Summary
In contrast to numerous enzymes involved in c-di-GMP synthesis and degradation in enterobacteria, only a handful of c-di-GMP receptors/effectors have been identified. In search of new c-di-GMP receptors, we screened the Escherichia coli ASKA overexpression gene library using the Differential Radial Capillary Action of Ligand Assay (DRaCALA) with fluorescently and radioisotope-labeled c-di-GMP. We uncovered three new candidate c-di-GMP receptors in E. coli and characterized one of them, BcsE. The bcsE gene is encoded in cellulose synthase operons in representatives of Gammaproteobacteria and Betaproteobacteria. The purified BcsE proteins from E. coli, Salmonella enterica and Klebsiella pneumoniae bind c-di-GMP via the domain of unknown function, DUF2819, which is hereby designated GIL, GGDEF I-site like domain. The RxGD motif of the GIL domain is required for c-di-GMP binding, similar to the c-di-GMP-binding I-site of the diguanylate cyclase GGDEF domain. Thus, GIL is the second protein domain, after PilZ, dedicated to c-di-GMP-binding. We show that in S. enterica, BcsE is not essential for cellulose synthesis but is required for maximal cellulose production, and that c-di-GMP binding is critical for BcsE function. It appears that cellulose production in enterobacteria is controlled by a two-tiered c-di-GMP-dependent system involving BcsE and the PilZ domain containing glycosyltransferase BcsA.
Introduction
The ubiquitous second messenger c-di-GMP controls various aspects of bacterial physiology. Most commonly, elevated levels of c-di-GMP are associated with inhibited motility and a sessile lifestyle, characterized by production of pili, protein adhesins and exopolysaccharides involved in biofilm formation. In addition, c-di-GMP signaling pathways affect, in various species, long-term survival, response to environmental stress, cell cycle progression, differentiation and the production of virulence factors (Römling et al., 2013). In Escherichia coli and Salmonella enterica ser. Typhimurium, the species discussed in this work, elevated c-di-GMP levels result in inhibition of flagellar motility, activation of synthesis of two extracellular polysaccharides, cellulose and poly-N-acetyl-D-glucosamine, increased formation of adhesive curli fimbriae (Povolotsky and Hengge, 2012), and affect various aspects of virulence (Hu et al., 2013; Römling et al., 2013).
Cyclic di-GMP synthesis is catalyzed by the GGDEF protein domains present in diguanylate cyclases (DGCs), while its hydrolysis is catalyzed by either the EAL or HD-GYP domains present in c-di-GMP phosphodiesterases (PDEs). These protein domains can be readily identified in silico. In contrast, c-di-GMP receptors/effector proteins bind this second messenger by diverse means (Römling et al., 2013), and our ability to predict c-di-GMP receptors in silico is limited. The only readily identifiable c-di-GMP receptor types include PilZ domains (Amikam and Galperin, 2006), enzymatically inactive EAL domains capable of c-di-GMP-binding, and enzymatically inactive GGDEF domains that contain I-sites, allosteric sites for product feedback inhibition (see Römling et al., 2013).
Of 29 proteins with GGDEF, EAL or GGDEF-EAL domains encoded in the E. coli K-12 genome, five (CsrD [Suzuki et al., 2006], YcgF [Tschowri et al., 2009], YdiV [Wada et al., 2011; 2012], YegE [Pesavento et al., 2008] and likely YeaI) are catalytically impaired. The remaining proteins function as DGCs and PDEs (Povolotsky & Hengge, 2012). The large number of enzymes involved in c-di-GMP metabolism contrasts with only a handful of c-di-GMP receptors thus far identified in E. coli K-12. These receptors include: (i) YcgR (Ryjenkov et al., 2006; Christen et al., 2007) that contributes to the motile-to-sessile transition (Boehm et al., 2010; Paul et al., 2010; Fang and Gomelsky, 2010), (ii) BcsA (Ross et al., 1987; Ryjenkov et al., 2006), a c-di-GMP-dependent glycosyltransferase responsible for cellulose synthesis; (iii) the PgaC-PgaD complex involved in poly-N-acetyl-D-glucosamine synthesis (Steiner et al., 2013); (iv) PnpA, a 3′-polyribonucleotide polymerase and a 3′-to-5′ exoribonuclease involved in RNA degradation (Tuckerman et al., 2011); (v) BdcA, a member of the short-chain oxidoreductase family, that increases biofilm dispersal when overexpressed (Ma et al., 2011); and (vi) YciR, an unusual PDE that also functions as a c-di-GMP receptor (Lindenberg et al., 2013). This situation resembles a dysfunctional army where officers (DGCs and PDEs controlling c-di-GMP levels) greatly outnumber soldiers (c-di-GMP receptors). While c-di-GMP receptors often respond to “commands” originating from multiple enzymes, the enzymes are not necessarily simultaneously expressed, and some of them are active only in the presence of specific environmental stimuli. The disparity in numbers remains puzzling. We hypothesize that E. coli K-12 contains additional, as yet unidentified c-di-GMP receptors.
To uncover new c-di-GMP receptors, we performed an E. coli genome-wide screen using a recently developed Differential Radial Capillary Action of Ligand Assay (DRaCALA) (Roelofs et al., 2011). In this assay, a mixture of a tested protein and a labeled ligand are spotted onto a nitrocellulose membrane. While the protein-ligand complex is immobilized by the membrane and remains in the center of the spot, the unbound ligand moves outward from the initial spot by capillary action. Therefore, the shape of the labeled spot differentiates between the bound and unbound ligand thus indicating the presence of a potential ligand receptor. DRaCALA is particularly attractive because it can be applied not only to purified proteins but also to cell lysates containing overexpressed proteins (Corrigan et al., 2013).
