistent with the inverse correlation between VL and disease duration (Figure 1C), the expansion of CD14+CD16+ cells correlated with early disease (data not shown).
Acute infection induced secretion of plasma pro-inflammatory chemokines IP-10 (CXCL-10), MCP-1 (CCL-2), MIP-1ß (CCL-4) and IL-1ra (Figure 4A), notably in patients with high VL and the early stage of disease (data not shown). Interestingly, levels of IP-10 and MIP-1ß correlated with the frequency and absolute numbers of the CD14+CD16+ cells in blood (Figure 4B). Additionally, we detected substantial quantity of IL-10, eotaxin (CCL-11), IL-6 and IL-8, however there was no significant correlation to VL (Figure 4A). In summary, acute dengue results in an expansion of the CD14+CD16+ monocytes, depletion of the BDCA-1+ mDCs and basophils and increase in plasma pro-inflammatory cytokines. This was most pronounced within patients with viral burden at the early stage of dengue.
Figure 4. Inflammatory cytokines in plasma of acute dengue patients correlate with the numbers of CD14+CD16+ monocytes.
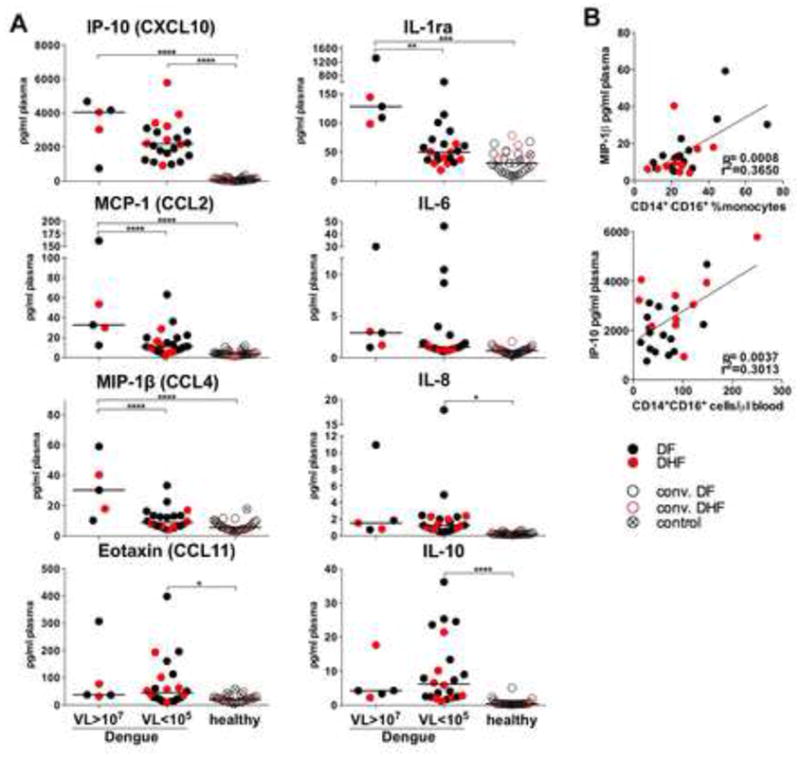
(A) Cytokines in plasma from patients with acute dengue (DF black dots, DHF red dots), convalescent (DF black circles, DHF red circles) and controls (crossed circles) compared between groups of dengue patients with high VL (VL>107, n=5), low VL (VL<105, n=22), and healthy (convalescent, n=19 and controls, n=9). (B) Correlation analysis of plasma concentration of MIP-1β (top) and IP-10 (bottom) with a proportion (top) and absolute counts of CD14+CD16+ monocytes in blood (bottom).
DENV-infected blood monocytes differentiate into CD14+CD16+ cells which induce the differentiation of resting B cells into plasmablasts
The rapid kinetics of expansion of the CD14+CD16+ monocyte subset in dengue raised the possibility that such cells may modulate the immune response to DENV infection. Infection of CD14+ monocytes from healthy individuals with DENV increased the fraction of CD14+CD16+ cells to over 70% of all monocytes (Figure 5A). Similar results were seen with the TLR7/8-ligand (R-848) but not TLR4-L (LPS) (Figure 5A). A fraction of the CD14+CD16+, DENV-infected monocytes demonstrated high expression of the MQ associated markers CD206 (MMR), CD115 (M-CSFR), and all of the DENV-infected monocytes expressed CCR5 as well as high CD163 and CD169 (Figure 5B). Moreover, electron microscopy imaging demonstrated presence of dendrites and large numbers of cytoplasmic vacuoles, similar to the cells cultured with M-CSF in monocytes infected with DENV (Figure 5C). In vitro infection of monocytes with DENV induced secretion of MCP-1 (CCL-2), IP-10 (CXCL-10), IL-6, IL-8 and IL-10, however no IL-1ß secretion was evident post infection with DENV (Figure 5D).
Figure 5. DENV-infected monocytes acquire CD14+CD16+ phenotype and stimulate plasmablast differentiation in vitro.
