Abstract
Atherosclerosis is a multi-focal disease; it is associated with arterial curvatures, asymmetries and branches/bifurcations where non-uniform arterial geometry generates patterns of blood flow that are considerably more complex than elsewhere, and are collectively referred to as disturbed flow. Such regions are predisposed to atherosclerosis and are the sites of ‘athero-susceptible’ endothelial cells that express regionally different cell phenotypes than endothelium in nearby athero-protected locations. The regulatory hierarchy of endothelial function includes control at the epigenetic level. MicroRNAs and histone modifications are established epigenetic regulators that respond to disturbed flow. However, very recent reports have linked transcriptional regulation by DNA methylation to endothelial gene expression in disturbed flow in vivo and in vitro. We outline these in the context of site-specific atherosusceptibility mediated by local hemodynamics.
Keywords: Endothelial gene expression, Hemodynamic disturbed flow, Differential methylation region, Methylome, Atherosclerosis, KLF4, HOX genes
Introduction
It is well established in vivo and in vitro that endothelial cells are highly sensitive to changes of flow/shear stress. The localization, susceptibility and initiation of atherosclerosis are intimately associated with flow-related mechanobiology and convective transport mechanisms affecting cells in the arterial wall, particularly the endothelium [1-4]. In the current issue of Vascular Pharmacology, Mao et al (5) demonstrate flow-induced changes in the expression of enzymes involved in the synthesis of gasotransmitters nitric oxide, carbon monoxide and hydrogen sulphide in cerebrovascular-derived endothelial cells in vitro. They identify phenotype similarities and differences in shear stress responses between the arteries of the brain and other vascular beds. Since the brain endothelium expresses critical morphological and functional specializations to protect the blood-brain barrier, these phenotypes highlight vascular bed-specific differences relevant to the epidemiology and mechanisms of cerebrovascular disease and its response to treatment. Phenotype identity is important because atherosclerosis of the carotid bifurcation and rupture of small arterial aneurysms are major causes of cerebral thrombosis and hemorrhagic stroke respectively, events that are strongly associated with the local hemodynamic environment. In a larger sense, however, the studies are a timely reminder of the diversity of phenotypes expressed by endothelial cells throughout the entire cardiovascular system and the potential contributions of the local flow to phenotype heterogeneity, particularly at atherosclerosis-susceptible sites in large and medium arteries. In this context, emerging epigenetics research is revealing regulatory mechanisms of flow-induced endothelial phenotype. Here we outline new evidence of a potentially important role for disturbed flow-mediated DNA methylation in the dynamic regulation of gene expression in the arterial endothelium.
Diversity of endothelial structure-function
In different vascular systems the diversity of endothelial structural specialization that has evolved to meet the functional needs of organs is largely established during embryological and early postnatal development. For example the different permeability properties of tight junctional endothelia of the blood-brain barrier, continuous endothelium in most tissue capillaries, fenestrated endothelia in the kidney and intestinal villi, and discontinuous endothelia in liver, bone marrow and spleen are all closely matched to critical tissue/organ functional requirements. However, endothelium is dynamically responsive to local flow-mediated biophysical/mechanical/chemical mechanisms (collectively mechanotransduction) [6]; indeed they are part of the global adaptive physiological homeostasis and remodeling capabilities of blood vessels. This subtle and dynamic form of endothelial phenotype diversity is prominent in atherogenesis which is associated with sites of disturbed blood flow (7-11). Atherosclerosis is associated with arterial curvatures, asymmetries, branches and bifurcations where disturbed flow is prevalent. Since traditional clinical risk factors for atherosclerosis are system-wide, whereas atherosclerotic lesions and aneurysms are regionalized, the disturbed flow environment is a contributing pre-lesional risk factor affecting endothelial phenotype. Lesion development at disturbed flow sites within coronary, carotid and other peripheral distributing arteries underlies most heart disease and stroke. Similarly, blood flow mechanics in smaller arteries of the brain plays an important role in both the initiation and rupture of intracranial aneurysms (12).