Here, we report the results of the screens of the gene overexpression library of E. coli K-12, the so-called ASKA library (Kitagawa et al., 2005), using fluorescently and radioisotope labeled c-di-GMP. These screens uncovered some known c-di-GMP binding proteins but missed others, thus highlighting advantages and limitations of DRaCALA in genome-wide screens. Ultimately, we uncovered three new candidate c-di-GMP binding proteins. In this study, we focus on one of these c-di-GMP receptors, the BcsE protein. We show that BcsE is involved in maximal cellulose synthesis. Via deletion and mutational analysis of BcsE we identified a novel protein domain in this protein dedicated to c-di-GMP binding, DUF2819, hereby designated GIL. We found residues in GIL critical for c-di-GMP binding and BscE function. We also determined that not only E. coli BcsE but the BcsE proteins from other enterobacteria bind c-di-GMP via the GIL domain.
Results
Assessment of fluorescently labeled c-di-GMP in DRaCALA
Because of the convenience associated with using fluorescently versus radioactively labeled c-di-GMP (e.g., commercially availability, lack of decay and ease of detection), we evaluated the performance of 2′-fluo-aminohexylcarbamoyl-c-di-GMP (2′-fluo-AHC-cdiGMP) in DRaCALA screening (Fig. S1 in Supporting Information). We found that E. coli lysates overexpressing YcgR produced a positive signal with 2′-fluo-AHC-cdiGMP, in contrast to the YcgR R118D mutant impaired in c-di-GMP binding (Ryjenkov et al., 2006) (Fig. 1A). Addition of excess unlabeled c-di-GMP prevented binding, whereas addition of excess GTP had no effect. These tests showed that 2′-fluo-AHC-cdiGMP binds YcgR specifically and performs on par with the radioactively labeled c-di-GMP tested earlier by Roelofs and colleagues (Roelofs et al., 2011).
Fig. 1.
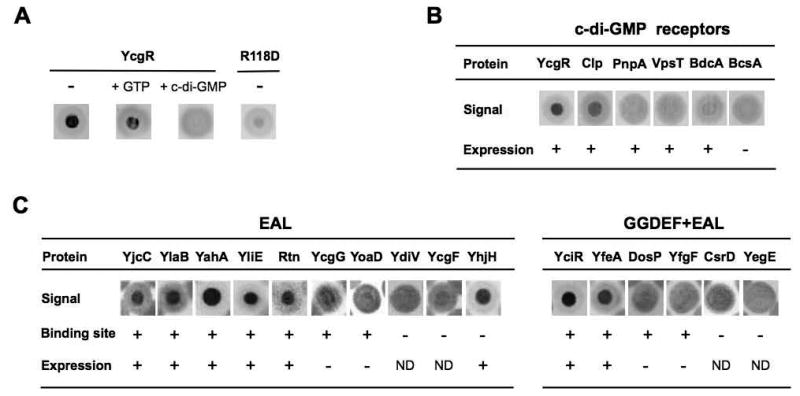
33P-c-di-GMP-DRaCALA of E. coli cell lysates expressing known c-di-GMP-binding proteins. (A) Testing c-di-GMP-binding specificity using lysates of cells overexpressing YcgR or the YcgR R118D mutant incapable of c-di-GMP binding. -, 33P-c-di-GMP-DRaCALA; + GTP, same as “-“ in the presence of 500 μM GTP; + c-di-GMP, same as “-“ plus 500 μM (unlabeled) c-di-GMP. (B) 33P-c-di-GMP-DRaCALA with cell lysates overexpressing known c-di-GMP receptors. Expression, visibly overexpressed protein band on SDS-PAGE. (C) 33P-c-di-GMP-DRaCALA with cell lysates overexpressing E. coli EAL and GGDEF-EAL domain proteins represented in the ASKA library.
Encouraged by these results, we proceeded to screen fifty-six 96-well microtiter plates of the ASKA library comprising 5,272 E. coli genes. In this library, each E. coli ORF is cloned on a plasmid downstream of the inducible T5 promoter (Kitagawa et al., 2005). The library screen produced an unexpectedly large number, approximately 150, of fluorescent spots. To investigate whether they correspond to overexpressed c-di-GMP receptors, we used four clones from a single screening membrane (Fig. S2). Each of the proteins that produced positive signals (AceE, PanC, SpeE, YadE) was purified using Ni2+ affinity chromatography and tested for binding of unlabeled c-di-GMP. None was found to bind c-di-GMP in equilibrium dialysis assays (Ryjenkov et al., 2006). We subsequently tested these overexpression clones with 33P-c-di-GMP (as described below) and again observed no positive signals (Fig. S2). We therefore had to conclude that 2′-fluo-AHC-cdiGMP is potentially suitable for verifying known c-di-GMP receptors but is prone to generating false-positives with high frequency when used in large-scale screening (see Discussion for possible reasons). These results prompted our switching from 2′-fluo-AHC-cdiGMP to 33P-c-di-GMP for DRaCALA screening.
E. coli overexpression library screening using 33P-c-di-GMP uncovers candidate c-di-GMP receptors
33P-c-di-GMP was prepared in house from α33P-GTP using Slr1143, a potent DGC from Synechocystis sp. PCC 6803 (Ryjenkov et al., 2005). To determine the sensitivity of the screening assay, we tested 33P-c-di-GMP binding using lysates from E. coli expressing several known c-di-GMP receptors including YcgR, BcsA, PnpA and BdcA from E. coli, VpsT from Vibrio cholerae (Krasteva et al., 2010) and Clp from Xanthomonas axonopodis (Leduc and Roberts, 2009). Only the YcgR and Clp lysates produced positive signals (Fig. 1B). The absence of a positive signal in the case of BcsA can be attributed to low expression of the BcsA protein, which was not detectable on SDS-PAGE (not shown). All other c-di-GMP receptors were expressed at high levels, therefore the lack of positive signals is not completely understood (see Discussion). Overall, these results suggest that DRaCALA can identify only a subset of c-di-GMP receptors.