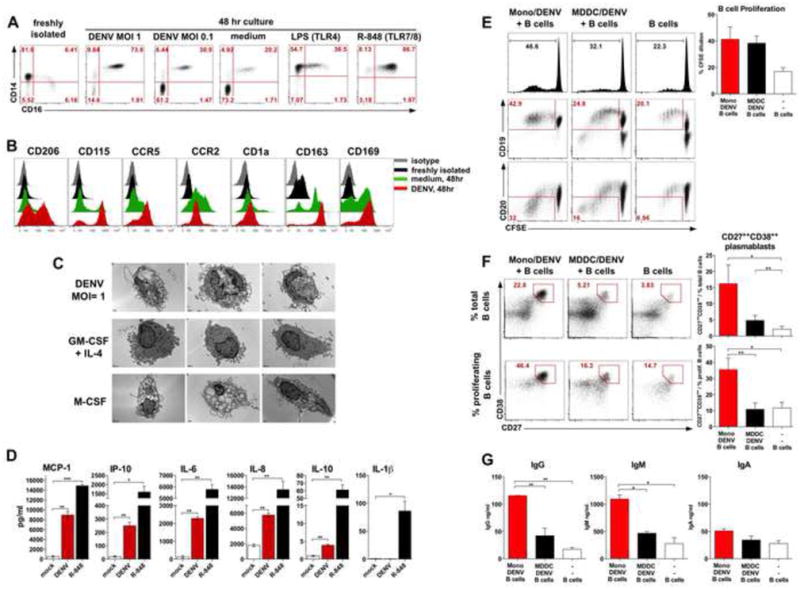
(A) CD14 and CD16 expression in monocytes infected with DENV-2 at the MOI=1, MOI=0.1, or stimulated with LPS or R-848. (B) Histograms show surface phenotype of unstimulated monocytes (black), or cultured in medium (green) or DENV-2 at MOI=1 (red), isotype (gray). (C) Electron microscopy images of representative cells from monocyte cultured for 3 d with DENV-2 at MOI=1, or with GM-CSF+IL-4, or M-CSF. Bars, 1μm. (D) Cytokines in supernatants of monocytes cultured in medium or with DENV-2 (MOI=1) or R-848 for 48 hr. (E) Histograms (top) show B cell proliferation measured by CFSE dilution after 6d. Expression of CD19 (center) or CD20 (bottom) in cultured B cells, numbers: % of proliferated cells. Bar-graph: % of proliferated B cells measured by CSFE dilution, mean +/- SEM (4 experiments). (F) CD27++CD38++ plasmablast differentiation within total (top) and proliferated (bottom) B cells after 6 d. Bar-graphs: % of CD27++CD38++ plasmablasts within total (top) and proliferated (bottom) B cells, mean +/- SEM (4 experiments). (G) Titers of IgG, IgM and IgA in cultures on day 6. (A, B, D and G) Data are shown from a one representative experiment out of 4 independent tests with four different blood donors. See also Figures S4 and S5.
To evaluate the potential functions of these cells, we tested if blood monocytes infected with DENV can stimulate B cells in vitro. We infected monocytes or monocyte-derived DC (MDDC) with DENV at the MOI=1 for 48 hours and cultured them with allogenic, resting CD19+ B cells in the presence of IL-2 and CpG (Figure 5E-G). After 6 days of co-culture monocytes infected with DENV stimulated robust proliferation of B cells as measured by CFSE dilution, however with comparable efficacy as MDDC (Figure 5E, top panel). In the Monocyte/DENV and B cells co-culture the dividing B cells expressed CD19 and down-regulated CD20, characteristic of differentiating plasmablasts (Wrammert et al., 2012) (Figure 5E, bottom). In fact, a 6-day co-culture of B cells with infected monocytes induced a major increase in the frequency of CD27++CD38++ plasmablasts (Figure 5F) predominantly within the proliferating B cells (Figure 5F, bottom). Differentiation of the CD27++CD38++ cells facilitated by monocytes was considerably larger than that stimulated by MDDC or B cells without any APC (Figure 5F). Commensurate with the increase in CD27++CD38++ plasmablast differentiation, DENV-infected monocytes stimulated secretion of IgG and IgM (but not IgA), significantly higher than MDDC or controls (Figure 5G).
We then determined the mechanism by which DENV induced CD14+CD16+ monocytes stimulate plasmablast differentiation. Interestingly, the expression of genes encoding BAFF and APRIL, molecules which induce B cell differentiation and survival (Litinskiy et al., 2002; Mackay and Schneider, 2009) was increased in the blood of patients with high viremia (Figure S4A) at the early stage of illness (Figure S4B) and correlated with the magnitude of the CD14+CD16+ monocytes in blood (Figure S4C). There was a trend for enhanced levels of APRIL protein in the plasma of dengue patients with high VL (Figure S4D), and consistent with this, monocytes secreted BAFF and APRIL in vitro after DENV infection (Figure S4E). We blocked BAFF and APRIL signaling to explore mechanisms of induction of Ab-producing plasmablast by monocytes (Figure S5). Anti-BAFF and TACI-Fc modestly diminished B cell proliferation and differentiation of plasmablasts (Figure S5A and B) and TACI-Fc significantly reduced production of IgM in vitro (Figure S5C). Furthermore, we also blocked IL-6, IL-10 and IP-10 in monocyte/B cell co-cultures (Figure S5). Both, α-IL-10 and α-IP-10 reduced B cell proliferation (Figure S5A), however only α-IL-10 significantly blocked plasmablast differentiation and IgM secretion (Figure S5B and C). These results demonstrated in-vitro that DENV induces differentiation of monocytes to CD14+CD16+ cells, with a distinct phenotype and morphology that possess a superior ability to stimulate resting B cells to differentiate into plasmablasts via BAFF/APRIL and IL-10.
