Abstract
Target of rapamycin (TOR) signaling is a nutrient-sensing pathway controlling metabolism and lifespan. Although TOR signaling can be activated by a metabolite of diacylglycerol (DAG), phosphatidic acid (PA), the precise genetic mechanism through which DAG metabolism influences lifespan remains unknown. DAG is metabolized to either PA via the action of DAG kinase or 2-arachidonoyl-sn-glycerol by diacylglycerol lipase (DAGL). Here, we report that in Drosophila and Caenorhabditis elegans, overexpression of diacylglycerol lipase (DAGL/inaE/dagl-1) or knockdown of diacylglycerol kinase (DGK/rdgA/dgk-5) extends lifespan and enhances response to oxidative stress. Phosphorylated S6 kinase (p-S6K) levels are reduced following these manipulations, implying the involvement of TOR signaling. Conversely, DAGL/inaE/dagl-1 mutants exhibit shortened lifespan, reduced tolerance to oxidative stress, and elevated levels of p-S6K. Additional results from genetic interaction studies are consistent with the hypothesis that DAG metabolism interacts with TOR and S6K signaling to affect longevity and oxidative stress resistance. These findings highlight conserved metabolic and genetic pathways that regulate aging.
Keywords: aging, diacylglycerol, diacylglycerol kinase, metabolism, phosphatidic acid, S6 kinase
Introduction
Longevity is regulated by conserved signaling pathways that modulate aging-associated stress responses (Haigis & Yankner, 2010; Lapierre & Hansen, 2012). The insulin/IGF-1 (IIS) and target of rapamycin (TOR) signaling pathways have been implicated in aging in diverse organisms including yeast, flies, worms, and mammals (Kapahi et al., 2010; Kenyon, 2010). TOR is a widely conserved serine/threonine kinase which acts as a nutrient sensor to regulate cell growth, translational control, ribosome biogenesis, autophagy, and metabolism (Wullschleger et al., 2006; Stanfel et al., 2009; Alic & Partridge, 2011; Zoncu et al., 2011). Reduction of TOR activity extends lifespan in many species (Vellai et al., 2003; Jia et al., 2004; Kapahi et al., 2004; Kaeberlein et al., 2005; Hansen et al., 2007; Harrison et al., 2009; Selman et al., 2009). Reduction in the activity of S6 protein kinase, a downstream signaling component in the TOR pathway, also leads to lifespan extension and resistance to age-related pathologies in mice (Selman et al., 2009), while the activation of Rheb-TOR signaling activity reduces oxidative stress tolerance and hastens emergence of age-related phenotypes in Drosophila (Patel & Tamanoi, 2006).
Diacylglycerol (DAG) is an important lipid metabolic intermediate involved in complex signaling pathways (Carrasco & Merida, 2007). DAG can be hydrolyzed by DAG lipase (DAGL) to become 2-arachidonoyl-sn-glycerol (2-AG) or modified by DAG kinase (DGK) resulting in its conversion to phophatidic acid (PA) for phosphoinositide turnover (Cai et al., 2009; Raghu & Hardie, 2009). PA, as well as the attenuation DAG levels in the cell membrane, affects numerous intracellular signaling pathways, including those regulating cell growth, differentiation, and membrane trafficking (Merida et al., 2008). PA can bind to mammalian TOR (mTOR) and promote mTORC1 and mTORC2 formation, which in turn induce the TOR signaling pathway (Toschi et al., 2009; Foster, 2013) and lead to elevated phosphorylation levels of S6K and 4EBP (Fang et al., 2001).
Previously, we have shown that a multi-stress screening strategy can be used to identify genes or mutants involved in the regulation of longevity (Wang et al., 2004, 2012; Liu et al., 2009). Here, we report the characterization of one such gene, identified in a Drosophila multi-stress resistant strain DAGL/inaEEP1101. This EP-element generated line is long-lived and resistant to oxidative stress. DAGL/inaEEP1101 shows upregulation of DAGL/inaE, a homolog of diacylglycerol lipase, and reduced levels of phosphorylated S6 kinase (p-S6K), consistent with the hypothesis that DAGL/inaE up-regulation causes a reduction in TOR signaling. Conversely, a second mutant with reduced DAGL/inaE expression, DAGL/inaEKG08585, displays shortened lifespan, reduced tolerance to oxidative stress and elevated levels of p-S6K. Genetic manipulation of DAGL/inaE, rdgA, or S6KKQ (a dominant-negative form of S6 kinase) also suggest that reduced TOR signaling mediates the effects of DAGL/inaE overexpression on lifespan and stress resistance. Using Caenorhabditis elegans, we show that, as in flies, the nematode ortholog of DAGL/inaE, F42G9.6 (herein named dagl-1), also regulates lifespan and oxidative stress response via TOR. We propose that DAGL/inaE and DGK regulate competing branches of pathways that metabolize DAG, ultimately resulting in altered PA levels, which in turn modulate TOR signaling. Collectively, our results show the modulation of longevity and oxidative stress response through conserved pathways that alter TOR signaling in Drosophila and C. elegans.
Results
Diacylglycerol lipase regulates longevity and oxidative stress response in Drosophila
In a screen for long-lived mutants with enhanced resistance to simultaneous oxidative stress and starvation, we identified an EP-element insertion mutant DAGL/inaEEP1101 with a 66% increase (P < 0.001) in mean survival time compared to that of the control fly w1118 (Fig. S1, Supporting information). The outcrossed DAGL/inaEEP1101 line was 72% longer lived than the control (Fig. 1A) and was similarly more resistant to oxidative stress induced by paraquat (Fig. 1B). To identify the target gene in DAGL/inaEEP1101 responsible for lifespan extension and stress resistance, we performed a plasmid rescue and verified that a single EP-element insertion was present in the 5′ un-translated region of DAGL/inaE. The EP-element insertion in DAGL/inaEEP1101 disrupts the binding site of a transcriptional repressor Tailless (Gui et al., 2011). Semi-quantitative RT-PCR analysis revealed a threefold increase of DAGL/inaE mRNA levels in DAGL/inaEEP1101 compared with the control (Fig. 1C). DAGL/inaE encodes diacylglycerol lipase (DAGL), which metabolizes DAG to 2-AG (Leung et al., 2008). Since increased DAGL/inaE expression extends lifespan and enhances resistance to oxidative stress (Fig. 1A–C), we asked whether DAGL/inaEKG08585, a mutant with reduced DAGL/inaE expression (Fig. 1G), would have the opposite phenotypes. As expected, DAGL/inaEKG08585 exhibited a 50% decrease (P < 0.001) in mean lifespan and a 34% reduction (P < 0.001) in mean survival time on oxidative stress compared to w1118 (Fig. 1E,F). Together, the results suggest that DAGL/inaE regulates lifespan and oxidative stress resistance in Drosophila.
Figure 1.