To further assess sensitivity of the DRaCALA screening, we tested lysates of cells overexpressing EAL domain proteins that use c-di-GMP as substrate. To prevent 33P-c-di-GMP hydrolysis, we supplemented the lysis buffer with EDTA, which was expected to scavenge Mg2+ essential for PDE activity (Schmidt et al., 2005). Among 16 EAL domain proteins represented in the ASKA library, four proteins CsrD (Suzuki et al., 2006), YegE (Pesavento et al., 2008), YcgF (Tschowri et al., 2009), and YdiV (Simm et al., 2009), were incapable of c-di-GMP binding. Of the remaining 12 EAL domain proteins, eight (YahA, YciR, YfeA, YhjH, YjcC, YlaB, YliE, Rtn) produced positive signals (Fig. 1C), while four others (DosP [Schmidt et al., 2005], YfgF [Lacey et al., 2010], YcgG, YoaD [Brombacher et al., 2006]) showed no signals, possibly due to poor expression. Interestingly, none of the GGDEF domain proteins (YaiC, YddV, YdeQ, YeaI, YeaJ) that contain the c-di-GMP-binding I-site (Chan et al., 2004) generated positive signals. This assessment also suggested that DRaCALA screening may uncover some but probably not all undiscovered E. coli c-di-GMP-binding proteins.
Following performance assessment, we screened the complete ASKA library using 33P-c-di-GMP-DRaCALA. In addition to the known c-di-GMP-binding proteins, we identified three new positive clones that overexpressed BcsE, IlvH and RimO proteins. Below, we present the characterization of one of these clones that overexpresses BcsE.
E. coli BcsE is a bona fide c-di-GMP binding protein
To ascertain whether BcsE binds c-di-GMP in vitro, we cloned and overexpressed this protein from two vectors, pET23a and pMAL-c2X, respectively, as BcsE::His6 and MBP::BcsE fusions, where MBP is maltose-binding protein. The tagged BcsE proteins were purified using affinity chromatography. The BcsE::His6 protein was found to quickly precipitate following its elution from the Ni2+ column. The MBP::BcsE fusion was also prone to precipitation but proved to be more stable than BcsE::His6, therefore all subsequent tests were done using MBP::BcsE.
Purified MBP::BcsE bound 2′-fluo-AHC-cdiGMP (Fig. 2A) and 33P-c-di-GMP (not shown). To test specificity of c-di-GMP binding, we investigated the ability of unlabeled c-di-GMP or other unlabeled nucleotides to outcompete 2′-fluo-AHC-cdiGMP. We found that while unlabeled c-di-GMP provided in 250- or 500-fold molar excess outcompeted 2′-fluo-AHC-cdiGMP, several other nucleotides (ATP, GTP, cAMP, cGMP or c-di-AMP) provided in 500-fold molar excess did not (Fig. 2A). Therefore, BcsE binds c-di-GMP specifically.
Fig. 2.
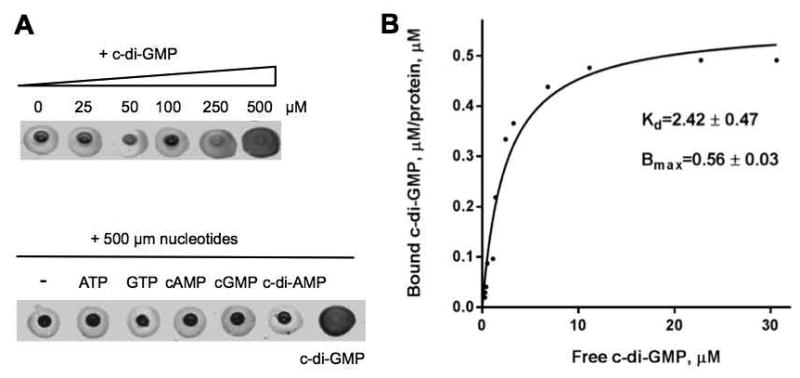
(A) 2′-fluo-AHC-cdiGMP-DRaCALA with purified MBP-BcsE. Upper panel, 2′-fluo-AHC-DRaCALA in the presence of increasing concentrations of unlabeled c-di-GMP as competitor. Lower panel, 2′-fluo-AHC-cdiGMP-DRaCALA in the presence of 500 μM nucleotides. (B) Saturation plot of equilibrium binding between c-di-GMP and MBP-BcsE. Plotted are concentrations of c-di-GMP in the chamber containing MBP-BcsE (bound c-di-GMP) versus the chamber lacking MBP-BcsE (free c-di-GMP).
Using equilibrium dialysis with unlabeled c-di-GMP, we estimated the dissociation constant, Kd, of MBP::BcsE for c-di-GMP to be 2.42 μM (Fig. 2B). This value is well within the physiologically relevant (sub-micromolar to low micromolar) range of intracellular c-di-GMP concentrations reported for various proteobacterial species (Römling et al., 2013). The maximum binding capacity of MBP::BcsE at saturation, Bmax, calculated based on the equilibrium dialysis experiments was 0.56 ± 0.03 (Fig. 2B). This Bmax value suggests that either two BcsE proteins bind a single molecule of c-di-GMP, or that a significant fraction of the protein is present in the form that is incapable of c-di-GMP binding. The latter possibility is plausible given the observed protein instability in solution. If true, then the actual Kd of BcsE may be lower than the observed value.
The DUF2819 domain of BcsE is involved in binding c-di-GMP
E. coli BcsE is composed of the 313-aa domain of unknown function, DUF2819 (PF10995 domain in the Pfam database; Punta et al., 2012), preceded by a 161-aa N-terminal fragment and followed by a 49-aa C-terminal fragment (Fig. 3A). Neither DUF2819 nor the N- and C-terminal fragments of BcsE show significant sequence similarity to any proteins of known function, other than BcsE homologs. Protein family searches using CD-Search, HHPred and GenThreader (McGuffin and Jones, 2003; Marchler-Bauer and Bryant, 2004; Söding et al., 2005) failed to identify any protein domains or 3D structures with statistically significant similarity to DUF2819, confirming that it represents a unique domain.
Fig. 3.