DENV infection in macaques results in expansion of the CD14+CD16+ monocyte subset in secondary lymphoid organs
Finally, we used an NHP model of dengue (Onlamoon et al., 2010) to test if DENV infection also induces expansion of the CD14+CD16+ monocyte subset in the secondary lymphoid organs, the sites where interaction with B cells might occur. We infected a cohort of 5 rhesus macaques intravenously with a high dose of DENV-2 (Figure 6A) and sampled blood at 7 days prior to the infection (baseline), at 5 hr and on days 1, 3, 5, 7, 14 and 28 after challenge.
Figure 6. DENV infection induces CD14+ CD16+ monocytes phenotype in rhesus macaques.
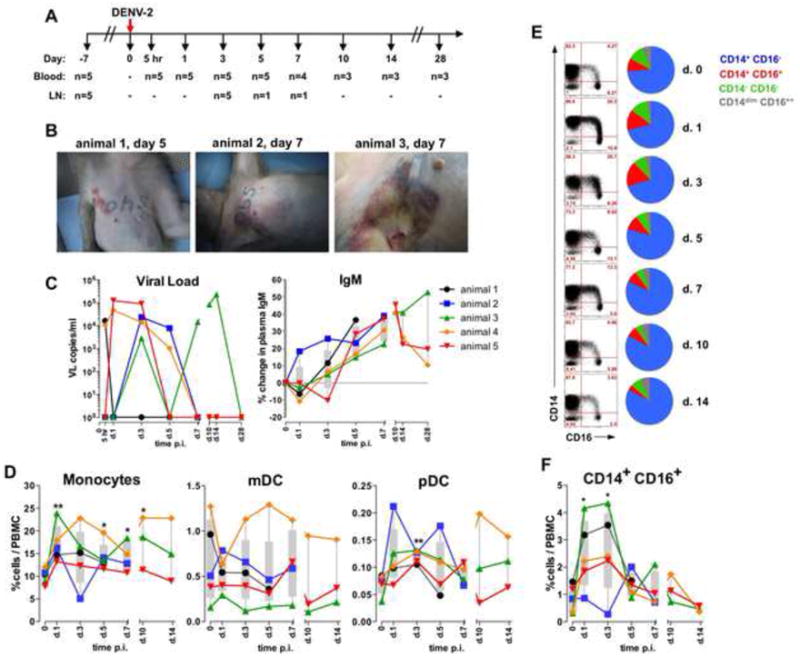
(A) NHP study design. (B) Images of hemorrhagic manifestation in three representative animals on days 5 and 7. (C) VL and IgM in plasma. (D) Proportions of innate cells within total PBMC. (E) Dot plots: CD14 and CD16 expression in blood monocytes. Pie charts: mean proportions of the CD14+CD16- (blue), CD14+CD16+ (red), CD14-CD16- (green) and CD14dimCD16++ (gray) cells within the total monocytes in all animals . (F) Kinetics of CD14+CD16+ monocytes within total PBMC. (C, D and F) Symbols and lines show individual animals in the cohort. (D and F) T-test. See also Figure S6.
Consistent with the previous report (Onlamoon et al., 2010) animals infected with DENV-2 developed skin hemorrhagic manifestations including coagulopathy (Figure 6B). Also, DENV infection in rhesus animals transiently reduced the absolute counts of WBC, predominantly within the PBMC (Figure S6A). In all the infected animals DENV was detectable in plasma between 5 hours and 14 days after infection, though the VL kinetics differed considerably among the individual monkeys (Figure 6C). However, all the macaques developed DENV-2-specific IgM antibody titer (Figure 6C). Hence, we were able to efficiently infect NHPs with DENV and induce viral replication, specific Ab response, loss of leucocytes and hemorrhagic manifestations.
Similar to the human disease, DENV infection augmented the frequency or total blood monocytes between days 1 and 10 (Figure 6D and E). Moreover, infection increased the proportion of blood pDC on day 3 after challenge (1.71-fold increase), but there was no significant difference in the frequency of mDC (Figure 6D and S6B). As seen with human dengue (Figure 3C-D), there was an increase in the fraction of the CD14+CD16+ monocytes at 1-3 days after DENV challenge in NHPs (Figure 6E, F and S6C).
Furthermore, infection with DENV induced an expansion of the CD14+CD16+ monocyte subset in the secondary lymphoid organs (Figure 7). At baseline (day -7) and 3 days after infection we collected inguinal and axillary LN respectively to verify the local innate cellular compartment (Figure 7A). Additionally, we analyzed axillary LN from two individual animals sacrificed on days 5 (n=1) and 7 (n=1) post infection (Figure 6A). We detected a substantial, 13-fold increase in the absolute numbers of CD14+CD16+ cells that comprised a total of 0.32 – 0.44 % of all cells in the axillary LNs of three animals with the highest proportion of total monocytes (Figure 7B). However, even in the two animals (no. 4 and 5) with lesser numbers of total tissue monocytes the CD14+CD16+ cells represented up to 11.1 % of all monocytes, significantly more than before the infection (Figure 7C).
Figure 7. DENV infection in rhesus macaques stimulates enhanced numbers of the CD14+CD16+ monocytes into LN.