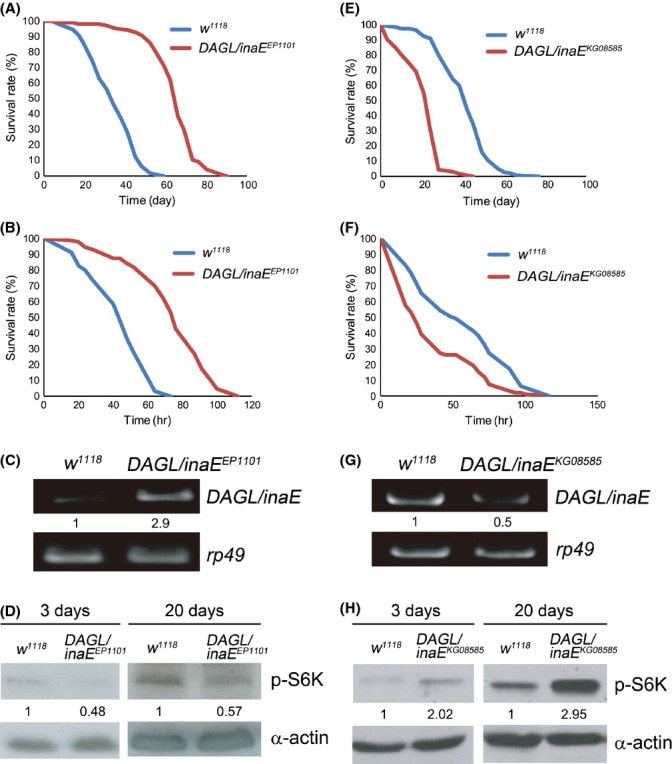
DAGL/inaE expression regulates lifespan and oxidative stress response, and negatively correlates with phosphorylated S6 kinase (p-S6K) levels in Drosophila (A–D). The Drosophila DAGL/inaEEP1101 mutant, which exhibits up-regulation of DAGL/inaE, shows extended lifespan, enhanced oxidative stress response and reduced levels of p-S6K. (A) Lifespan of DAGL/inaEEP1101 (mean = 64 d, n = 306, red line) and w1118 (mean = 37 d, n = 273, blue line). (B) Survival under 10 mm paraquat-induced oxidative stress of DAGL/inaEEP1101 (mean = 74 h, n = 87, red line) and w1118 (mean = 44 h, n = 69, blue line). (C) RT-PCR analysis shows a nearly 200% increase in DAGL/inaE levels in DAGL/inaEEP1101 compared to that of w1118. (D) Levels of p-S6K are decreased in DAGL/inaEEP1101 compared to that of w1118 in 3-d and 20-d old flies. (E–H) The DAGL/inaEKG08585 mutant, which exhibits down-regulation of DAGL/inaE, shows shortened lifespan, reduced oxidative stress resistance and increased levels of p-S6K. (E) Lifespan of DAGL/inaEKG08585 (mean = 21 d, n = 254, red line) and w1118 (mean = 41 d, n = 300, blue line). (F) Survival under 10 mm paraquat-induced oxidative stress of DAGL/inaEKG08585 (mean = 37 h, n = 100, red line) and w1118 (mean = 55 h, n = 100, blue line). (G) RT-PCR analysis shows a 50% decrease in DAGL/inaE levels in DAGL/inaEKG08585 compared to that of w1118. RT-PCR results are normalized to rp49 as an internal control (C, G). (H) Levels of p-S6K are increased in DAGL/inaEKG08585 compared to that of w1118 in 3-d and 20-d old flies. α-actin was used as an internal control (D, H).
Overexpression of DAGL/inaE and knockdown of rdgA similarly extend lifespan
To determine whether overexpression of DAGL/inaE is sufficient to extend lifespan and increase oxidative stress resistance, we generated transgenic flies expressing either the 2214-nt long isoform DAGL/inaE-PD cDNA (UAS-DAGL/inaE-PD) or the 1935-nt short isoform DAGL/inaE-PA cDNA (UAS-DAGL/inaE-PA). Since DAGL/inaE expresses mainly in adult fly brain, eye, and thoracic-abdominal ganglion according to the data from FlyAtlas (Chintapalli et al., 2007), thus we used GMR-Gal4 (eye and thoracic-abdominal ganglion Gal4 driver), Appl-Gal4 (neuronal Gal4 driver), hs-Gal4, and da-Gal4 (ubiquitous Gal4 drivers) to express either UAS-DAGL/inaE-PD or UAS-DAGL/inaE-PA to determine if overexpression of DAGL/inaE would also enhance lifespan and oxidative stress response. In all cases, expression of either UAS-DAGL/inaE-PD or UAS-DAGL/inaE-PA by those drivers extended mean lifespan (Table S1, Supporting information) and enhanced oxidative stress resistance (Table S2, Supporting information). These results suggest that neurons are a target tissue for lifespan extension and oxidative stress resistance by DAGL/inaE overexpression. Since overexpression of both isoforms resulted in similar outcomes, we used only UAS-DAGL/inaE-PD in all subsequent experiments and hereafter refer to it as UAS-DAGL/inaE.
Diacylglycerol can be converted to 2-AG by DAGL or metabolized to form phosphatidic acid (PA) by DAG kinase (encoded by retinal degeneration A (rdgA) in Drosophila (Hardie, 2003). In mammalian systems PA is reported to activate target of rapamycin (mTOR) kinase resulting in elevated levels of 4EBP and phosphorylated S6K (Fang et al., 2001). Thus, we hypothesize that the enhanced longevity of DAGL/inaEEP1101 resulted from reduced TOR signaling, since DAGL/inaE overexpression shunts more DAG into 2-AG and it should also lower PA levels (Fig. S2, Supporting information). To examine this possibility, we measured the levels of phosphorylated S6 kinase (p-S6K), a downstream molecular marker of TOR signaling. Levels of p-S6K were reduced by 50% and 40% in young and old DAGL/inaEEP1101 flies, respectively, relative to levels in w1118 (Fig. 1D). Conversely, in the short-lived DAGL/inaEKG08585 p-S6K levels were elevated by 1.5- and threefold in young and old flies, respectively (Fig. 1H).
If longevity resulting from DAGL/inaE overexpression is due to reduced PA formation and TOR signaling, then the knockdown of rdgA (DAG kinase) should produce similar phenotypes. Overexpression of DAGL/inaE resulted in a 41% increase (P < 0.001) in mean lifespan compared to Gal4 alone and 16% (P < 0.001) compared to UAS alone (Fig. 2A). Knockdown of rdgA also significantly increases mean lifespan by 44% (P < 0.001) compared to Gal4 alone or 12% (P < 0.01) compared to UAS alone (Fig. 2B). Simultaneous DAGL/inaE overexpression and rdgA knockdown did not further extend lifespan of that achieved by either manipulation independently (Fig. 2C). Similar to DAGL/inaE overexpression, rdgA mutants rdgABL33306 and rdgABL20320 also displayed an increase of 53% (P < 0.001) and 48% (P < 0.001) in mean lifespan and 43% reduction for both in p-S6K levels compared to those in control w1118 (Fig. S3A,B, Supporting information). Together, these results are consistent with the idea that DAGL/inaE and rdgA modulate lifespan via a common pathway.
Figure 2.