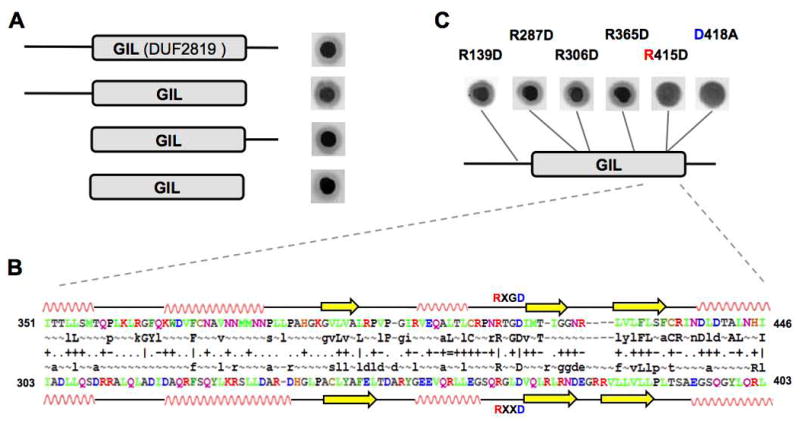
GIL is a novel c-di-GMP-binding domain. (A) Deletion analysis of the MBP-BcsE fusions. Shown are results of 2′-fluo-AHC-cdiGMP-DRaCALA with cell lysates expressing truncated BcsE derivatives. (B) Alignment of primary and secondary structures of the approximately 100-aa fragment of BcsE (top) and P. aeruginosa PelD (bottom). Residues (colored according to their properties) and secondary structures (spirals, α-helices; arrows, β-strands) are as predicted by HHPred (Soding et al., 2005). The RxGD and RxxD motifs conserved in the GIL domains and in the I-site of GGDEF domains and shown. (C) Identification of the RxGD motif at the C-terminal fragment of GIL as a c-di-GMP-binding site. Shown are results of 2′-fluo-AHC-cdiGMP-DRaCALA with cell lysates expressing MBP-BcsE point mutants.
To identify the minimal fragment responsible for c-di-GMP binding, we made several N- and C-terminal deletion constructs and tested E. coli lysates overexpressing MBP-fusions of these constructs for binding 2′-fluo-AHC-cdiGMP. We found that the DUF2819 domain alone was sufficient for c-di-GMP binding (Fig. 3A). We also noticed that the conserved 415RxGD418 motif of BcsE is similar to the c-di-GMP-binding RxxD motif present in the I-sites of many DGCs (Chan et al., 2004; Schirmer and Jenal, 2009) (Fig. 3B; see also Fig. S3 for fragments of the logos of the DUF2819 and GGDEF domains). Further, the predicted secondary structure around this RxGD site was found to be remarkably similar to the secondary structures of the I-sites in structurally characterized GGDEF domains (Fig. 3B). While this local similarity did not appear statistically significant (as scored by HHPred) to be taken as evidence of the common origin of DUF2819 and GGDEF domains, it prompted us to investigate the role of Arg415 and Asp418 of E. coli BcsE in c-di-GMP binding.
The RxGD motif of BcsE is required for c-di-GMP binding
To test the possibility that the RxGD motif of DUF2819 is involved in c-di-GMP binding, we performed site-directed mutagenesis of the Arg415 and Asp418 residues of this motif. According to 33P-c-di-GMP-DRaCALA, point mutations in Arg415 (R415D) or Asp418 (D418A) abolished c-di-GMP binding (Fig. 3C). To diminish the possibility that these mutations affected protein conformation nonspecifically, we mutated four additional Arg residues of BcsE. Arginines were chosen because these residues are most commonly involved in binding to negatively charged phosphates of c-di-GMP in all kinds of c-di-GMP receptors (Römling et al., 2013). The Arg residues were chosen both within and outside of the DUF2819 domain (R139D, R287D, R306D and R365D) (Fig. 3C). None of the mutations in the Arg residues outside of the RxGD motif impaired c-di-GMP binding (Fig. 3C) despite the fact that, according to SDS-PAGE, all mutants were expressed at comparable levels (Fig. S4A). This result strengthens the possibility that the RxGD motif of BcsE is involved in c-di-GMP binding.
The BcsE proteins from Enterobacteriacea bind c-di-GMP
To test whether BcsE proteins from species other than E. coli bind c-di-GMP, we overexpressed (as MBP-fusions) and purified DUF2819 domains from S. Typhimurium and Klebsiella pneumoniae (see Fig. S5 for multiple sequence alignment of the BcsE proteins from these species). The domain fusions from both species gave positive signals in 33P-c-di-GMP-DRaCALA (Fig. 4). Further, mutations in the Arg and Asp residues of the RxGD motif in the K. pneumoniae BcsE also impaired its c-di-GMP binding, despite the sequence divergence between the K. pneumoniae and E. coli proteins (Fig. 4). Note that the mutations did not significantly affect protein abundance (Fig. S4B). These data suggest that the BcsE proteins of Enterobacteriacea are c-di-GMP receptors, and by extension, that their function and mode of action are likely to be conserved. Notably, the RxGD motif is highly conserved in the DUF2819 domains not only from Enterobacteriacea but from the species beyond this group (Fig. S5). These results support the scenario that the RxGD motif is directly involved in c-di-GMP binding. We therefore designated the DUF2819 domain GIL for GGDEF I-site-like domain.
Fig. 4.
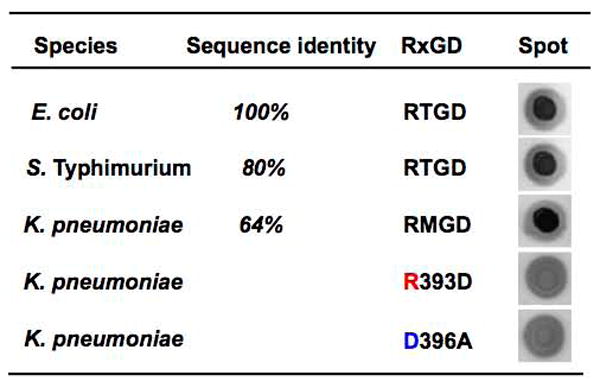
Conservation of the c-di-GMP-binding RxGD motif of the GIL domain in enterobacteria. Shown are results of 2′-fluo-AHC-cdiGMP-DRaCALA with E. coli cell lysates expressing MBP::GIL domain fusions from S. Typhimurium, K. pneumoniae as well as point mutants in the RxGD motif of the K. pneumoniae GIL domain.