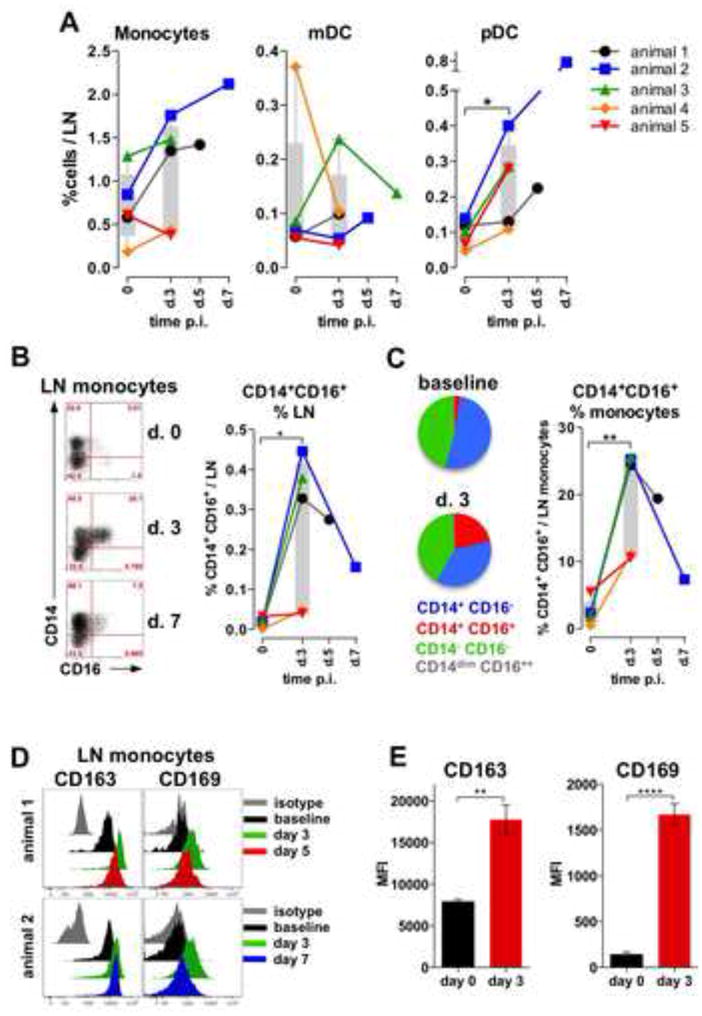
(A) Proportions of innate cells in LNs. (B) Dot plot representation of CD14 and CD16 expression in monocyte subsets in LN. Graph represents frequency of CD14+CD16+ monocytes within all cells in LN. (C) Pie charts show mean proportions of the CD14+CD16- (blue), CD14+CD16+ (red), CD14-CD16- (green) and CD14dimCD16++ (gray) cells within the total monocytes at baseline and day 3, n=5. Graph indicates proportion of CD14+CD16+ cells within all monocytes in LN. (A-C) Symbols represent individual animals in the cohort. (D) Histograms: monocytes expression of CD163 (left) and CD169 (right) at baseline (black), d 3 (green), d 5 (red, animal 1), 7 (blue, animal 2) and isotype (gray) in 2 representative animals. (E) Mean Fluorescent Intensity of expression of CD163 (left) and CD169 (right) at baseline (black) and d 3 (red) post infection, mean +/- SEM (n=5), t-test.
We analyzed the phenotype of the monocytes in LN by assessing surface expression of CD163 and CD169, distinct markers for tissue subcapsular sinus macrophages (SSM) (Junt et al., 2007; Martinez-Pomares and Gordon, 2012) (Figure 7D). At baseline, all resident monocytes expressed surface CD163, however it was significantly up-regulated on day 3 after challenge, at the peak expansion of the CD14+CD16+ subset in the axillary LN (Figure 7D and E). Similarly, all tissue-resident monocytes up-regulated expression of CD169 at 3 days after infection as measured by MFI (Figure 7D and E). The surface expression of CD163 and CD169 was diminished at later time points (days 5 and 7), consistent with decreasing quantity of the CD14+CD16+ in the LNs as tested in the necropsy tissues collected from 2 animals (Figure 7D). Taken together, these results demonstrate that DENV infection of NHPs result in an expansion of the CD14+CD16+ subset, in secondary lymphoid organs and in blood.
DISCUSSION
In this study, we performed a systems biological approach, similar to that used to study immune responses to vaccination in humans (Nakaya et al., 2011; Querec et al., 2009) to investigate the early host response to dengue in humans. Previous studies have used transcriptomic approaches to evaluate immune responses to DENV infections (Devignot et al., 2010; Hoang et al., 2010; Loke et al., 2010; Long et al., 2009; Popper et al., 2012; Simmons et al., 2007; Ubol et al., 2008), however, to the best of our knowledge to date, there has been no integrated study of the cellular, transcriptional and cytokine responses that underlie the innate response during acute dengue. We demonstrate that the transcriptional response to dengue is associated with both the VL and duration of illness. This is not surprising, since the VL and NS-1 correlated with the days of illness (fever days) reported by the patient at the time of sample collection (Figure 1B and C). However, since patients’ reports on the illness duration can be inaccurate we classified our analyses on laboratory-tested values of VL and NS-1. Nevertheless, an alternative analysis based on days of illness (Figure S1) revealed almost identical results as that based on the viral burden.