Flow characteristics predict site-specific endothelial phenotype heterogeneity
Following the immunocytochemical demonstration of inflammatory marker expression in endothelium of the inner aortic arch (disturbed flow region) of C57BL/6 mice when compared with nearby undisturbed flow locations, Cybulsky, Collins and colleagues (2000) [13] suggested that the arch endothelial phenotype may be ‘primed’ for pathological change and indeed found evidence of occasional activation of pro-inflammatory nuclear factor-kappa-B (NFκB). This was considerably extended by Passerini et al (2004)[2] who profiled the endothelial transcriptome from susceptible and protected locations in normal adult swine. They reported the coexistence of enhanced expression of pro-pathological inflammatory genes and protective genes within the same athero-susceptible cells when referenced to protected regions. Thus the adaptation of multiple pathways to a different equilibrium state in athero-susceptible cells in a disturbed flow environment permitted normal function but conferred regional susceptibility (reviews 11,14). Many investigations have since addressed disturbed flow-related endothelial phenotype identifying characteristics of the atherosusceptible state that include the chronic activation of low level endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) (15,16), important roles for localized reactive oxygen and nitrosyl species (ROS and RNS) (5,17,18), and the activation of the mitogen-activated protein kinase (19) and proinflammatory NF-κB signaling pathways (2,20).
Complementary to in vivo phenotype profiling, it is well-established that endothelial cells in vitro display dynamic gene transcriptional and translational sensitivity to changes in flow characteristics (9). A common hemodynamic element of disturbed flow in vivo is the presence of a transient flow reversal component within each cardiac cycle. When incorporated into disturbed flow experiments in vitro, the cells recapitulate morphologies and phenotype profiles that approximate the atherosusceptible endothelium in vivo (21). This general convergence permits regulatory mechanisms, including epigenetic contributions, to be investigated under controlled flow conditions in vitro and the mechanisms can be referenced to measurements of the steady state phenotype in vivo.
Epigenetic regulation of endothelial atherosusceptibility in disturbed flow
Epigenetics encompasses regulatory mechanisms by which covalent modification of the blueprint of DNA (or protein or RNA) promotes change of function without changes in the primary sequence. DNA epigenetics is a potent regulatory mechanism of transcription. Epigenetic modifications may be stable and inheritable, for example defining the specificity of cell differentiation and ensuring the fidelity of development, or they may be responses to changes in the physical and chemical environment. The principal mechanisms in DNA epigenetics are DNA methylation, histone modification and chromatin remodeling, and post-transcriptional regulation by short and long non-coding RNAs (primarily microRNAs and lncRNAs). There is a developing literature demonstrating histone modification/chromatin remodeling (22-24) and microRNA mechanisms (25-29) in the regulation of endothelial responses to fluid flow, especially disturbed flow. Very recently, however, the relative contribution of DNA methylation to transcriptional regulation is receiving scrutiny under well-defined, controlled conditions of flow disturbance (30-33), a relationship that has not previously been investigated.
DNA methylation is the methylation of the 5-carbon of cytosine residues (5-mC) in DNA usually at cytosine-phosphate-guanine dinucleotides (CpGs) sites. Clusters of dinucleotides, known as CpG islands are associated with approx 50% of gene promoters; they tend to be unmethylated, allowing transcription. Methylation of cytosines at or near the promoter region results in gene silencing by suppression of gene transcription (34) (Figure 1A). During early embryonic development/cell differentiation, methylation status is tightly regulated because it establishes properties of cell identity important for broad areas of development. The loss of DNA methylation as well as hypermethylation of growth repressors are leading epigenetic characteristics of cell proliferation in cancer (35), and changes in DNA methylation patterns have been linked with oxidative stress. Dysregulation of gene expression in various disease states such as lupus, multiple sclerosis, neuropsychiatric disorders, homocysteinemia and senescence have been linked to aberrant methylation profiles (36). DNA methylation can be evaluated in different genomic contexts of functional purposes that range from cell identity to transcriptional activation, mRNA splicing and dynamic regulation (37), including the transcriptional regulation of a limited number of genes as part of the normal management of physiological homeostasis. As the dynamic mechanisms regulating DNA methylation/demethylation have become better understood, their regulatory role in vascular physiology and transitional pathology warrants further investigation.
Figure 1. DNA methylation dynamics and gene regulation.
(A) Gene silencing by DNA hypermethylation. In vertebrates, DNA methylation occurs at carbon 5 of cytosine in CpG dinucleotides (mC). When occurring within the promoter regions of genes, it dramatically suppresses transcription by direct inhibition of transcription factor binding and recruitment of methyl-CpG-binding proteins, which further hinder access to the recognition site of transcription factors. (B) Heritable and reversible epigenomic DNA methylation. DNA methylation status is a net outcome of methylation and demethylation. DNA (cytosine-5-)-methyltransferase 1 (DNMT1) maintains DNA methylation patterns via methylation of a hemi-methylated nascent DNA strand during cell proliferation. DNMT3A and DNMT3B are required for genome-wide de novo methylation and play crucial roles in the establishment of DNA methylation patterns. Methylation by DNMTs is counterbalanced by DNA demethylation. TET (ten-eleven-translocation) has been suggested to play a central role in oxidizing 5mC to 5-hydroxymethylcytosine (hmC) and further to 5-formylcytosine (fC) and 5-carboxylcytosine (caC). Carboxyl group of caC is excised by thymine-DNA glycosylase (TDG).