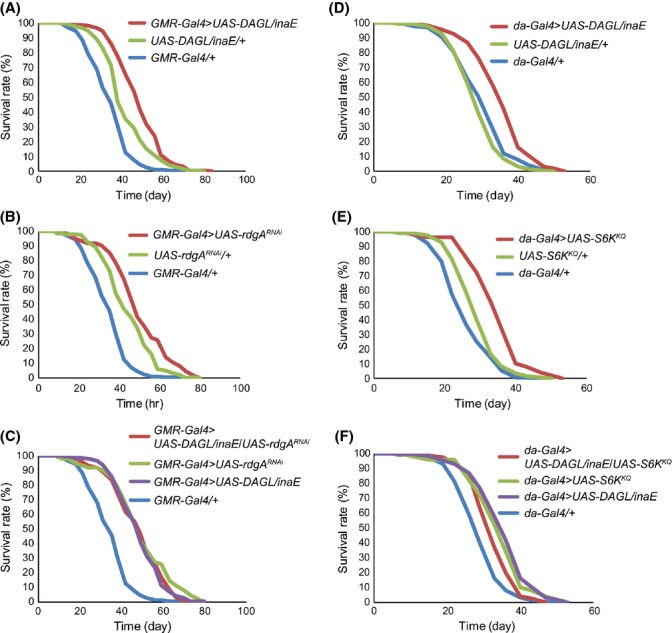
Genetic interaction between DAGL/inaE, rdgA, and S6K on lifespan. (A) Overexpression of DAGL/inaE driven by GMR-Gal4 (GMR-Gal4 > UAS-DAGL/inaE, mean = 49 d, n = 206) extends lifespan compared to controls harboring either genetic element alone (GMR-Gal4/+, mean = 34 d, n = 258; UAS-DAGL/inaE/+, mean = 42 d, n = 292). (B) Knockdown of rdgA by RNAi (GMR-Gal4>UAS-rdgARNAi, mean = 50 d, n = 62) also extends lifespan compared to controls (GMR-Gal4/+, mean = 34 d, n = 258; UAS-rdgARNAi/+, mean = 44, n = 173). (C) Lifespan extension by simultaneous overexpression of DAGL/inaE and knockdown of rdgA (GMR-Gal4>UAS-DAGL/inaE /UAS-rdgARNAi, mean = 48 d, n = 203) is similar to either manipulation alone (GMR-Gal4>UAS-DAGL/inaE, mean = 49 d, n = 206; GMR-Gal4>UAS-rdgARNAi, mean = 50 d, n = 62). (D) Overexpression of DAGL/inaE driven by da-Gal4 (da-Gal4 > UAS-DAGL/inaE, mean = 36 d, n = 210) extends lifespan compared to controls (da-Gal4/+, mean = 29 d, n = 209; UAS-DAGL/inaE/+, mean = 30 d, n = 232) at 29 °C. (E) Overexpression of S6KKQ, a dominant-negative form of S6K, (da-Gal4 > UAS-S6KKQ, mean = 35 d, n = 50) also extends lifespan compared to controls (da-Gal4/+, mean = 29 d, n = 209; UAS-S6KKQ/+, mean = 26 d, n = 129) at 29 °C. (F) Lifespan extension by simultaneous overexpression of both DAGL/inaE and S6KKQ (da-Gal4 > UAS-DAGL/inaE/UAS-S6KKQ, mean = 33 d, n = 76) does not further extend lifespan compared to either manipulation alone (da-Gal4 > UAS-DAGL/inaE, mean = 36 d, n = 210) or S6KKQ (da-Gal4 > UAS-S6KKQ, mean = 35 d, n = 50) at 29 °C.
To examine whether overexpression of DAGL/inaE extends lifespan via reduced TOR signaling, we overexpressed DAGL/inaE (UAS-DAGL/inaE) and the dominant-negative form of S6K (UAS-S6KKQ) individually and simultaneously. Overexpression of DAGL/inaE (UAS-DAGL/inaE) increases mean lifespan by 22% (P < 0.001) compared to Gal4 alone (Fig. 2D). Overexpression of the dominant-negative form of S6K (UAS-S6KKQ) extends mean lifespan by 18% (P < 0.001) compared to Gal4 alone (Fig. 2E). Overexpression of both the dominant-negative form of S6K and DAGL/inaE (UAS-S6KKQ; UAS-DAGL/inaE) simultaneously extends mean lifespan by 17% (P < 0.001) compared to Gal4 alone, which is similar to the longevity observed by overexpression of either transgene individually. Hence, the effects of the individual manipulations on lifespan are non-additive (Fig. 2F); similar results were also observed in the oxidative stress assay (Fig. S4), supporting the notion that DAGL/inaE -mediated lifespan extension and oxidative stress resistance are the result of lowered TOR signaling.
Expression of DAGL/inaE ortholog dagl-1 also regulates lifespan and is required for oxidative stress response in C. elegans
To determine whether the role of DAGL/inaE in extending longevity is conserved across species, we performed similar experiments using C. elegans. Overexpression of the DAGL/inaE ortholog dagl-1 from C. elegans driven by a ubiquitous promoter dpy-30 was achieved using two independent overexpression transgenic lines, N2; Ex[Pdpy-30::dagl-1::GFP](3) and N2; Ex[Pdpy-30::dagl-1::GFP](4) and comparing the results obtained to the control line N2; Ex[Pdpy-30::GFP]. Overexpression of dagl-1 extends mean lifespan by 12% (P < 0.01) and 13% (P < 0.001) (Fig. 3A, and Table S3, Supporting information).
Figure 3.
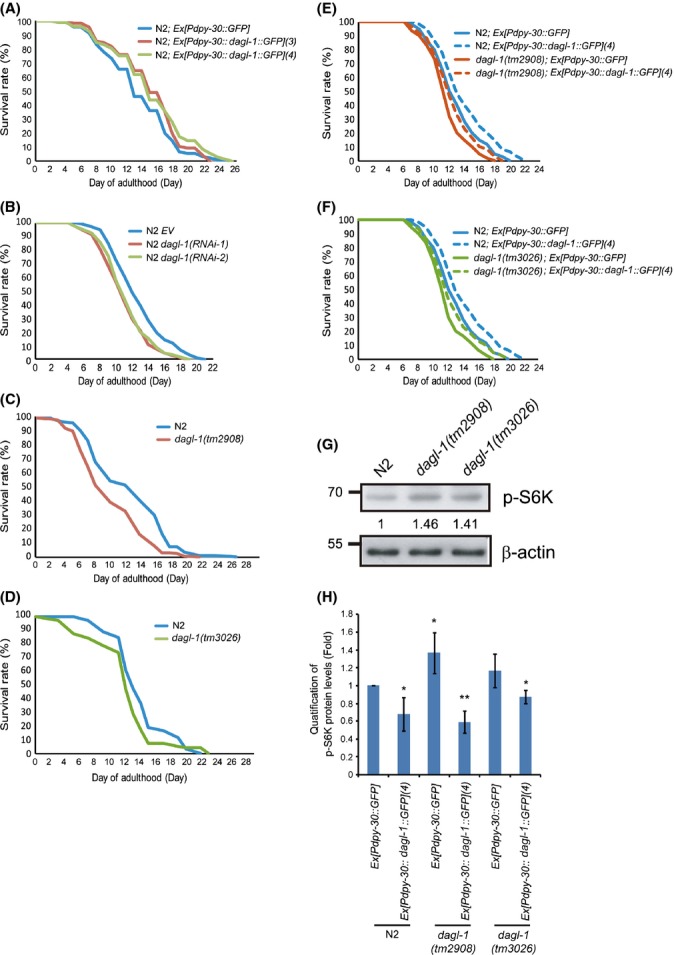
dagl-1 expression regulates lifespan and negatively correlates with levels of p-S6K in C. elegans. (A) Two independent transgenic lines that overexpress dagl-1 (N2; Ex[Pdpy::dagl-1::GFP](3), red line, and N2; Ex[Pdpy::dagl-1::GFP](4), green line) show extended lifespan compared to the control (N2; Ex[Pdpy::GFP], blue line). (B) N2 worms treated with RNAi against either 5′ (dagl-1(RNAi-1)) or 3′ (dagl-1(RNAi-2)) coding sequence of dagl-1 display shortened lifespan compared to the empty vector (EV) control. (C, D) The two dagl-1 deletion mutants, dagl-1(tm2908) and dagl-1(tm3026), exhibit reduced lifespan compared to the control N2. (E, F) Shortened lifespan of dagl-1(tm2908) and dagl-1(tm3026) can be rescued by transgenic overexpression of dagl-1. See also Table S3. (G) Western blot shows elevated levels of p-S6K in dagl-1(tm2908) and dagl-1(tm3026). β-actin was used as an internal control. (H) Elevated levels of p-S6K in dagl-1(tm2908) and dagl-1(tm3026) are reduced by transgenic overexpression of dagl-1. Western blots are shown in Fig. S8A.