Consistent with other reports (Devignot et al., 2010; Loke et al., 2010) we did not detect any difference in the transcriptional signatures between patients with DF and DHF (Figure 1D and S1A), however our cohort did not contain DSS cases which show most variability in gene expression profiles among patients experiencing different grades of dengue disease severity (Devignot et al., 2010; Loke et al., 2010; Simmons et al., 2007). Moreover, the lack in discrepancy between distinct dengue phenotypes might also be attributed to the variation in viral burden and duration of the disease. The reports of reduced IFN-mediated response in severe shock (DSS) patients suggest that DENV can inhibit type I IFN signaling (Devignot et al., 2010; Popper et al., 2012). Blockade of the efficient IFN response by DENV (Schoggins et al., 2012) might be one of the mechanisms of progression into severe DHF/DSS symptoms. In fact, the NS-4B and NS-5 proteins of DENV efficiently impede STAT signaling and subsequent IFN-mediated response to infection (Ashour et al., 2010; Munoz-Jordan et al., 2003). Our experimental cohort comprised uncomplicated cases of DF and DHF and we focused our analysis on potential mechanisms of immune response to dengue rather than on pathogenesis of DF/DHF and DSS.
A robust antibody production and plasmablast response is one of the hallmarks of the immune response during dengue (Beltramello et al., 2010; Wrammert et al., 2012), though the mechanisms orchestrating such a response remain unclear. Our analysis identified the “intermediate” CD14+CD16+ monocytes as a one of most abundant blood phagocytes during acute dengue in humans, and blood and LN of infected NHP. However, the fact that numbers of CD14+CD16+ cells did not correlate with Ab titers (data not shown), and numbers of plasmablasts were higher in patients with low VL at late symptomatic disease (Figure S2A and B) (Wrammert et al., 2012) is due to the duration of symptomatic disease and different kinetics of these cell subsets in blood.
Blood monocytes and tissue MQ were previously suggested as a major reservoir for DENV replication (Durbin et al., 2008). Moreover, monocytes are broadly decorated with Fc-γ receptors (Nimmerjahn and Ravetch, 2008; Wong et al., 2011), a recognized feature that contributes to ADE during secondary dengue (Halstead et al., 2010). In fact, both the CD14+ and the non-classical CD14dimCD16+ cells can respond to DENV with a comparable efficiency and are equally susceptible for infection in vitro (Wong et al., 2012). Though long considered as the transitional population between the CD14+CD16- and CD14dimCD16++ cells, the intermediate, CD14+CD16+ monocytes represent a separate cell population with distinct phenotype and molecular signature (Cros et al., 2010; Wong et al., 2011). Of interest, we detected a considerable proportion of the CD14+CD16+ cells in the LNs of rhesus monkeys after DENV infection (Figure 7B and C), similarly as in the animals immunized with TLR ligands (Kwissa et al., 2012). These cells expressed high surface CD163 and CD169 (siglec-1), a feature of the subcapsular sinus macrophages (SSM) (Cyster, 2010; Martinez-Pomares and Gordon, 2012). The subset of CD169+ SSM reside in the LN sinus in the close proximity of migratory B cells (Carrasco and Batista, 2007; Cyster, 2010), can capture and present viral particles to antiviral B cells across the subcapsular sinus floor (Junt et al., 2007) and drive antibody affinity maturation (Phan et al., 2009). Relocation of the activated CD14+CD16+ cells demonstrating characteristics of the CD163+CD169+ SSM may contribute to the mechanisms driving a robust humoral response despite the depletion of classical BDCA-1+ myeloid DCs during acute dengue.
DENV stimulates IFN-I response in APC by inducing RIG-I and MDA-5 (Loo et al., 2008; Nasirudeen et al., 2011), nucleic acid receptors commonly expressed by myeloid cells including monocytes. Moreover, DENV has been previously shown to signal through TLR3 (Nasirudeen et al., 2011; Tsai et al., 2009), tough monocytes weakly express TLR3 (Cros et al., 2010) and do not response to TLR3 agonists (Cros et al., 2010; Kadowaki et al., 2001). However, distinct monocytes subsets express TLR7 and 8 (Cros et al., 2010), sensors of ssRNA that might be involved in DENV recognition. Hence, DENV-mediated activation of monocytes can be mediated by both RIG-I and MDA-5, and possibly, endogenous TLR signaling.
Dendritic cells and monocytes can stimulate B cells with the BAFF and APRIL dependent mechanism (Litinskiy et al., 2002; Mackay and Schneider, 2009). However, in contrast to the study by Xu et al. (Xu et al., 2012) where CD163+ human MQ efficiently stimulated CD138++CD38++ plasma cells through the IP-10 (CXCL-10) and IL-6 dependent mechanism, we did not observe IL-6 and IP-10-dependent plasmablast stimulation and Ab production (Figure S5). On the other hand, secretion of IgM and plasmablast differentiation were substantially reduced with α-BAFF, TACI-Fc and α-IL-10 treatment, similarly to the blood monocytes conditioned with the serum from patient with systemic lupus erythematosus (SLE) (Joo et al., 2012). Hence, we attribute the mechanism of plasmablast differentiation by DENV-infected monocytes to BAFF/APRIL as well as IL-10 signaling.