Central role of DNA methyltransferases (DNMTs)
Consideration of adaptive differential methylation in response to physiological stimuli has emerged from a better understanding of the mechanisms of the cytosine methylation and demethylation cycle (Figure 1B). Three active mammalian DNA methyltransferases DNMT1, 3A and 3B promote methylation which is counterbalanced by DNA demethylation in which the TET (tet methylcytosine dioxygenase) gene pathway has been suggested to play a central role (38). DNMT1 is primarily a maintenance enzyme to ensure fidelity of methylation pattern during somatic cell division. DNMT3A and 3B are closely related and, with their molecular partners, regulate de novo DNA methylation. Another methyltransferase DNMT3L lacks a catalytic activity and is inactive but may be important for the stabilization of DNMT3A and associated assembly complexes during de novo methylation in germ cells (39). Changes in the expression, targeting and activities of DNMTs and/or TET and their associated factors may quantitatively shift the net methylation status for selective genes.
Endothelial DNA methylation and arterial hemodynamics
The remainder of this emergent topic-review outlines several recent papers (30-33) in the cardiovascular literature that in 4 species - human, swine, mouse, rat - have independently demonstrated (i) the association of differential endothelial DNA methylation with cell phenotypes in atherosusceptible and disturbed flow sites in vivo, and (ii) the plasticity of endothelial DNA methylation mechanisms in response to disturbed flow characteristics in vitro. Collectively, the studies demonstrate that DNA methylation differences are associated with differential gene expression coincident with sites susceptible to, or protected from, atherogenesis in vivo and that disturbed flow induces (primarily) hypermethylation of important transcription factors in vitro with consequences for pro-inflammatory downstream targets related to atherosusceptibility. The central role of DNMT expression and DNMT promoter enrichment resulting in hypermethylation is a recurrent finding in the studies.
(i). Differential Methylated Regions (DMRs) of the endothelial methylome in vivo
Mining the Methylome
Whole and partial genome sequencing of the endothelial DNA methylome in swine (30) and mice (31), respectively, provides a high-level view of site-specific differential methylation regions (DMRs) in endothelium in vivo without discriminating between developmentally ‘fixed’ and physiological ‘dynamic’ methylation. Comparative genome sequencing of endothelium from well-characterized arterial regions of disturbed (susceptible) and undisturbed (protected) blood flow was performed to create a DMR atlas. Although only a snapshot, the association of DMRs with specific cytosines/CpG in the promoters of important annotated genes provides insightful guidance for mechanistic experiments. Jiang et al (30) examined in detail the genome-wide DNA methylation pattern in endothelial cells from aortic arch (AA; athero-susceptible) and descending thoracic aorta (DT; athero-resistant) of adult swine by methylated DNA immunoprecipitation sequencing (MeDIP-seq). A total of 5517 DMRs were identified in somatic chromosomes with an average length of 804 ± 45 bp representing about 0.2% of the swine genome. 4019 regions were hypomethylated while 1498 regions were hypermethylated in AA relative to DT. Methylated regions and DMRs predominantly overlapped with gene- and CGI-rich regions. DMRs were enriched in the 5′UTR suggesting a functional role of DMRs in contributing to the transcriptional activity of genes. DMR-associated genes were related to ER stress and superoxide radical degradation pathways that have been identified as common features in disturbed flow athero-susceptible regions of swine arteries (16,17). Dunn et al (31) created disturbed flow by ligation of carotid arteries of mice (3) to demonstrate an upregulation of the maintenance methyltransferase DNMT1 leading to hypermethylation of a subset of potentially important molecules, including transcription factors, that were identified by cross-referencing transcriptomic and DNA methylomic datasets. Several DMRs in the promoters of the gene subset contained cyclic AMP Response Elements (CRE) sites that were hypermethylated suggesting a possible cooperative regulatory role for CRE binding protein. Using in situ immunostaining protocols in rats, Zhou et al (32) also reported increased endothelial DNMT1 expression and DNA methylation in partially ligated carotid arteries (to induce disturbed flow).