BcsE is required for maximal cellulose expression
The bcsE gene is part of the bcsEFG operon, one of two operons responsible for cellulose synthesis in E. coli and S. Typhimurium (Zogaj et al., 2001; Solano et al., 2002). Insertional inactivation of the bcsE gene has been previously reported to affect biofilm formation in a clinical isolate of S. Typhimurium, i.e. the mutant colonies formed a fragile pellicle in LB and exhibited the bdar (brown, dry and rough) morphotype (Solano et al., 2002). More recently, deletion of the entire bcsEFG operon has been shown to abrogate cellulose synthesis in the cellulose producing E. coli strain (Serra et al., 2013). Identification of BcsE as a new c-di-GMP receptor warranted a reassessment of its role in cellulose production.
To investigate the role of BcsE, we created a conditional bcsE mutant in the chromosome of S. Typhimurium strain UMR1 by replacing the coding region of bcsE with the tetAR cassette (ΔbcsE101::tetAR). The outward promoters of the tetAR genes in the cassette can activate expression of downstream genes in the presence of the inducer, tetracycline, which relieves the polar effect of the tetAR cassette (Fig. 5A). We assessed cellulose production using Congo red, the dye that binds to cellulose fibers and curli fimbriae. While the wild-type strain UMR1 showed a rdar (red, dry and rough) colony morphotype indicative of cellulose and curli fimbriae production (Römling, 2005), the bcsE deletion mutant lacked red pigmentation in the absence of tetracycline (Fig. S6A). In the presence of tetracycline, the morphotype of the bcsE mutant was mainly bdar, but turned reddish after 48 h (Fig. 5B). To verify the bcsE mutant phenotype and detect even minor levels of cellulose, we tested binding of the fluorescent dye Calcoflour, which binds to 1,4-β-glucosides of cellulose but not to curli fimbriae. In the presence of tetracycline, the bcsE mutant showed greatly reduced, compared to strain UMR1, yet detectable Calcofluor binding thus confirming the requirement of bcsE for maximal cellulose production (Fig. 5B). Calcoflour binding increased in intensity from 24 to 48 h of growth, but did not reach wild type levels. Either the S. Typhimurium bcsE gene or the E. coli bcsE gene provided in trans could restore cellulose production in the bcsE mutant (Fig. 5B and data not shown). Quantification of cellulose levels in the UMR1 background confirmed highly reduced cellulose levels in the bcsE mutant and mutation complementation by bcsE provided in trans (Fig. S6C and data not shown). This shows that the defect in cellulose production in the mutant was due to the lack of bcsE, as opposed to a polar effect of the bcsE mutation.
Fig. 5.
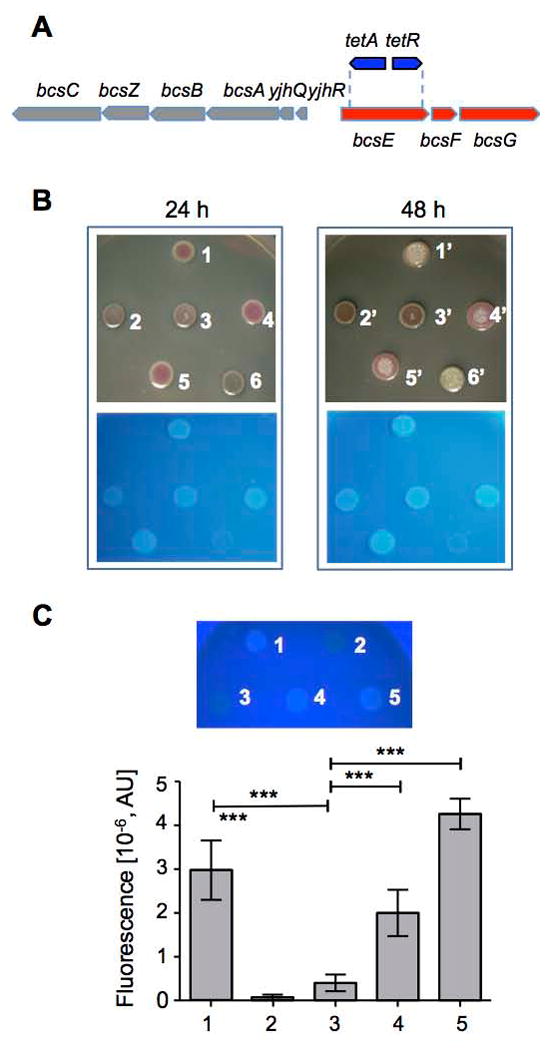
Characterization of BcsE function in vivo. (A) Organization of the bcs operons in E. coli and S. Typhimurium. The structure of the S. Typhimurium bcsE101::tetAR chromosomal mutant involving insertion of the tetAR gene cassette is also shown. (B) Requirement of BcsE for maximal cellulose production in S. Typhimurium UMR1. Rdar morphotype observed after growth on Congo red (top panels) and Calcofluor (lower panels) agar plates at 28°C for 24 and 48 h. Reduced cellulose levels in the bcsE mutant (spot 2) compared to the wild type (spot 1) can be complemented by expressing in trans the wild-type bcsE gene (spots 3 and 4), the bcsE R139D mutant (spot 5), but not the bcsE R415D mutant (spot 6). 1, UMR1 (pBAD28; pACYC184); 2, UMR1 bcsE101::tetAR (pBAD28); 3, UMR1 bcsE101::tetAR (pBcsE); 4, UMR1 bcsE101::tetAR (pMycBcsESt); 5, UMR1 bcsE101::tetAR (pMAL::BcsE R139D); 6, UMR1 bcsE101::tetAR (pMAL::BcsE R415D). (C) Quantification of cellulose expression in S. Typhimurium MAE97. Qualitative Calcofluor binding by agar-grown colonies of MAE97 and derivatives (top) and quantification of Calcofluor binding (bottom). The bcsE mutant showed reduced Calcofluor binding compared to the wild type, which could be complemented by bcsE in trans. 1, MAE97 (pBAD28; pSRKTc); 2, MAE52 ΔbcsA102 csgBA102 (pBAD28; pSRKTc); 3, MAE97 bcsE101::tetAR (pBAD28); 4, MAE97 bcsE101::tetAR (pBcsE); 5, MAE97 bcsE101::tetAR (pMycBcsESt). A representative experiment performed with seven technical replicates is shown. ***, p<0.0001.