In summary (Figure S7), we present a set of findings for dissecting the immunity to DENV infection. We determined the early host response to DENV infection is highly dependant on the viral burden with inflammatory and IFN genes expression, loss of myeloid DCs and increase of the CD14+CD16+ monocytes in blood and LN at early phase of symptomatic disease, followed by robust expansion of plasmablasts afterward. We suggest a role of the CD14+CD16+ cells in supporting the humoral response to DENV infection by driving plasmablast differentiation via BAFF/APRIL and IL-10. These results may impact further understanding of dengue pathogenesis and help in discovering of new therapeutic and vaccination strategies.
EXPERIMENTAL PROCEDURES
Human subjects and specimen collection
All studies were pre-approved by the Faculty of Medicine at the Siriraj Hospital and the Emory University institutional review boards. Single blood sample was collected from patients diagnosed clinically with DENV infection admitted to Siriraj Hospital in Bangkok, Thailand. The DENV infection was confirmed by a serotype specific RT-PCR from freshly isolated specimens as well as other diagnostic tests including NS1 test, dengue-specific IgG and IgM ELISA or dip-stick tests and DENV antigen specific ELISPOT (Wrammert et al., 2012). Days of febrile illness were reported by patients at the admission. Clinical and laboratory patient data are described in the supplemental materials and summarized in Tables S1 and S2.
RNA specimens and gene array analysis
Whole blood was preserved on RNALater buffer (Ambion). RNA was isolated using the RiboPure-Blood kit (Ambion) and alpha and beta globin mRNA was depleted by GLOBINclear™ Kit (Ambion). Samples were checked for purity and hybridized on Human U133 Plus 2.0 Arrays (Affymetrix). Microarray intensity data of probe sets were normalized by RMA, which includes global background adjustment and quantile normalization. Probe sets representing the same gene were collapsed by taking the probe set with highest expression across all samples. Hierarchical clustering and PCA analyses were performed using Spotfire (TIBCO Software Inc.). Pathway analyses were performed using Gene Set Enrichment Analysis (GSEA) and Ingenuity Pathway Analysis (Ingenuity Systems) software. Detailed description of analyses in supplemental materials.
Rhesus animals and infection
Five adult, Indian rhesus macaques (macaca mulatta) were maintained at the Yerkes NPRC. Animals were cared for under guidelines established by the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals” with protocols approved by the Emory University IACUC. NHP were age and weight matched and infected by an intra-venous injection (i.v.) with 2 × 107 pfu of DENV serotype 2, strain 16681 as described before (Onlamoon et al., 2010). Animals were monitored daily for the onset of disease symptoms. Two individual animals were euthanized on days 5 and 7 respectively for necropsy.
Flow cytometry analysis
For human samples, whole blood from patients was collected in CPT tubes (BD) and 100-200μl of was stained with an appropriate antibody cocktail. Cells were washed in PBS with 5% FBS and RBC were lysed using the FACS lysis buffer (BD). Cells were washed and fixed with the Cytofix buffer (BD) and analyzed on the FACSAria machine (BD). Cell populations were defined within whole blood as represented in the Figure S3A and the CD27++CD38++ plasmablast B cells were defined as described before (Wrammert et al., 2012). Absolute cell counts were calculated using the complete blood counts (CBC). For NHP, PBMC were isolated from CPT tubes (BD) and LN were processed as described before (Kwissa et al., 2012). The detailed staining procedures and list of antibodies are represented in the supplemental materials.
Viral load and NS-1 antigen
Plasma viral loads in human and NHP specimens were determined by the quantitative RT-PCR and the NS-1 antigen was measured by ELISA from the cryopreserved samples. Detailed VL and NS-1 quantification are described in the supplemental materials.
Cytokines
Human plasma cytokines were analyzed using the Bio-Plex Pro Human Cytokine 27-plex Assay (Bio-Rad) and are represented as pg/ml. APRIL and BAFF concentrations were assessed by ELISA (eBioscience). Cytokines from cell cultures were measured in supernatants in duplicates by BD™ Cytometric Bead Array (BD) and analyzed on an LSRII (BD) instrument.
In-vitro cell assays
Studies with human blood cells were pre-approved by the Emory University institutional review board. PBMC from total of 10 healthy donors were purified from blood by a standard technique using LSM medium (MPBiomedicals). Monocytes were isolated by the positive selection using CD14 beads (Miltenyi) and B cells were isolated using the CD19 beads (Miltenyi) to a purity of ≥95%. CD14+ monocytes or MDDC were infected with the DENV-2 (strain 16681) at MOI=1 for 48hr. Resting, allogenic CD19+ B cells were stained with CFSE (Invitrogen) and co-cultured in 96-well plates with the MDDC or monocytes at the ratio of 5:1 in the presence of 20U/ml of IL-2 (Peprotech) and 50nM CpG type B 2006 (TriLink Biotech) and blocking Ab. After 6 days sups were collected for IgG/A/M ELISA and cells were stained with the appropriate Ab cocktail and analyzed for CFSE dilution and B cell phenotype on a LSRII flow cytometer (BD). Detailed conditions of assays are described in the supplemental materials.
Electron microscopy
Plates were coated with Poly-L-Lysine (Sigma) according to manufacturers protocol and 5 × 105 of freshly isolated CD14+ monocytes were infected with DENV-2 at MOI=1 or cultured with GM-CSF (1000U/106 cells, Miltenyi) and IL-4 (500U/106, Peprotech) or M-CSF (200ng/ml, Peprotech) for 3 days. Preparation of the EM slides and imaging is describe in the supplemental materials.