DMR and HOX genes
DMR comparisons between athero-susceptible and protected sites in normal swine methylome revealed a sharp distinction in the HOX locus (31), which is associated with previously identified differentially expressed HOX genes, including HOXA4/5/7/10/11, HOXB5/6/7/13, HOXD10 (2) and the regulatory microRNA mir-10a/b (26). HOX transcriptional factors, master regulators of body patterning (40) have been reported to specify positional identities in arterial blood vessels (41,42). The HOX genes regulate endothelial cell proliferation, migration, differentiation, morphogenesis and permeability during development and vascular remodeling in adults. HOXA3/9, HOXB3/5 and HOXD3 regulate endothelial cell activation, whereas HOXA5 and HOXD10 sustain quiescent endothelial phenotype (41,43,44). HOXA3 and HOXD3, which promote a proliferative and migrative phenotype, are induced in early phase of endothelial differentiation; HOXA5 and HOXD10, which maintain a quiescent EC phenotype, are increased during the maturation of endothelial cells (41). A link between DMR and HoxA5 gene expression in vivo was reported by Dunn et al (31) who studied DNA methylation patterns in endothelium harvested from apoE-/- mice carotid artery. Disturbed flow was created in the left common carotid artery by ligation, the contralateral artery serving as control. DNMT1 expression was induced by disturbed flow and was rescued by the DNMT inhibitor 5-Aza-2-deoxycytidine (5Aza). Of 11 mechanosensitive genes whose promoters were hypermethylated under disturbed flow conditions, HoxA5 was robustly silenced by DNA methylation and rescued by 5-aza. Thus these separate studies identified endothelial Hox genes as DMR-sensitive transcription targets associated with site-specific disturbed flow in vivo. The role of DNA methylation in regulating endothelial HOX gene expression is likely of high relevance for understanding the mechanisms underlying regional physiological and phenotype diversities in the cardiovascular system.
(ii). Mechanisms: DNA methylation reductionist experiments under controlled flow conditions in vitro
Changes in DNMT1 and DNMT3A by disturbed flow in vitro
The application of different flow characteristics to cultured human arterial and umbilical vein endothelial cells (HAEC and HUVEC respectively) allows mechanisms of disturbed and undisturbed flow to be addressed under controlled and accessible conditions. Flow waveforms that capture the dominant characteristics of human arterial hemodynamics were generated in vitro. All flow in large arteries is unsteady (pulsatile). The defining feature of disturbed flow regions is the presence of a flow reversal phase during the cardiac cycle – sometimes referred to as an oscillating shear index OSI - whereas in undisturbed flow the blood accelerates and decelerates while constantly traveling in the antegrade direction. Dunn et al (31) reported that HUVEC DNMT1 transcript and protein expressions were significantly upregulated by disturbed flow and monocyte adhesion, a functional in vitro assay for inflammation, was significantly enhanced. Inhibition of DNMTs by 5-Aza and siRNA (directed to DNMT1) inhibited monocyte adhesion to HUVEC. Other DNMTs were unchanged by disturbed flow. Zhou et al (32) also exposed HUVEC to disturbed flow; DNMT1 mRNA was increased 1.9-fold and immunocytochemistry revealed enhanced nuclear localization of DNMT1 in disturbed flow. Immuno-slot blot indicated an overall increased methylation by disturbed flow at 24h. In contrast, in HAEC subjected to disturbed and undisturbed flow waveforms for 48h, no significant changes in mRNA expression of the methyltransferases DNMT1, 3A, and 3B, nor enzymes involved in cytosine demethylation – TET1, 2, 3; TDG1; GADD45B; MBD4; SMUG1 - were induced by disturbed flow as determined by qPCR (33); DNMT3L expression was undetectable. However, a significant increase (1.7-fold) was detected in DNMT3A protein. An increase in DNMT3A protein without changing the mRNA levels may suggest a potential post-transcriptional and/or post-translational mechanism (eg. sumoylation) (45) in regulating DNMT3A by flow. These global measurements are useful in linking methylome data to controlled flow conditions and both approaches are important for identifying specific gene subsets that may play a dominant role in site-specific phenotype regulation. However, once specific genes are identified to be methylated by disturbed flow, it is necessary to conduct detailed studies assessing the methylation status of individual CpG sites in the promoter and gene body sequences. This approach has been taken for the important endothelial transcription factor Kruppel-like Factor 4 (KLF4) (33)