To negate the effect of curli fimbriae completely, we constructed the bcsE mutation in strain S. Typhimurium MAE97 lacking csgBA encoding curli structural proteins. In addition, this strain produces temperature-independent high amounts of cellulose due to a mutation in the csgD promoter (Römling et al., 1998) in a designed pdar (pink, dry and rough) morphotype. In accordance to the results in UMR1, the bcsE deletion in strain MAE97 resulted in a nearly white phenotype on a Congo red plate (Fig. S6B) in the absence of tetracycline even after prolonged (72 h) growth. However, addition of tetracycline to the medium turned colonies first light and then deep pink and smooth and subsequently rough (Fig. S6B), which indicated that cellulose production was reduced, but not abolished as long as the bcsFG genes downstream of bcsE were expressed (Fig. 5A, S6B). Quantification of cellulose production in the MAE97 background after 24 h confirmed highly reduced cellulose levels in the bcsE mutant, which can be complemented by the S. Typhimurium and E. coli bcsE genes in trans (Fig. 5C and data not shown). Based on these observations, we conclude that bcsE is not essential for cellulose production in S. Typhimurium, however it is required for maximal cellulose production. This conclusion more precisely defines the role of bcsE in cellulose production compared to the conclusions reached earlier.
BcsE does not affect BcsA protein levels
Poly-β-1,6-N-acetylglucosamine synthase is activated and stabilized by protein-protein interactions upon c-di-GMP binding (Steiner et al., 2013). To investigate the possibility that a similar, proteolysis-based mechanism is involved in controlling cellulose synthesis, we tested the level of cellulose synthase BcsA protein in the bcsE mutant. To monitor BcsA, we tagged the bcsA gene with a 3xFLAG tag leading to expression of a BcsA-3XFLAG fusion protein. The strain expressing the BcsA-3xFLAG derivative produced cellulose similarly to UMR1 suggesting that the C-terminal 3xFLAG-tag did not negatively affect BcsA function (data not shown).
The level of BcsA-3xFLAG protein, assessed via Western blotting with the anti-FLAG antibody, in the bcsE deletion mutant in the absence of tetracycline was significantly decreased compared to the level of BcsA-3xFLAG in strain UMR1 (Fig. S7). However, BcsA was restored in the presence of tetracycline, which allowed expression of the genes downstream of bcsE (Fig. S7). This result indicates that BcsE does not significantly affect BcsA protein abundance, therefore the mechanism of regulation of cellulose production by BcsE remains unclear and will have to be elucidated in the future studies.
Cyclic d-GMP is essential for BcsE function in vivo
To test the role of c-di-GMP in BcsE function, we expressed in the S. Typhimurium bcsE mutant background the E. coli BcsE(R139D) and BcsE(R415D) proteins, the former of which is capable of c-di-GMP binding and the latter is not. We found that BcsE(R139D) complemented the bcsE mutation, whereas the BcsE(R415D) mutant did not. Furthermore, expression of BcsE(R415D) in the bcsE mutant lead to an additional decline in cellulose production from the already low level (Fig. 4B). These results suggest that c-di-GMP binding is important for BcsE function. Because activity of the cellulose synthase BcsA is also c-di-GMP-dependent (Ross et al., 1987), we could not assess the effect of c-di-GMP by analyzing cellulose levels in the S. Typhimurium strains with different intracellular c-di-GMP levels.
Discussion
Potentially biased view of c-di-GMP signaling in bacteria
Cyclic di-GMP differs from other mono- and dinucleotide bacterial second messengers (cAMP, cGMP, (p)ppGpp and c-di-AMP) by having a more diverse set of receptor/effector proteins. At present, our understanding of these receptors lags behind the understanding of enzymes involved in c-di-GMP synthesis and degradation. In most bacteria, including E. coli, the number of DGCs and PDEs, which are readily predictable by sequence analysis, by far exceeds the number of known c-di-GMP receptors, only a fraction of which can be predicted. This creates an appearance of a top-heavy signaling scheme (analogous to a dysfunctional army). Since such schemes are unprecedented, we expected that the lists of c-di-GMP receptors in most bacteria are likely incomplete, as is our knowledge about the functions of this second messenger. To test the hypothesis of potentially undiscovered c-di-GMP receptors in E. coli, we used DRaCALA (Roelofs et al., 2011) to screen the ASKA overexpression library (Kitagawa et al., 2005) and, as a result, we uncovered three new candidate c-di-GMP receptors, BcsE, IlvH and RimO. This and an earlier study (Corrigan et al., 2013) proved that DRaCALA is a powerful approach for a genome-wide identification of new ligand-binding proteins. To date, DRaCALA has been used scarcely. Below we discuss some of the lessons that we learned in applying this technique.
Caveats of library screening using DRaCALA
We first tested the performance of DRaCALA with a fluorescent c-di-GMP derivative, 2′-fluo-AHC-cdiGMP. While this compound worked well for testing c-di-GMP binding to known receptors, in a library screen it produces a high number of false positives. We envision at least two reasons responsible for this phenomenon. One reason concerns autofluorescence of some E. coli cell lysates observed even in the absence of added 2′-fluo-AHC-cdiGMP. A closer look at the autofluorescent spots revealed that many of them had a light-yellow color indicative of yellow pigments accumulated as a result of protein overexpression. One reason for this would be if overexpressed proteins contained yellow pigments, e.g., flavoproteins or proteins that form complexes with flavoproteins. This observation suggests that conjugating c-di-GMP to a fluorophore with a fluorescence emission outside of the emission of flavins would make such a compound more useful for library screening.