Statistical analyses
Analyses were done with GraphPad Prism (La Jolla, CA). For clinical samples analyses between the groups were performed by ANOVA with Bonferroni correction. For NHP analysis shows significant change relevant to the baseline (day -7), mean ± SEM, t-test. Box bars show values between the first and the 99th %, min/max. Correlation p-values were calculated for efficiency. Spearman’s rank correlation coefficient values are represented (r2). Diagonal lines indicate linear regression. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Material
Highlights.
Transcriptional response to dengue depends on viral burden and duration of illness
Expansion of CD14+CD16+ monocyte population in acute dengue infection in humans
Enhanced CD14+CD16+ monocyte numbers in lymph nodes of dengue infected macaques
Dengue infected CD14+CD16+ monocytes induce differentiation of plasmablasts
Acknowledgments
We acknowledge the expertise help of the clinical and supporting staff at the Siriraj Hospital, Bangkok, Thailand. We warmly acknowledge the exceptional assistance of Stephanie Ehnert, Elizabeth Strobert and all Yerkes NPRC personnel, for their excellent care of the animals and sample collections. We are very grateful to Jean-Francois Paré from Yerkes NPRC for the outstanding expertise in EM specimen preparation and imaging. We appreciate a great help of Mohan Maddur Satyanarayana from EVC with MDDC preparation, and Hui-Mien Hsiao from Emory University for the RT-PCR and NS-1 ELISA. We also thank Kiran Gill and Barbara Cervasi from the EVC Flow Cytometry Core for technical support.
This work was supported by grants from the NIH (R37 DK057665, R37 AI048638, U19 AI090023, U19 AI057266, U54 AI057157, N01 AI50019 and N01 AI50025) and from the Bill & Melinda Gates Foundation to) to Bali Pulendran and PS10D11132 to Yerkes.
Footnotes
Accession codes. GEO: microarray data, GSE51808
Authors declare no financial conflict of interest that might be construed to influence the results or interpretation of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, et al. Mouse STAT2 restricts early dengue virus replication. Cell host & microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annual review of immunology. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- Balakrishnan T, Bela-Ong DB, Toh YX, Flamand M, Devi S, Koh MB, Hibberd ML, Ooi EE, Low JG, Leo YS, et al. Dengue virus activates polyreactive, natural IgG B cells after primary and secondary infection. PloS one. 2011;6:e29430. doi: 10.1371/journal.pone.0029430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell host & microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonnak K, Slike BM, Burgess TH, Mason RM, Wu SJ, Sun P, Porter K, Rudiman IF, Yuwono D, Puthavathana P, et al. Role of dendritic cells in antibody-dependent enhancement of dengue virus infection. Journal of virology. 2008;82:3939–3951. doi: 10.1128/JVI.02484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch I, Xhaja K, Estevez L, Raines G, Melichar H, Warke RV, Fournier MV, Ennis FA, Rothman AL. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. Journal of virology. 2002;76:5588–5597. doi: 10.1128/JVI.76.11.5588-5597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. The American journal of tropical medicine and hygiene. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- Carrasco YR, Batista FD. B cells acquire particulate antigen in a macrophagerich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 2007;27:160–171. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, Lei HY, Lee CK, Chiou TW, Wong CH, Hsieh SL. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453:672–676. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, Puel A, Biswas SK, Moshous D, Picard C, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster JG. B cell follicles and antigen encounters of the third kind. Nature immunology. 2010;11:989–996. doi: 10.1038/ni.1946. [DOI] [PubMed] [Google Scholar]
- Devignot S, Sapet C, Duong V, Bergon A, Rihet P, Ong S, Lorn PT, Chroeung N, Ngeav S, Tolou HJ, et al. Genome-wide expression profiling deciphers host responses altered during dengue shock syndrome and reveals the role of innate immunity in severe dengue. PloS one. 2010;5:e11671. doi: 10.1371/journal.pone.0011671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376:429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bates TM, Cordeiro MT, Nascimento EJ, Smith AP, Soares de Melo KM, McBurney SP, Evans JD, Marques ET, Jr, Barratt-Boyes SM. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. Journal of immunology. 2013;190:80–87. doi: 10.4049/jimmunol.1103350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Current opinion in infectious diseases. 2006;19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- Guzman MG, Kouri G, Valdes L, Bravo J, Alvarez M, Vazques S, Delgado I, Halstead SB. Epidemiologic studies on Dengue in Santiago de Cuba, 1997. American journal of epidemiology. 2000;152:793–799. doi: 10.1093/aje/152.9.793. discussion 804. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. The Lancet infectious diseases. 2010;10:712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB, O’Rourke EJ. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. The Journal of experimental medicine. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LJ, Wang JJ, Shaio MF, Kao CL, Chang DM, Han SW, Lai JH. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. Journal of immunology. 2001;166:1499–1506. doi: 10.4049/jimmunol.166.3.1499. [DOI] [PubMed] [Google Scholar]