DMR and Kruppel-like factor 4
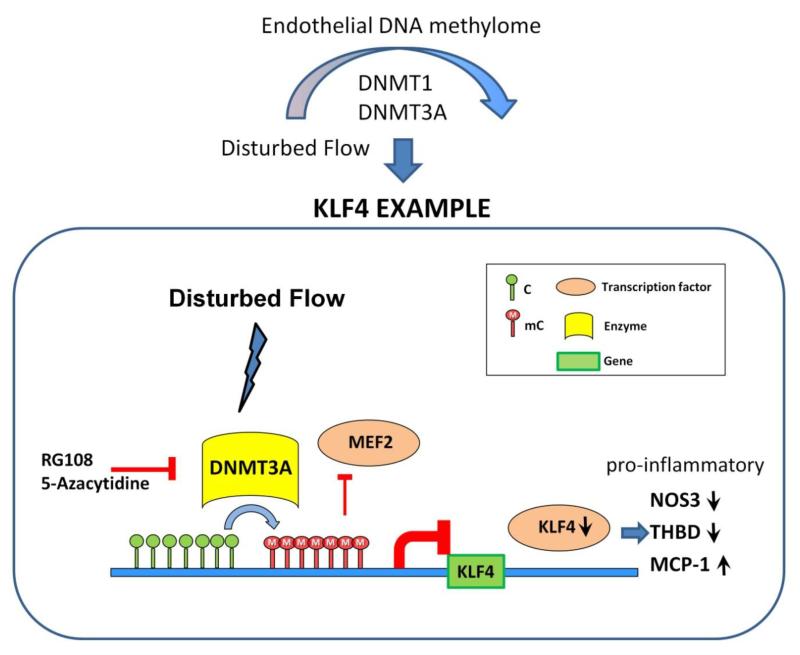
KLF4 is a member of the zinc-finger regulatory transcription factor family; it targets gene networks that confer atheroprotective (46), anti-inflammatory (47) and anti-thrombotic (48) properties to the endothelium. Localized dysfunction and suppression of KLF4 are therefore pro-pathological. Since nitric oxide synthase 3 (NOS3; endothelial NOS), monocyte chemoattractant protein-1 (MCP-1), and thrombomodulin (THBD) are downstream targets of KLF4, its suppression in endothelium may have important downstream pro/anti-inflammatory consequences. MeDIP studies identified an endothelial DMR site close to the promoter of KLF4 which was hypermethylated in the atherosusceptible aortic arch of swine. Exposure of HAEC to disturbed flow in vitro for 48h showed regional hypermethylation of the human KLF4 promoter (33). Differential CpG site methylation was measured by methylation specific PCR (MSP), bisulfite pyrosequencing and methylation-sensitive restriction enzyme-PCR. Disturbed flow increased DNA methylation of CpG islands within the KLF4 promoter that significantly contributed to suppression of KLF4 transcription. The DNMT inhibitors 5-Aza and the longer-lived RG108 completely prevented disturbed flow-induced methylation of the KLF4 promoter extending 1.5kb upstream of the transcription start site (TSS). As noted above, disturbed flow induced a modest increase of DNMT3A protein in HAEC. However, when chromatin loading of DNMT3A enzyme at the KLF4 promoter was examined by ChIP-PCR assay, a specific 11-fold DNMT3A enrichment of the KLF4 promoter was identified in disturbed flow; in contrast, TET1 enrichment was equivalent in disturbed and undisturbed flow.
The human KLF4 promoter contains a single myocyte enhancer factor-2 (MEF2) binding site. To test if hemodynamic forces could regulate the methylation status of CpG in and near the MEF2 binding sequence, bisulfite pyrosequencing was used to quantify the methylation levels at individual CpG sites. Disturbed flow was shown by MSP analysis to further enhance CpG methylation close to the MEF2 binding sequence (33). Consistent with robust enhanced methylation near the MEF2 binding site, disturbed flow reduced the chromatin loading of MEF2 protein to the KLF4 promoter by 80%, confirming that MEF2-enhanced KLF4 gene transcription is impeded by methylation of the KLF4 promoter. Sustained knock-down with shDNMT3A blocked methylation of the MEF2 region demonstrating DNMT3A specificity. Thus competitive inhibition by DNMT3A-mediated hypermethylation of a MEF2 binding region in the KLF4 promoter near the TSS significantly contributes to transcriptional suppression of KLF4 in disturbed flow. Figure 2 includes a general outline of this mechanism.
Figure 2. Hemodynamic forces and endothelial DNA methylome.
Disturbed flow (DF) globally and gene-specifically influences endothelial DNA methylation via DNMT1 [31,32] and DNMT3A [33] pathways. A mechanism regulating KLF4 promoter methylation that contributes to suppression of transcription (and described in the text) is illustrated. DF-induced DNMT3A enrichment of endothelial KLF4 promoter near the TSS increased CpG methylation. The resulting hypermethylation of KLF4 promoter induced gene silencing by preventing the chromatin binding of MEF2 to the KLF4 promoter. Decreased KLF4 expression inhibited its transcription targets Thrombomodulin (THBD) and Nitric Oxide Synthase 3 (NOS3) and de-repressed expression of Monocyte Chemoattractant Factor-1 (MCP-1) leading to a pro-inflammatory, pro-atherosclerosis phenotype. Intervention by DNMT inhibitors (RG108; 5-Azacytidine) rescued this pathway. Adapted from Reference 33 with permission.