Conversely to test whether reduced dagl-1 expression decreased lifespan, we constructed two RNAi clones (dagl-1(RNAi-1) and dagl-1(RNAi-2)) targeting the 5′ and 3′ fragment of dagl-1 coding sequence, respectively. The levels of dagl-1 expression were reduced by approximately 45% and 24% by feeding N2 worms E. coli HT115 harboring dagl-1(RNAi-1) and dagl-1(RNAi-2), respectively (Fig. S5, Supporting information). The N2 nematodes treated with dagl-1(RNAi-1) and dagl-1(RNAi-2) were 14% and 12% shorter lived, relative to the control (Fig. 3B, and Table S3, Supporting information). These results were in agreement with a second approach in which dagl-1 mutants, dagl-1(tm2908) and dagl-1(tm3026), were used. Mean lifespan was reduced by 20% and 13% (Fig. 3C,D, and Table S3, Supporting information). Thus, lower level of dagl-1 expression is associated with reduced longevity.
To determine whether the effects on longevity in the mutant strains resulted from the reduced expression of dagl-1, we generated transgenic lines which overexpressed dagl-1 (Pdpy-30::dagl-1). The Pdpy-30::dagl-1, but not Pdpy-30::GFP, transgene significantly rescued the lifespan of the dagl-1(tm2908) mutant to the level similar to that seen in the control N2 [Pdpy-30::GFP] worms (Fig. 3E, and Table S3, Supporting information). Similar rescue results using the Pdpy-30::dagl-1 transgene were also observed in the dagl-1(tm3026) mutants (Fig. 3F, and Table S3, Supporting information). Together, the data indicate that dagl-1 expression regulates lifespan in C. elegans.
As expected the effects of modulating dagl-1 expression were similar for both longevity and stress resistance assays. Paraquat-induced oxidative stress was applied to either mutant or RNAi-knockdown dagl-1 strains. In both cases, the lines with reduced activity were less resistant, dagl-1(tm2908 or tm3026) worms 21–26% less (Fig. S6A, and Table S4, Supporting information) and worms treated with either dagl-1(RNAi-1) or dagl-1(RNAi-2) 22-27% less resistant (Fig. S6B, and Table S4, Supporting information). Together, the results suggest that dagl-1 expression is required to respond to oxidative stress in C. elegans.
dagl-1 modulates lifespan and oxidative stress response through reduced TOR signaling in C. elegans
To confirm that dagl-1 also modulates TOR signaling in C. elegans, we first inspected the levels of p-S6K in the dagl-1 mutant worms compared to N2. In the mutants, p-S6K levels were 41–46% higher than in control worms (Fig. 3G), a result similar to those seen in Drosophila (Fig. 1H). The transgenic worms overexpressing dagl-1 showed significantly lower levels of p-S6K relative to that of the control (Fig. 3H). Moreover, the elevated levels of p-S6K in both dagl-1 mutant worms were reduced after introducing transgenic dagl-1 (Fig. 3H). Thus as in Drosophila, a clear correlation exists between the effects on TOR signaling and the resulting longevity and stress resistance phenotypes.
Knockdown of dgk-5 rescues the shortened lifespan and reduced oxidative stress tolerance in dagl-1 mutants
Since dagl-1 mutation may enhance TOR signaling by converting more DAG to PA by diacylglycerol kinase (DGK) (model in Fig. S2), we predicted that the enhancement of TOR signaling in dagl-1 mutants could be blocked by using RNAi knockdown of the other branch of the pathway, DGK/dgk-5. In both dagl-1 mutants p-S6K levels were significantly reduced by RNAi knockdown of DGK/dgk-5 (Fig. 4A). In addition, DGK/dgk-5 RNAi knockdown fully rescued the shortened lifespan (Fig. 4B, and Table S5, Supporting information) and improved the response to oxidative stress in both the dagl-1 mutant worms (Fig. S6C, D, and Table S4, Supporting information). These results showed that knockdown of DGK/dgk-5 can rescue the shortened lifespan and the reduced oxidative stress tolerance in both dagl-1 mutants.
Figure 4.
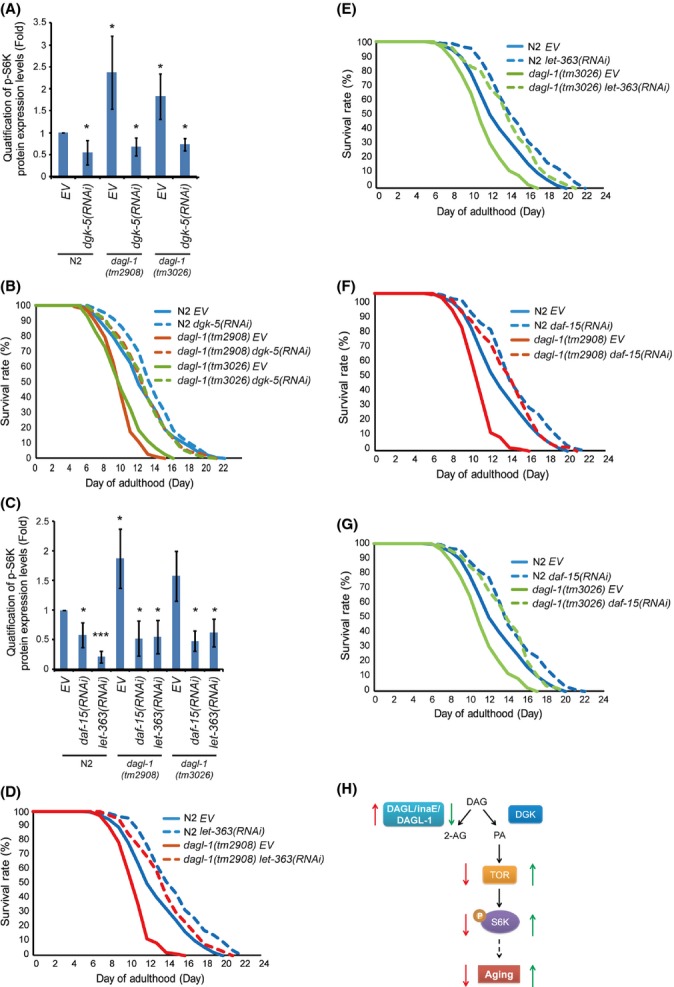
Knockdown of dgk-5, daf-15, or let-363 rescues the shortened lifespan and elevated p-S6K levels in dagl-1 mutants. (A) Elevated levels of p-S6K in dagl-1(tm2908) and dagl-1(tm3026) are reduced by RNAi knockdown of dgk-5. (B) Shortened lifespan of dagl-1(tm2908) and dagl-1(tm3026) is rescued by RNAi knockdown of dgk-5. (C) RNAi knockdown of daf-15 or let-363 also reverts the elevated levels of p-S6K observed in dagl-1(tm2908) and dagl-1(tm3026). (D–G) Shortened lifespan of dagl-1 mutants is also rescued by RNAi knockdown of daf-15 or let-363. See also Table S5. All western blots are shown in Fig. S8B and C. (H) Model for DAGL/inaE/dagl-1 in regulation of lifespan in Drosophila and C. elegans. DAGL/inaE/dagl-1 overexpression reduces TOR signaling and p-S6K levels to slow aging (red arrows). Hypomorphs of DAGL/inaE/dagl-1 increase TOR signaling and p-S6K levels to accelerate aging (green arrows).
As rdgA (Drosophila DGK) mutants demonstrated increased lifespan and lowered p-S6K levels (Fig. S3A,B, Supporting information), we examined whether C. elegans dgk-5 mutants also present the similar phenotypes. Intriguingly, C. elegans dgk-5 mutants dgk-5(ok2366) and dgk-5(gk631) had an 8% (P < 0.05) and 11% (P < 0.01) increase in mean lifespan and 31% and 50% decline in p-S6K levels compared to those in control N2, respectively (Fig. S3D,E, Table S6, Supporting information). These results are consistent with the rdgA mutant analysis in Drosophila and, together they bolster our hypothesis (Fig. S2, Supporting information).