The second potential problem is likely associated with protein binding to the fluorescein and/or aminohexylcarbamoyl moieties of 2′-fluo-AHC-cdiGMP (Fig. S1). We suspect that the undesired protein binding to functional moieties on c-di-GMP derivatives is not unique to 2′-fluo-AHC-cdiGMP. Two recent studies pulled out potential c-di-GMP receptors from cell lysates of P. aeruginosa and S. Typhimurium (Düvel et al., 2012; Nesper et al., 2012) using nonfluorescent c-di-GMP derivatives bound to inert beads via linkers containing additional moieties. Numerous c-di-GMP-binding candidates have been reported but not verified. Although BcsE was found in these screens, our analysis of many other proteins pulled out using nonfluorescent c-di-GMP derivatives coupled to beads, failed to verify their c-di-GMP binding (not shown). Therefore, screening with c-di-GMP derivatives requires a lot of caution because of the high frequency of false positives.
DRaCALA screening with 33P-c-di-GMP proved to be much more specific compared to 2′-fluo-AHC-cdiGMP. However, as with all screening approaches, it has its own limitations. For example, 33P-c-di-GMP-DRaCALA failed to detect several known c-di-GMP receptors (Fig. 1B, 1C). We envision several possible reasons to account for this observation. The first reason is that a significant fraction of E. coli proteins of the ASKA library were not overexpressed at levels sufficient for detection with 33P-c-di-GMP as judged by the intensity of protein bands on SDS-PAGE (e.g., BcsA and four EAL domain PDEs, see Fig. 1B, 1C). Based on a random sample of the ASKA library clones (not shown), we estimate that only approximately 60-70% of clones would be reasonably expected to produce positive signals, if they were c-di-GMP receptors. Second, the lysis buffer applied in our screen may have been too stringent for preserving some receptor-c-di-GMP interactions. Here, we used buffer containing 300 mM NaCl. While decreasing salt concentration may have improved c-di-GMP receptor discovery, lower salt concentrations also produced more false-positives. For example, the YcgR R118D mutant produced positive signals in 100- or 200-mM (but not in 300-mM) NaCl lysis buffers. Third, proteins expressed in the insoluble form (inclusion bodies) may be incapable of c-di-GMP binding. Fourth, proteins that bind c-di-GMP at the homo- or heterodimer interfaces, may not be detectable by DRaCALA unless the homodimers are stable (e.g., the case of homodimeric V. cholerae VpsT [Fig. 1B]) or unless overexpression of one c-di-GMP-binding component induces overexpression of its partner protein (heterodimer PgaC-PgaD). Fifth, there may be limits in c-di-GMP binding affinities detectable by DRaCALA. For example, BdcA has low affinity for c-di-GMP (Kd, ∼11 mM [Ma et al., 2011]), which is possibly below the detection level of this assay. Furthermore, there are instances where the lack of c-di-GMP binding is difficult to explain, e.g., PnpA in our hands showed no c-di-GMP binding, whether E. coli lysates (Fig. 1B) or pure protein (not shown) were tested. This analysis leads us to conclude that more c-di-GMP receptors likely exist in E. coli K-12 than what we have uncovered.
GIL, a c-di-GMP-binding domain in Gammaproteobacteria and Betaproteobacteria
In this study, we focused on one of the newly identified c-di-GMP receptors, BcsE. This protein was found to bind c-di-GMP via the DUF2819 domain, which we designated GIL. The region in the vicinity of the apparent c-di-GMP-binding RxGD motif of the GIL domain has the same predicted secondary structure as the corresponding fragment of the GGDEF domains (Fig. 3B), and the RxGD motif is perfectly aligned with the RxxD motif involved in c-di-GMP binding in the I-sites of the GGDEF domains. Notably, the primary sequence similarity in this region is low (∼12.5 % identity). While striking, this similarity is not statistically significant. Therefore, it remains unclear whether it reflects a common origin of the two domains or is coincidental and reflective of convergent evolution. We anticipate that the knowledge of the three-dimensional structure of the GIL domain will help resolve this issue. What is clear, however, is that GIL represents only the second, after PilZ, protein domain dedicated specifically to c-di-GMP binding. However, unlike PilZ, that has a broad phylogenetic distribution, GIL has a relatively narrow distribution among the BcsE homologs of Gammaproteobacteria and Betaproteobacteria.
BcsE role in cellulose production
In this study, we clarified the role of the BcsE protein in cellulose synthesis. Gluconacetobacter xylinus and other Alphaproteobacteria produce cellulose via a single operon, bcsABCD (Wong et al., 1990; Saxena et al., 1990). In these organisms, the BcsAB proteins are sufficient for cellulose synthesis in vitro, and together with BcsC and BcsD, they comprise a complete cellulose synthase apparatus necessary for synthesis and secretion of cellulose across cytoplasmic and outer membranes (Ross et al., 1987; Morgan et al., 2013). BcsZ, a cellulose hydrolase, is an additional component needed to balance cellulose synthesis and hydrolysis (Standal et al., 1994). However, most Gammaproteobacteria contain two divergent bcs operons, yhjR-bcsQABZC and bcsEFG (Zogaj et al., 2001; Solano et al., 2002; Le Quere et al., 2009; Serra et al., 2013). The alphaproteobacterial and gammaproteobacterial cellulose gene clusters have recently been referred to as group A and group B, respectively (Jahn et al., 2011). Members of the families Vibrionaceae, Pseudomonadaceae and Aeromonadaceae (Gammaprotobacteria) and Burkholderiaceae, Comamonadaceae and Neisseriaceae (Betaprotobacteria) show a variety of gene arrangements of the yhjR-bcsQABZC and bcsEFG operons, or even a single operon, as in some Burkholderia spp. (Fig. S8). Still, all these operons display remarkably consistent gene content, and the bcsE gene is a constant feature in group B operons.