The above mechanisms have downstream consequences for KLF4 transcription targets. Disturbed flow-induced KLF4 hypermethylation in HAEC inhibited the athero-protective expression of NOS3, THBD and upregulated pro-inflammatory MCP-1 (which is normally inhibited by KLF4) (33). All 3 genes are KLF4-targets and appear to be resistant to changes of DNA methylation For example, the resilience of the human NOS3 gene to hypermethylation is attributed to its hypomethylated status being a critical determinant of endothelial cell identity established during development (50). The expression of such genes and their physiological expression may be more readily regulated by pleiotropic transcription factors such as KLF4 that are sensitive to transcriptional regulation by DNA methylation, in this case by hemodynamic characteristics.
How common is flow-related DNA methylation plasticity in influencing endothelial phenotype?
Genome-wide MeDIP studies identified over 5000 DMRs in swine atherosusceptible sites (30). However, it is unclear whether the DNA methylation status of most genes is responsive to changes in the cellular environment, whether it is a widespread mechanism of physiological regulation, or whether the phenomenon is readily reversible and /or inheritable. The vast majority of DMRs may represent a ‘fixed’ methylation status determined during embryological (and ongoing stem cell) development. To date, only a small number of gene subsets responsive to hemodynamic characteristics have been identified from in vivo manipulations and MeDIP studies of the methylome (30-33).
How do hemodynamic characteristics modify endothelial DNA methylation?
The ability of flow to induce changes of endothelial DNA methylation patterns sheds new light on how biophysical forces and/or the flow-dependent transport of metabolites locally influence gene expression and disease susceptibility. Together with other epigenetic mechanisms that relate to endothelial flow responses, e.g. chromatin remodeling and miRNA regulation, DNA methylation appears to contribute to genome-environment mechanisms important in the spatial distribution of endothelial phenotypes. For transcription factors such as KLF4 and KLF2, previous studies demonstrated their flow-related regulation by miR-92a (28,29) and by histone modification (23). Hox genes are also targets of multiple miRNAs including flow-sensitive miR10a (26). Thus multiple mechanisms involving trans- and cis-acting elements (transcription factors and DNA methylation respectively) coordinate endothelial transcriptional regulation in response to disturbed flow. Left out of this consideration of gene expression is a complex hierarchy of post-translational modifications that have yet to be evaluated in flow-related endothelial phenotype regulation. They include phosphorylation, ubiquitination, ribosylation, fatty acylation, glycosylation and SUMOylation (35).
The ability of the endothelium to respond to disturbed flow by sensing different hemodynamic forces may reside in subcellular spatio-temporal mechanotransduction criteria transmitted throughout the cell by the cytoskeleton and converted to chemical/metabolic responses (mechanotransduction) at multiple decentralized subcellular locations (6,50). An important additional consideration is that convective transport in disturbed flow is radically different than in undisturbed flow; flow disturbance is created by flow separation to form vortices of average low flow velocity (and low shear stress) within which solute and particle retention times are significantly longer than in undisturbed flow. The arterial wall is constantly producing ROS and other potent intermediary metabolites. Active peptides and hormones generated from precursors in the blood are rapidly metabolized at the vessel surface. In the disturbed flow region of the swine aortic arch the concentrations of these molecules may be much higher than in nearby undisturbed flow regions (51) creating an unbalanced local environment near the cell surface that may influence DNA methylation. In support of this mechanism, oxidative damage is known to target assembly complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands (36). Since the NFkB pathway is also more active in disturbed flow (2) and Rel/p65 is able to recruit DNMTs to specific genome loci (52), these pathways may influence DNMT enrichment.