RNAi knockdown of Tor/let-363 or raptor/daf-15 reduces the elevated p-S6K levels, rescues the shortened lifespan and improves the oxidative stress response in dagl-1 mutants
To further verify that TOR signaling plays a role in dagl-1-mediated lifespan and oxidative stress response in C. elegans, we examined whether RNAi knockdown of the Tor kinase, Tor/let-363, blocks the increase in p-S6K levels in the dagl-1 mutants. As expected, p-S6K levels were dramatically reduced in dagl-1 mutant worms treated with Tor/let-363 RNAi-containing bacteria (Fig. 4C). Raptor binds to TOR to form TOR complex 1 and regulates TOR downstream signaling (Wullschleger et al., 2006). Therefore, we checked whether RNAi knockdown of raptor/daf-15 expression could also diminish the elevated levels of p-S6K in the dagl-1 mutants and found that the enhanced p-S6K levels were also significantly reduced in both dagl-1(tm2908) and dagl-1(tm3026) mutants treated with raptor/daf-15 RNAi (Fig. 4C). Moreover, treatment of either Tor/let-363 or raptor/daf-15 RNAi to dagl-1(tm2908) and dagl-1(tm3026) mutant worms also rescued their shortened lifespan (Fig. 4D–G, and Table S5, Supporting information).
To determine whether comparable results would be obtained in oxidative stress response, we conducted similar experiments in dagl-1(tm2908) and dagl-1(tm3026) worms treated with or without Tor/let-363 or raptor/daf-15 RNAi under paraquat-induced oxidative stress. In these experiments the reduced survival rates in both dagl-1(tm2908) and dagl-1(tm3026) mutants were almost completely rescued by of the treatment of either Tor/let-363 or raptor/daf-15 RNAi (Fig. S6E,F, and Table S4, Supporting information). Together, the results reveal that the RNAi knockdown of Tor/let-363 or raptor/daf-15 not only lowers the elevated p-S6K levels but also rescues the shortened lifespan and partially improves the oxidative stress response in the dagl-1 mutant worms.
To exclude the possibility that 2-AG itself may also reduce TOR signaling, we exogenously supplemented 2-AG into NIH3T3 and Hep3B cell lines and examined the levels of p-S6K. 2-AG did not cause any reduction in the levels of p-S6K in both NIH3T3 and Hep3B cell lines, while rapamycin treatment dramatically reduced the levels of p-S6K (Fig. S7, Supporting information).
In summary, our parallel analysis using Drosophila and C. elegans demonstrate that DAGL/inaE/dagl-1 regulates lifespan and modulates oxidative stress response through inversely modulating TOR signaling (Fig. 4H).
Discussion
Genetic studies in model organisms have led to the discovery of many genes that can modulate aging. In addition many of these studies suggest that the pathways that control aging have been evolutionarily conserved. TOR signaling is one of the conserved nutrient sensor pathways involved in metabolism, growth, and nutrient sensing, and plays an important role in the regulation of aging from yeast to mammals including humans (Kapahi et al., 2010). TOR is proposed to be a lipid sensor that modulates cell growth and proliferation (Foster, 2013). However, we did not observe any changes in the eye and wing sizes (Fig. S9, Supporting information) neither the body size (data not shown) upon DAGL/inaE overexpression, suggesting that DAGL/inaE overexpression does not affect developmental growth. Accumulated evidence has shown that lipid metabolism is linked to lifespan regulation (Oldham, 2011; Ackerman & Gems, 2012). In this study, we demonstrated that diacylglycerol lipase (DAGL/inaE/dagl-1) regulates lifespan and oxidative stress response through TOR signaling in both Drosophila and C. elegans. Overexpression of DAGL/inaE/dagl-1 may shunt more DAG toward the production of 2-AG, thereby leaving less DAG available to produce PA, and consequently resulting in reduced TOR signaling. Both in flies and worms, DAGL/inaE/dagl-1-mediated lifespan is negatively correlated with levels of p-S6K. Both the shortened lifespan and the elevated levels of p-S6K can be rescued and reverted by the RNAi knockdown of dgk-5, daf-15, or let-363 in the dagl-1 mutants, suggesting that TOR signaling plays a role in DAGL/inaE/dagl-1 mediated lifespan. In addition, we also showed that both RNAi knockdown of DGK/rdgA/dgk-5 and their mutants extend lifespan and exhibit reduced level of pS6K in Drosophila and C. elegans. This is the first demonstration showing that reduced rdgA and dgk-5 expression extend lifespan in Drosophila and C. elegans. Together, it suggests that genetically altered DAG metabolism may influence PA levels to affect TOR signaling mediated lifespan and stress response.
Lipid homeostasis is critical to aging. Several genes involved in lipid metabolism control lifespan (Ackerman & Gems, 2012; McCormick et al., 2012). DAG is a lipid metabolic intermediate as a second messenger involved in complex signaling (Carrasco & Merida, 2007). DAG can activate protein kinase D (PKD). It has been suggested that DGK functions upstream of PKD in the regulation of oxidative-induced intestinal cell injury (Song et al., 2008). Thus, genetic manipulation of DGK and PKD should produce similar phenotypes. Indeed, it was reported that PKD/DFK-2 deficiency increases adult lifespan by 40% in C. elegans (Feng et al., 2007), implying that a lower level of DAG may extend lifespan in C. elegans. This is in agreement with our idea that lower DAG levels results in less PA formation, reduced TOR signaling, and thus to an extension of lifespan – an effect mimicked by knockdown of DGK/rdgA/dgk-5 both in Drosophila and C. elegans. It was reported that Drosophila microRNA mir-14 inhibits reaper-dependent cell death and is required for lipid metabolism (Xu et al., 2003). Depletion of mir-14 results in reduced lifespan and lowered stress tolerance and is accompanied with increased levels of triacylglycerol and diacylglycerol and the above phenotypes are reverted upon increasing mir-14 copy number in Drosophila. This suggests that lifespan negatively correlates with DAG level. DAG activation of protein kinase C (PKC) is linked to hepatic insulin resistance, a risk for type 2 diabetes (Jornayvaz & Shulman, 2012). In addition, PKC activity is associated with prefrontal cortical decline in aging and pharmacological inhibition of PKC rescues working memory malfunction in aged rat and increased working memory in aged rhesus monkeys (Brennan et al., 2009), indicating accumulated DAG is deleterious to lifespan and health. DAG is a second messenger triggering activation of PKC to enhance calcium influx for the activation of mTORC1. Overexpression of DAGL/inaE in neurons may result in less DAG levels for lowered PKC activity leading to reduced calcium influx and hence diminished mTORC1 activity to account for the extended lifespan and oxidative stress resistance. Thus, altered lipid metabolism achieved by lowering DAG levels is beneficial to lifespan and stress response.
Phophatidic acid is implicated in the activation of mammalian target of rapamycin (mTOR) and the control of cell growth and differentiation (Fang et al., 2001; Merida et al., 2008). Overexpression of DAGL/inaE/dagl-1 may result in lower level of PA for reduced TOR signaling in extending lifespan. It was reported that the expression of a specific isoform DGKζ, which modulates PA levels, regulates the levels of serum-induced phosphorylation of S6K for mTOR signaling in HEK293 cells (Avila-Flores et al., 2005). Interestingly, the closest homologs of DGKζ in Drosophila and C. elegans are rdgA and dgk-5, respectively. Our data showed that not only the mutants of rdgA and dgk-5 but also both knockdown of rdgA in fly and knockdown of dgk-5 in worm extend lifespan and reduce the levels of p-S6K. This provides the first in vivo evidence that reducing DGK extends lifespan via its effect on TOR signaling in both Drosophila and C. elegans. As we hypothesized that DAGL overexpression may result in more 2-AG formation, and 2-AG can be further metabolized to become arachidonic acid, also known as omega-6 polyunsaturated fatty acids, and glycerol. Omega-6 polyunsaturated fatty acids recently have been reported to extend C. elegans lifespan via activation of autophagy (O’Rourke et al., 2013). Therefore, it is also possible that DAGL/inaE/dagl-1 overexpression may result in more 2-AG for increased levels of omega-6 polyunsaturated fatty acids to activate autophagy for lifespan extension.