- Hoang LT, Lynn DJ, Henn M, Birren BW, Lennon NJ, Le PT, Duong KT, Nguyen TT, Mai LN, Farrar JJ, et al. The early whole-blood transcriptional signature of dengue virus and features associated with progression to dengue shock syndrome in Vietnamese children and young adults. Journal of virology. 2010;84:12982–12994. doi: 10.1128/JVI.01224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingersoll MA, Platt AM, Potteaux S, Randolph GJ. Monocyte trafficking in acute and chronic inflammation. Trends in immunology. 2011;32:470–477. doi: 10.1016/j.it.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H, Coquery C, Xue Y, Gayet I, Dillon SR, Punaro M, Zurawski G, Banchereau J, Pascual V, Oh S. Serum from patients with SLE instructs monocytes to promote IgG and IgA plasmablast differentiation. The Journal of experimental medicine. 2012;209:1335–1348. doi: 10.1084/jem.20111644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, Henrickson SE, Shayakhmetov DM, Di Paolo NC, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. The Journal of experimental medicine. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwissa M, Nakaya HI, Oluoch H, Pulendran B. Distinct TLR adjuvants differentially stimulate systemic and local innate immune responses in nonhuman primates. Blood. 2012;119:2044–2055. doi: 10.1182/blood-2011-10-388579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nature immunology. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke P, Hammond SN, Leung JM, Kim CC, Batra S, Rocha C, Balmaseda A, Harris E. Gene expression patterns of dengue virus-infected children from nicaragua reveal a distinct signature of increased metabolism. PLoS neglected tropical diseases. 2010;4:e710. doi: 10.1371/journal.pntd.0000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HT, Hibberd ML, Hien TT, Dung NM, Van Ngoc T, Farrar J, Wills B, Simmons CP. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. The Journal of infectious diseases. 2009;199:537–546. doi: 10.1086/596507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. Journal of virology. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay F, Schneider P. Cracking the BAFF code. Nature reviews Immunology. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- Martinez-Pomares L, Gordon S. CD169+ macrophages at the crossroads of antigen presentation. Trends in immunology. 2012;33:66–70. doi: 10.1016/j.it.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell host & microbe. 2012;12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JL, de Wet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, Gordon S. The mannose receptor mediates dengue virus infection of macrophages. PLoS pathogens. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS neglected tropical diseases. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nature reviews Immunology. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Onlamoon N, Noisakran S, Hsiao HM, Duncan A, Villinger F, Ansari AA, Perng GC. Dengue virus-induced hemorrhage in a nonhuman primate model. Blood. 2010;115:1823–1834. doi: 10.1182/blood-2009-09-242990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nature immunology. 2009;10:786–793. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper SJ, Gordon A, Liu M, Balmaseda A, Harris E, Relman DA. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLoS neglected tropical diseases. 2012;6:e1966. doi: 10.1371/journal.pntd.0001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Saha P, Geissmann F. Toward a functional characterization of blood monocytes. Immunol Cell Biol. 2011;89:2–4. doi: 10.1038/icb.2010.130. [DOI] [PubMed] [Google Scholar]
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. American journal of epidemiology. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A, Rice CM. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14610–14615. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina NV, Cherny M, Shi C, Bleau SA, Collins NH, Young JW, Pamer EG. Distinct responses of human monocyte subsets to Aspergillus fumigatus conidia. Journal of immunology. 2009;183:2678–2687. doi: 10.4049/jimmunol.0803398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CP, Farrar JJ, Nguyen v V, Wills B. Dengue. N Engl J Med. 2012;366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, Dung NT, Long TH, Hoang DM, Chau NV, Thao le TT, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. The Journal of infectious diseases. 2007;195:1097–1107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Garcia J, Comach G, Vahey MT, Wang Z, Forshey BM, Morrison AC, Sierra G, Bazan I, Rocha C, et al. Sequential waves of gene expression in patients with clinically defined dengue illnesses reveal subtle disease phases and predict disease severity. PLoS neglected tropical diseases. 2013;7:e2298. doi: 10.1371/journal.pntd.0002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. The Journal of experimental medicine. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YT, Chang SY, Lee CN, Kao CL. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cellular microbiology. 2009;11:604–615. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, Kasisith J. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. The Journal of infectious diseases. 2008;197:1459–1467. doi: 10.1086/587699. [DOI] [PubMed] [Google Scholar]
- Wong KL, Chen W, Balakrishnan T, Toh YX, Fink K, Wong SC. Susceptibility and response of human blood monocyte subsets to primary dengue virus infection. PloS one. 2012;7:e36435. doi: 10.1371/journal.pone.0036435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, Kourilsky P, Wong SC. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood. 2011;118:e16–31. doi: 10.1182/blood-2010-12-326355. [DOI] [PubMed] [Google Scholar]
- Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, Kwissa M, Pulendran B, Wilson PC, Wittawatmongkol O, et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. Journal of virology. 2012;86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, et al. Human skin Langerhans cells are targets of dengue virus infection. Nature medicine. 2000;6:816–820. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- Xu W, Joo H, Clayton S, Dullaers M, Herve MC, Blankenship D, De La Morena MT, Balderas R, Picard C, Casanova JL, et al. Macrophages induce differentiation of plasma cells through CXCL10/IP-10. The Journal of experimental medicine. 2012;209:1813–1823. S1811–1812. doi: 10.1084/jem.20112142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L, Hofer TP. Toward a refined definition of monocyte subsets. Front Immunol. 2013;4:23. doi: 10.3389/fimmu.2013.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