In summary, endothelial phenotypes exhibit spatial and temporal diversity throughout the arterial circulation. A case is presented from newly emerging studies that epigenetic DNA methylation is responsive to local arterial flow characteristics and can influence gene expression through transcriptional regulation, primarily in disturbed flow-specific gene silencing through hypermethylation of CpG sites at or close to promoter regions. The mechanisms are highly DNMT-dependent, may differ in detail from gene to gene and their frequency is yet to be determined. The studies provocatively suggest a physiological regulatory role for DNA methylation in addition to its well-studied functions in development and differentiation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Davies PF. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nature Clin Prac Cardiovasc Med. 2009;6:16–26. doi: 10.1038/ncpcardio1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Passerini AG, Polacek DC, Shi C, Francesco NM, Manduchi E, Grant GR, et al. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proc Natl Acad Sci U S A. 2004;101:2482–2487. doi: 10.1073/pnas.0305938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni CW, Qiu H, Rezvan A, Kwon K, Nam D, Son DJ, et al. Discovery of novel mechanosensitive genes in vivo using mouse carotid artery endothelium exposed to disturbed flow. Blood. 2010;116:e66–73. doi: 10.1182/blood-2010-04-278192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng C, van Haperen R, de Waard M, van Damme LC, Tempel D, Hanemaaijer L, et al. Shear stress affects the intracellular distribution of eNOS: direct demonstration by a novel in vivo technique. Blood. 2005;106:3691–3698. doi: 10.1182/blood-2005-06-2326. [DOI] [PubMed] [Google Scholar]
- 5.Mao XO, Xie L, Greenberg R, Greenberg J, Peng B, Mieling I, et al. Flow-induced regulation of brain endothelial cells in vitro. Vascul Pharmacol. 2014 doi: 10.1016/j.vph.2014.02.003. Editor to complete the citation: Manuscript Number: VPH-D-14-00006R1. [DOI] [PubMed] [Google Scholar]
- 6.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ku DN, Giddens DP, Zarins CK, Glagov S. Pulsatile flow and atherosclerosis in the human carotid bifurcation. Positive correlation between plaque location and low oscillating shear stress. Arteriosclerosis. 1985;5:293–302. doi: 10.1161/01.atv.5.3.293. [DOI] [PubMed] [Google Scholar]
- 8.Davies PF, Polacek DC, Handen JS, Helmke BP, DePaola N. A spatial approach to gene expression profiling: mechanotransduction and the focal origin of atherosclerosis. Trends Biotechnol. 1999;17:347–351. doi: 10.1016/s0167-7799(99)01348-7. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Cardena G, Comander J, Anderson KR, Blackman BR, Gimbrone MA. Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proc Natl Acad Sci USA. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nerem RM, Levesque MJ. Fluid dynamics as a factor in the localization of atherogenesis. Ann NY Acad Sci. 1983;416:709–719. doi: 10.1111/j.1749-6632.1983.tb35222.x. [DOI] [PubMed] [Google Scholar]
- 11.Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol. Rev. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shojima M, Oshima M, Takagi K, Torii R, Hayakawa M, Katada K, et al. Magnitude and role of wall shear stress on cerebral aneurysm: computational fluid dynamic study of 20 middle cerebral artery aneurysms. Stroke. 2004;35:2500–2505. doi: 10.1161/01.STR.0000144648.89172.0f. [DOI] [PubMed] [Google Scholar]
- 13.Hajra L, Evans AI, Chen M, Hyduk SJ, Collins T, Cybulsky MI. The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc Natl Acad Sci USA. 2000;97:9052–9057. doi: 10.1073/pnas.97.16.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies PF, Civelek M, Fang Y, Fleming I. The atherosusceptible endothelium: Endothelial phenotypes in complex hemodynamic shear stress regions in vivo. Cardiovasc Res. 2013;99:315–327. doi: 10.1093/cvr/cvt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gargalovic PS, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, et al. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:2490–2496. doi: 10.1161/01.ATV.0000242903.41158.a1. [DOI] [PubMed] [Google Scholar]
- 16.Civelek M, Manduchi E, Riley R, Stoeckert CJ, Davies PF. Chronic endoplasmic reticulum (ER)-stress activates unfolded protein response (UPR) in arterial endothelium in regions of susceptibility to atherosclerosis. Circ Res. 2009;105:453–461. doi: 10.1161/CIRCRESAHA.109.203711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies PF, Civelek M. Endoplasmic reticulum stress, redox, and a proinflammatory environment in athero-susceptible endothelium in vivo at sites of complex hemodynamic shear stress. Antioxid Redox Signal. 2011;15:1427–32. doi: 10.1089/ars.2010.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oemar BS, Tschudi MR, Godoy N, Brovkovich V, Malinski T, Luscher TF. Reduced endothelial nitric oxide synthase expression and production in human atherosclerosis. Circulation. 1998;97:2494–2498. doi: 10.1161/01.cir.97.25.2494. [DOI] [PubMed] [Google Scholar]
- 19.Hoefen RJ, Berk BC. The role of MAP kinases in endothelial activation. Vascul Pharmacol. 2002;38:271–273. doi: 10.1016/s1537-1891(02)00251-3. [DOI] [PubMed] [Google Scholar]