Insulin signaling is a well-studied and conserved pathway that also regulates lifespan (Kenyon, 2010). The interplay between insulin and TOR signaling pathways is well characterized (Hay, 2011). Interestingly, we found that the longevity and oxidative stress resistance of daf-2 can be partially inhibited by knockdown of dagl-1, and increased dagl-1 expression was detected in a daf-2 mutant. Two putative daf-16 binding sites were identified in the regulatory region of dagl-1 (Liu et al., 2012). In addition, we also found increased levels of phosphorylated Akt (p-Akt) in the two dagl-1 mutants compared to N2 (Lin and Wang, unpublished data), suggesting that dagl-1 plays a role in the lifespan and oxidative stress response of the daf-2 mutant and insulin signaling may also modulate dagl-1 expression in C. elegans. However, we did not detect any changes in the levels of p-Akt in DAGL/inaEEP1101 and DAGL/inaEKG08585 compared to w1118 (data not shown). It suggests that there is a discrepancy between Drosophila and C. elegans in insulin signaling for the DAGL/inaE/dagl-1-mediated lifespan regulation.
In summary, our study shows that DAGL/inaE/dagl-1 regulates lifespan and oxidative stress response via negatively modulating TOR signaling in both Drosophila and C. elegans. Since TOR signaling is a conserved pathway among different species regulating nutrient sensing, cell growth, and aging, our discovery may be relevant in mammals. Our results provide new insights on how the altered genetic regulation of DAG metabolism affects lifespan and stress response and may help in developing therapies to DAG imbalance-related diseases.
Experimental procedures
Drosophila and C. elegans strains and RNAi-expressing bacteria clones
The fly line DAGL/inaEEP1101 (Rorth, 1996) was initially identified in a double stress screen in Dr. Seymour Benzer’s laboratory (Caltech, Pasadena, CA, USA). DAGL/inaEKG08585, rdgABL33306, and rdgABL20320 were later obtained from the Bloomington Drosophila stock center. All were outcrossed with w1118 for at least six or ten generations to eliminate background effects and the resultant homozygous lines were used for lifespan and oxidative stress assays. UAS-rdgARNAi (VDRC #3024) was obtained from the Vienna Drosophila RNAi Center (VDRC). All flies were raised on standard Caltech fly food at 25 °C with 65%-humidity and a 12-hour light/dark cycle (Liu et al., 2009). The dagl-1 frame-shift mutant strains, dagl-1(tm2908) and dagl-1(tm3026), were obtained from the National Bioresource Project. The dgk-5(ok2366) and dgk-5(gk631) strains were provided by Dr. Chang-Shi Chen from Taiwan C. elegans Core. All nematodes were grown at 20 °C on Nematode Growth Medium (NGM) plates seeded with OP50 for regular culture or with HT115 for RNAi treatment. The RNAi clones targeting daf-15 and let-363 were kindly provided by Dr. Ao-Lin Hsu at University of Michigan. Two RNAi plasmids, dagl-1(RNAi-1) and dagl-1(RNAi-2), were constructed using the L4440 vector that express double-stranded RNA targeting either the 5′ or 3′ end of dagl-1 cDNA upon IPTG induction. The 517-nt amplicon of dagl-1 for dagl-1(RNAi-1) and the 570-nt amplicon of dagl-1 for dagl-1(RNAi-2) were PCR amplified by the primer sets (RNAi-1 forward: 5′-GGCAAGTCAATGGTAGTGGA-3′ and RNAi-1 reverse: 5′- CGAAACAACGCTCATCACAT-3′; and RNAi-2 forward: 5′- TTCCGCTTGCCTGTTCTACT-3′ and RNAi-2 reverse: 5′-CCTGCAACAACATCACTTGG-3′) and subcloned into L4440 vector.
Generation of DAGL/inaE transgenic flies and dagl-1 transgenic worms
To generate the DAGL/inaE transgenic flies, the 2214-nt long isoform (inaE-PD, FlyBase) and the 1935-nt short isoform (inaE-PA, FlyBase) of DAGL/inaE cDNAs based on the information of FlyBase were PCR-amplified using LD44686 and GH19816 plasmids as templates and subcloned into the XhoI/BglII sites of pINDY6 transgenic vector (Wang et al., 2004). The resultant transgenic constructs were verified by DNA sequencing to confirm no mutations in the cDNAs derived from PCR, and later micro-injected into w1118 embryos to generate the transgenic flies, UAS-DAGL/inaE-PD and UAS-DAGL/inaE-PA, expressing either the long or the short isoforms of DAGL/inaE upon Gal4 induction. For dagl-1 transgenic nematodes, F42G9.6b isoform full-length cDNA – which is the most homologous to fly DAGL/inaE-PD gene – was subcloned and fused with GFP driven by the dpy-30 ubiquitous promoter in the ps235 vector (Hsu et al., 1995). The resultant plasmid, Pdpy-30::dagl-1::GFP, was verified by DNA sequencing and micro-injected at a concentration of 20 ng/μl into N2 young adult worms to generate the two independent transgenic worms, N2; Ex[Pdpy-30::dagl-1::GFP](3) and N2; Ex[Pdpy-30::dagl-1::GFP](4). The control worms, N2; Ex[Pdpy-30::GFP], were obtained by micro-injecting the control plasmid Pdpy-30::GFP into N2. The progeny of the injected animals were screened for GFP expression to establish independent lines.
Lifespan and oxidative stress assays in Drosophila and C. elegans
For Drosophila, the lifespan assay and paraquat-induced oxidative stress assay for the progeny from specific crosses were carried out as described previously (Liao et al., 2008; Liu et al., 2009; Wang et al., 2012). We found female flies in DAGL/inaEEP1101 and DAGL/inaEKG08585 showed similar results to males in the lifespan and stress assays and thus only results from male flies were used in this paper. Most experiments were carried out at 25 °C unless otherwise stated. For C. elegans, lifespan assays were performed at 20 °C as described previously (Liu et al., 2009) but without adding 5′ flourodeoxyuridine (FUdR). N2, dagl-1(tm2908), dagl-1(tm3026), N2; Ex[Pdpy-30::GFP], N2; Ex[Pdpy-30::dagl-1::GFP](3) and N2; Ex[Pdpy-30::dagl-1::GFP](4) worms were grown on NGM plates seeded with E. coli OP50 bacteria. For RNAi treatment, worms were placed on NGM plates with E. coli HT115 containing the control L4440 plasmid or L4440 expressing dsRNA targeting the specific gene. All the worms were initially transferred daily for the first seven days and later every 2 or 3 days. Dead worms not responding to gentle prodding were scored until all were dead. The oxidative stress assay for worms was conducted at 20 °C. Young adult hermaphrodites were immersed in S-media containing either 10 or 40 mm of paraquat (1,1-dimethyl-4,4-bipyridinium dichloride, Sigma-Aldrich, St. Louis, MO, USA). The number of dead worms was scored every hour until all worms were dead. All experiments were repeated at least three times. Gene expression changes were monitored by RT-PCR and real-time PCR. Statistical differences in survival were calculated by the log-rank test. Differences in oxidative stress resistance were determined by Student’s t-test.
Western blot
Fly heads of specific age for each strain were collected and homogenized in lysis buffer containing protease inhibitor (Cat#: 04693159001, Roche, Indianapolis, IN, USA) and phosphatase inhibitor (Cat#: 04906837001, Roche). Synchronized four-day-old adult worms of each strain, with or without RNAi treatment, were collected in 15ml centrifuge tubes, washed three times by M9 buffer, transferred to new microfuge tubes, and homogenized by lysis buffer containing protease inhibitor and phosphatase inhibitor. In the cell lines, NIH3T3 and Hep3B cells were treated with DMSO as a mock, 10 or 20 μm of 2-AG (Cat#: 1298, TOCRIS, Bristol, UK), or 10 nm Rapamycin as a positive control (Cat#: 553210, Millipore, Billerica, MA, USA). After 24-h incubation, the treated cells were collected in 15-mL centrifuge tubes and the cell pellets were lysed in lysis buffer containing protease inhibitor and phosphatase inhibitor as mentioned above. After homogenization, 2% SDS was added to each sample again and then the sample was vortexed and incubated 5 min at 70 °C. The sample was centrifuged at 13 000 rpm for 10 min at room temperature and the supernatant was transferred into new tube to measure protein concentration. Equal amounts of protein for each sample were loaded and separated in a 12% SDS-PAGE gel, and transferred to a nitrocellulose membrane. The membrane was blocked with 5% BSA in TBST for 1 h, and later incubated with anti-pS6K (Cell Signaling, Billerica, MA, USA, #9209, 1:500 dilution in 5% BSA /1XTBST or Abbomax Inc., #601-030, 1:1000 dilution in 5% BSA /1XTBST), anti-pERK (Epitomics, #1481-1, 1:1000 in 5% BSA/1XTBST), anti-α−actin or β-actin or tubulin (α−actin, Santa Cruz, Dallas, Texas, USA, #SC-1616; β-actin, Spring, #E4554, 1:10 000; tubulin, Epitomics, #1871-1, 1:1000 dilution, in 5% BSA/1XTBST), or anti-GAPDH (Epitomics, #S0011, 1:2000 in 5% BSA/1XTBST) at 4 °C overnight. The membrane then was washed three times with TBST, and incubated with the secondary antibody (goat anti-rabbit IgG, 1:10 000 in 5% BSA/1XTBST) for 2 h at 4 °C, again washed three times with TBST, incubated with ECL reagent (Cat#: RPN 2132, Amersham, GE Healthcare, Fairfield, CT, USA) and exposed to the X-ray film (Kodax, Rochester, NY, USA). The protein image was quantified by ImageJ® to calculate the fold of changes by normalizing each measurement to its control.
Semi-quantitative RT-PCR and quantitative real-time PCR assays
Drosophila total RNA extraction and reverse-transcription following by semi-quantitative polymerase chain reaction (RT-PCR) were described in Wang et al. (2004). For C. elegans, worms with or without RNAi treatment were collected into 1.5-mL microfuge tube, washed three times by M9 buffer, and lysed by using 1 ml TRIzol® reagent (Life Technologies, Grand Island, NY, USA) to extract RNA. Subsequent procedures were similar to those used for Drosophila. Each gene was amplified by gene specific primers (sequences available upon request). The genes rp49 and actin were used as internal controls in the PCR reactions for Drosophila and C. elegans, respectively. The fold changes for gene expression were calculated, normalized to the internal control, by quantification of the image of the DNA in agarose gel by ImageJ® software. Alternatively, the cDNAs were used as templates in quantitative real-time PCR utilizing SYBR Green PCR Master Mix in the Applied Biosystems 7900HT Fast Real-Time PCR System (7900HT Fast System, Life Technologies). Each gene was amplified with the specific real-time PCR primer set, and was normalized to the control (rp49 for Drosophila and actin for C. elegans). The relative transcriptional levels of the genes were presented as fold of 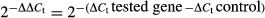 . Ct is the threshold cycle value clarified as the fractional cycle number at the time of target fluorescent signal passed a threshold above baseline.
. Ct is the threshold cycle value clarified as the fractional cycle number at the time of target fluorescent signal passed a threshold above baseline.
Acknowledgments
We thank Drs. Jui-Chou Hsu, Ao-Lin Hsu, and Ms. Sany Hoxha for comments on the manuscript. We are indebted to the facilities of Fly Core in Taiwan and C. elegans Core Facility Taiwan, both supported by the National Science Council.
Author contributions
Designed the experiments: YHL, YCC, HDW. Performed the experiments: YHL, YCC, TYK, YCL, TEH, LKY, ZHL, RJY, and YTJ. Analyzed the data: YHL, YCC, TYK, YCL, TEH, YCW, WWJ, TJB, PK, LKY, ZHL, CHY, and HDW. Contributed reagents and materials: YCW, PK. Wrote the manuscript: YHL, YCC, YCW, WWJ, TJB, PK, CHY, and HDW.
Conflict of interest
None declared.
Funding
This study was supported by the National Science Council (NSC 100-2311-B-007-006- and 96-2311-B-007-003-, H.-D.W.), the National Tsing Hua University Brain Research Center (102N2061E1, H.-D.W.), a NTHU President scholarship (Y.-H.L.), and a scholarship from Apex Biotechnology Corp. (Y.-C.C.). We also acknowledge grants from the National Institutes of Health, R00AG030493 (W.W.J.), R15AG027749 (T.J.B.), and R01 AG031337-01A1 (P.K.), and the National Science Council (NSC102-2321-B-400-016, C.-H.Y.).
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
Fig. S1. DAGL/inaEEP1101 mutant is resistant to the combination of oxidative stress and starvation.
Fig. S2. Model for DAGL/inaE/dagl-1 regulation of lifespan in Drosophila and C. elegans.
Fig. S3. Drosophila rdgA and C. elegans dgk-5 mutants exhibit lower p-S6K levels and extended lifespan.
Fig. S4. Overexpression of both DAGL/inaE and S6KKQ, a dominant-negative form of S6K, does not further enhance resistance to oxidative stress compared to the overexpression of DAGL/inaE or S6KKQ individually.
Fig. S5. Real-time quantitative PCR analysis shows reduced dagl-1 expression upon RNAi knockdown in N2 worms.
Fig. S6. C. elegans dagl-1 mutants show reduced tolerance to oxidative stress that can be rescued by RNAi knockdown of dgk-5, daf-15, or let-363.
Fig. S7. Exogenous supplementation of 2-AG does not reduce levels of phosphorylated-S6K (p-S6K) in NIH3T3 and Hep3B cell lines.
Fig. S8. Western blots for p-S6K. Independent experiments are shown for the results in Figs.
Fig. S9. No smaller eye or wing size was detected upon DAGL/inaE overexpression.
Table S1. Lifespan of DAGL/inaE transgenic overexpression flies by different Gal4 drivers.
Table S2. Oxidative stressg response of DAGL/inaE transgenic overexpression flies by different Gal4 drivers.
Table S3. Effect of dagl-1 expression on lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans.
Table S4. Effect of dagl-1 expression and knockdown of dgk-5, let-363, daf-15 on oxidative stress response of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans.
Table S5. Effect of dgk-5, let-363, and daf-15 RNAi knockdown on the lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans.
Table S6. The lifespan of N2, dgk-5(ok2366) and dgk-5(gk631) in C. elegans.
Table S7. Effect of dagl-1 expression on lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans. (the three separate data for Table S3).
Table S8. Effect of dagl-1 expression and knockdown of dgk-5, let-363, daf-15 on oxidative stress response of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans. (the three separate data for Table S4).
Table S9. Effect of dgk-5, let-363, and daf-15 RNAi knockdown on the lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans. (the three separate data for Table S5).
Table S10. The lifespan of N2, dgk-5(ok2366) and dgk-5(gk631) in C. elegans. (the three separate data for Table S6).