- 20.Enesa K, Zakkar M, Chaudhury H, Luong le A, Rawlinson L, Mason JC, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283:7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 21.De Keulenaer GW, Chappell DC, Ishizaka N, Nerem RM, Alexander RW, Griendling KK. Oscillatory and steady laminar shear stress differentially affect human endothelial redox state: role of a superoxide-producing NADH oxidase. Circ Res. 1998;82:1094–1101. doi: 10.1161/01.res.82.10.1094. [DOI] [PubMed] [Google Scholar]
- 22.Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res. 2003;93:155–161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Ha CH, Jhun BS, Wong C, Jain MK, Jin ZG. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood. 2010;115:2971–2979. doi: 10.1182/blood-2009-05-224824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee DY, Lee CI, Lin TE, Lim SH, Zhou J, Tseng YC, et al. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc Natl Acad Sci USA. 2012;109:1967–1972. doi: 10.1073/pnas.1121214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci USA. 2011;108:10355–10360. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Jr., Wang KC, Geary GG, et al. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation. 2011;124:633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Y, Davies PF. Site-Specific MicroRNA-92a Regulation of Kruppel-Like Factors 4 and 2 in Atherosusceptible Endothelium. Arterioscler Thromb Vasc Biol. 2012;32:979–987. doi: 10.1161/ATVBAHA.111.244053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y-Z, Manduchi E, Stoeckert CJ, Davies PF. Epigenomic code in arterial endothelial methylome: Differential DNA methylation patterns of athero-susceptible and protected endothelial phenotypes in vivo. Circulation Cardiovascular Genetics. 2014 In press. [Google Scholar]
- 31.Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, et al. Flow alters genome-wide methylation, regulating gene expression and atherosclerosis. J Clin Invest. 2014 doi: 10.1172/JCI74792. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Li Y-S, Wang K-C, Chien S. Epigenetic mechanism in regulation of endothelial function by disturbed flow: Induction of DNA hypermethylation by DNMT1. Cellular Molecular Bioeng. 2014 doi: 10.1007/s12195-014-0325-z. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y-Z, Jiménez JM, Ou K, McCormick ME, Davies PF. Differential DNA methylation of endothelial Kruppel-like Factor 4 (KLF4) promoter in response to hemodynamic disturbed flow in vitro and in vivo. Circulation Research. 20142014 doi: 10.1161/CIRCRESAHA.115.303883. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 35.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Reports. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Rev Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 38.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of DNMT3A bound to DNMT3L suggests a model for de novo DNA methylation. Nature. 449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 41.Bahrami SB, Veiseh M, Dunn AA, Boudreau NJ. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adh Migr. 2011;5:133–141. doi: 10.4161/cam.5.2.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruett ND, Visconti RP, Jacobs DF, Scholz D, McQuinn T, Sundberg JP, Awgulewitsch A. Evidence for Hox-specified positional identities in adult vasculature. BMC Dev Biol. 2008;8:93. doi: 10.1186/1471-213X-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rössig L, Urbich C, Brühl T, Dernbach E, Heeschen C, Chavakis E, et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douville JM, Wigle JT. Regulation and function of homeodomain proteins in the embryonic and adult vascular systems. Can J Physiol Pharmacol. 2007;85:55–65. doi: 10.1139/y06-091. [DOI] [PubMed] [Google Scholar]
- 45.Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32:598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villarreal G, Jr, Zhang Y, Larman HB, Gracia-Sancho J, Koo A, García-Cardeña G. Defining the regulation of KLF4 expression and its downstream transcriptional targets in vascular endothelial cells. Biochem Biophys Res Comm. 2010;391:984–989. doi: 10.1016/j.bbrc.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, et al. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 48.Zhou G, Hamik A, Nayak L, Tian H, Shi H, Lu Y, et al. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J Clin Invest. 2012;122:4727–4731. doi: 10.1172/JCI66056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan Y, Fish JE, D’Abreo C, Lin S, Robb GB, Teichert AM, et al. The cell-specific expression of endothelial nitric oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 50.Davies PF, Helmke BP. Endothelial mechanotransduction. In: Mofrad MRK, Kamm RD, editors. Cellular Mechanotransduction. Cambridge Univ. Press; London: 2009. [Google Scholar]
- 51.Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Davies PF. Coronary artery endothelial transcriptome in vivo: identification of endoplasmic reticulum stress and enhanced reactive oxygen species by gene connectivity network analysis. Circulation Cardiovasc Genet. 2011;4:243–252. doi: 10.1161/CIRCGENETICS.110.958926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y1, Mayo MW, Nagji AS, Smith PW, Ramsey CS, Li D, Jones DR. Phosphorylation of RelA/p65 promotes DNMT-1 recruitment to chromatin and represses transcription of the tumor metastasis suppressor gene BRMS1. Oncogene. 2012;31:1143–1154. doi: 10.1038/onc.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]