References
- Ackerman D, Gems D. The mystery of C. elegans aging: an emerging role for fat. Distant parallels between C. elegans aging and metabolic syndrome? BioEssays. 2012;34:466–471. doi: 10.1002/bies.201100189. [DOI] [PubMed] [Google Scholar]
- Alic N, Partridge L. Death and dessert: nutrient signalling pathways and ageing. Curr. Opin. Cell Biol. 2011;23:738–743. doi: 10.1016/j.ceb.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Flores A, Santos T, Rincon E, Merida I. Modulation of the mammalian target of rapamycin pathway by diacylglycerol kinase-produced phosphatidic acid. J. Biol. Chem. 2005;280:10091–10099. doi: 10.1074/jbc.M412296200. [DOI] [PubMed] [Google Scholar]
- Brennan AR, Yuan P, Dickstein DL, Rocher AB, Hof PR, Manji H, Arnsten AF. Protein kinase C activity is associated with prefrontal cortical decline in aging. Neurobiol. Aging. 2009;30:782–792. doi: 10.1016/j.neurobiolaging.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Abramovici H, Gee SH, Topham MK. Diacylglycerol kinases as sources of phosphatidic acid. Biochim. Biophys. Acta. 2009;1791:942–948. doi: 10.1016/j.bbalip.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco S, Merida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Feng H, Ren M, Chen L, Rubin CS. Properties, regulation, and in vivo functions of a novel protein kinase D: Caenorhabditis elegans DKF-2 links diacylglycerol second messenger to the regulation of stress responses and life span. J. Biol. Chem. 2007;282:31273–31288. doi: 10.1074/jbc.M701532200. [DOI] [PubMed] [Google Scholar]
- Foster DA. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab. 2013;24:272–278. doi: 10.1016/j.tem.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui H, Li ML, Tsai CC. A tale of tailless. Dev. Neurosci. 2011;33:1–13. doi: 10.1159/000321585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA. The aging stress response. Mol. Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Hardie RC. TRP channels in Drosophila photoreceptors: the lipid connection. Cell Calcium. 2003;33:385–393. doi: 10.1016/s0143-4160(03)00051-4. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N. Interplay between FOXO, TOR, and Akt. Biochim. Biophys. Acta. 2011;1813:1965–1970. doi: 10.1016/j.bbamcr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DR, Chuang PT, Meyer BJ. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development. 1995;121:3323–3334. doi: 10.1242/dev.121.10.3323. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Jornayvaz FR, Shulman GI. Diacylglycerol activation of protein kinase Cepsilon and hepatic insulin resistance. Cell Metab. 2012;15:574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Lapierre LR, Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 2012;23:637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H-T, Tseng-Crank J, Kim E, Mahapatra C, Shino S, Zhou Y, An L, Doerge RW, Pak WL. DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron. 2008;58:884–896. doi: 10.1016/j.neuron.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PC, Lin HY, Yuh CH, Yu LK, Wang HD. The effect of neuronal expression of heat shock proteins 26 and 27 on lifespan, neurodegeneration, and apoptosis in Drosophila. Biochem. Biophys. Res. Commun. 2008;376:637–641. doi: 10.1016/j.bbrc.2008.08.161. [DOI] [PubMed] [Google Scholar]
- Liu YL, Lu WC, Brummel TJ, Yuh CH, Lin PT, Kao TY, Li FY, Liao PC, Benzer S, Wang HD. Reduced expression of alpha-1,2-mannosidase I extends lifespan in Drosophila melanogaster and Caenorhabditis elegans. Aging Cell. 2009;8:370–379. doi: 10.1111/j.1474-9726.2009.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Kao TY, Ge Q, Wu ZX, Yuh CH, Shan G. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 2012;3:1073. doi: 10.1038/ncomms2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging Cell. 2012;11:192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merida I, Avila-Flores A, Merino E. Diacylglycerol kinases: at the hub of cell signalling. Biochem J. 2008;409:1–18. doi: 10.1042/BJ20071040. [DOI] [PubMed] [Google Scholar]
- Oldham S. Obesity and nutrient sensing TOR pathway in flies and vertebrates: functional conservation of genetic mechanisms. Trends Endocrinol. Metab. 2011;22:45–52. doi: 10.1016/j.tem.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke EJ, Kuballa P, Xavier R, Ruvkun G. omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013;27:429–440. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Tamanoi F. Increased Rheb-TOR signaling enhances sensitivity of the whole organism to oxidative stress. J. Cell Sci. 2006;119:4285–4292. doi: 10.1242/jcs.03199. [DOI] [PubMed] [Google Scholar]
- Raghu P, Hardie RC. Regulation of Drosophila TRPC channels by lipid messengers. Cell Calcium. 2009;45:566–573. doi: 10.1016/j.ceca.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl Acad. Sci. USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Li J, Mourot JM, Evers BM, Chung DH. Diacylglycerol kinase regulation of protein kinase D during oxidative stress-induced intestinal cell injury. Biochem. Biophys. Res. Commun. 2008;375:200–204. doi: 10.1016/j.bbrc.2008.07.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim. Biophys. Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol. Cell. Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Wang H-D, Kazemi-Esfarjani P, Benzer S. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc. Natl Acad. Sci. USA. 2004;101:12610–12615. doi: 10.1073/pnas.0404648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Chen YC, Wang YY, Huang MH, Yen TL, Li H, Liang CJ, Sang TK, Ciou SC, Yuh CH, Wang CY, Brummel TJ, Wang HD. Reduced neuronal expression of ribose-5-phosphate isomerase enhances tolerance to oxidative stress, extends lifespan, and attenuates polyglutamine toxicity in Drosophila. Aging Cell. 2012;11:93–103. doi: 10.1111/j.1474-9726.2011.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. DAGL/inaEEP1101 mutant is resistant to the combination of oxidative stress and starvation.
Fig. S2. Model for DAGL/inaE/dagl-1 regulation of lifespan in Drosophila and C. elegans.
Fig. S3. Drosophila rdgA and C. elegans dgk-5 mutants exhibit lower p-S6K levels and extended lifespan.
Fig. S4. Overexpression of both DAGL/inaE and S6KKQ, a dominant-negative form of S6K, does not further enhance resistance to oxidative stress compared to the overexpression of DAGL/inaE or S6KKQ individually.
Fig. S5. Real-time quantitative PCR analysis shows reduced dagl-1 expression upon RNAi knockdown in N2 worms.
Fig. S6. C. elegans dagl-1 mutants show reduced tolerance to oxidative stress that can be rescued by RNAi knockdown of dgk-5, daf-15, or let-363.
Fig. S7. Exogenous supplementation of 2-AG does not reduce levels of phosphorylated-S6K (p-S6K) in NIH3T3 and Hep3B cell lines.
Fig. S8. Western blots for p-S6K. Independent experiments are shown for the results in Figs.
Fig. S9. No smaller eye or wing size was detected upon DAGL/inaE overexpression.
Table S1. Lifespan of DAGL/inaE transgenic overexpression flies by different Gal4 drivers.
Table S2. Oxidative stressg response of DAGL/inaE transgenic overexpression flies by different Gal4 drivers.
Table S3. Effect of dagl-1 expression on lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans.
Table S4. Effect of dagl-1 expression and knockdown of dgk-5, let-363, daf-15 on oxidative stress response of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans.
Table S5. Effect of dgk-5, let-363, and daf-15 RNAi knockdown on the lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans.
Table S6. The lifespan of N2, dgk-5(ok2366) and dgk-5(gk631) in C. elegans.
Table S7. Effect of dagl-1 expression on lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans. (the three separate data for Table S3).
Table S8. Effect of dagl-1 expression and knockdown of dgk-5, let-363, daf-15 on oxidative stress response of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans. (the three separate data for Table S4).
Table S9. Effect of dgk-5, let-363, and daf-15 RNAi knockdown on the lifespan of N2 and dagl-1(tm2908) and dagl-1(tm3026) in C. elegans. (the three separate data for Table S5).
Table S10. The lifespan of N2, dgk-5(ok2366) and dgk-5(gk631) in C. elegans. (the three separate data for Table S6).