Deletion of bcsE has been suggested to abolish cellulose biosynthesis in S. Typhimurium and E. coli (Solano et al., 2002; Serra et al., 2013). However, we found that BcsE is actually not essential for cellulose synthesis, but it is required for maximal cellulose production in S. Typhimurium. Our results suggest that BcsE affects the temporal initiation and abundance of secreted cellulose fibers. In addition, the occurrence of the deep pink and smooth morphotype of the MAE97 bcsE mutant, which subsequently turned rough (Fig. S6B) suggests that the structure of the cellulose fibers may be altered in the absence of BcsE. However, the mechanism of BcsE action has yet to be deciphered. Importantly, according to our analysis (Fig. 5), c-di-GMP binding to the GIL domain plays a critical role in enabling BcsE activity.
It is noteworthy that BcsA, the glycosyltransferase involved in cellulose synthesis, is a c-di-GMP-dependent enzyme itself (Ross et al., 1987; Ryjenkov et al., 2006; Pultz et al., 2012). It therefore appears that cellulose synthesis in Enterobacteriaceae and probably other BcsE-containing bacteria, involves c-di-GMP-dependent control at two levels, i.e., BcsE and BcsA. It is noteworthy that another exopolysaccharide common to Enterobacteriaceae, poly-N-acetyl-D-glucosamine, is also regulated at two levels, one involves proteolytic instability of the PgaD protein in the absence of c-di-GMP, another involves c-di-GMP-dependent regulation of glycosyltransferase activity (Steiner et al., 2013). We wonder whether a two-tiered c-di-GMP-dependent control may be necessary to ensure an orderly transition between the motile planktonic bacterial state and the sessile, surface-attached state. Since c-di-GMP concentrations in planktonic bacteria change very quickly by as much as several-fold (Russell et al., 2013), an insurance against premature exopolysaccharide production, which could commit cells to switching to the sessile lifestyle seems reasonable. We anticipate that new insights into the physiological significance of the two-tiered c-di-GMP-mediated regulation of exopolysaccharide production will emerge from the better understanding of BcsE function.
Experimental Procedures
Strains, plasmids and growth conditions
E. coli and S. Typhimurium strains (Table 1) were cultured in liquid or solid Luria-Bertani (LB) media (Sambrook et al., 1989) at 37 °C, unless indicated otherwise, with appropriate antibiotic supplementation for plasmid maintenance (100 μg/mL ampicillin; 25 μg/mL chloramphenicol). To induce read-out from the tetR promoter, 15 μg/mL tetracycline was added to the medium.
Table 1.
Strains and plasmids used in this study.
| Strain / plasmid | Description | Reference |
|---|---|---|
| E. coli | ||
| AG1 | ASKA collection host (T5 Pol) | Kitagawa et al., 2005 |
| BL21[DE3] | Strain for protein overexpression (T7 Pol) | NEB |
| DH5α | Strain for plasmid maintenance and overexpression of MBP-protein fusions | NEB |
| XL-Blue | Strain for plasmid maintenance | Stratagene |
| S. enterica ser. Typhimurium | ||
| UMR1 | ATCC1 14028 Nalr, cellulose28°C, curli fimbriae28°C | Römling et al., 1998 |
| MAE97 | UMR1 pcsgD1 ΔcsgBA102, cellulose28/37°C | Römling et al., 2000 |
| MAE299 | UMR1 ΔbcsA102 | Grantcharova et al., 2010 |
| MAE775 | UMR1 ΔbcsA102 csgBA::Km | Grantcharova et al., 2010 |
| MAE777 | MAE52 ΔbcsA102 csgBA::Km | Grantcharova et al., 2010 |
| MAE1261 | MAE97 bcsAΔStop::tetRA | This study |
| MAE1264 | MAE97 BcsA-3xFLAG | This study |
| MAE1268 | MAE97 bcsA104::tetRA | This study |
| MAE1574 | UMR1 BcsA-3xFLAG | This study |
| MAE2100 | UMR1 bcsE101::tetAR | This study |
| MAE2103 | MAE97 bcsE101::tetAR | This study |
| MAE2101 | MAE1574 bcsE101::tetAR | This study |
| Plasmids | ||
| pACYC184 | Tetr vector | NEB |
| pBAD/Myc-HisB | Arabinose-inducible expression vector; Para, Apr | Invitrogen |
| pBAD28 | Arabinose-inducible expression vector; Para, Apr, Cmr | Guzman et al., 1995 |
| pMycBcsEEc | pBAD/Myc-HisB::bcsE from E. coli MG1655 | This study |
| pMycBcsESt | pBAD/Myc-HisB::bcsE from S. Typhimurium | This study |
| pBcsE | pBAD28::bcsE-6xHis from S. Typhimurium | This study |
| pET23a | His tag protein overexpression vector; PT7, Apr | Novagen |
| pET::bcsE1 | pET23a::bcsE from E. coli MG1655 | This study |
| pET::bcsE2 | pET23a::bcsE (153-523 aa) | This study |
| pET::bcsE3 | pET23a::bcsE (153-492 aa) | This study |
| pET::bcsE4 | pET23a::bcsE (1-492 aa) | This study |
| pET::ycgR | pET23a::ycgR from E. coli MG1655 | Ryjenkov et al., 2006 |
| pET::ycgR(R118D) | pET23a::ycgR R118D | Ryjenkov et al., 2006 |
| pEXT20::clp | pEXT20::clp from X. axonopodis | Leduc and Roberts, 2009 |
| pMAL-c2x | MBP fusion overexpression vector; Plac, Apr | NEB |
| pMAL::bcsE | pMAL-c2X::bcsE from E. coli MG1655 | This study |
| pMAL::bcsE(R139D) | pMAL-c2X::bcsE R139D from E. coli | This study |
| pMAL::bcsE(R287D) | pMAL-c2X::bcsE R287D from E. coli | This study |
| pMAL::bcsE(R306D) | pMAL-c2X::bcsE R306D from E. coli | This study |
| pMAL::bcsE(R345D) | pMAL-c2X::bcsE R345D from E. coli | This study |
| pMAL::bcsE(R415D) | pMAL-c2X::bcsE R415D from E. coli | This study |
| pMAL::bcsE(D418A) | pMAL-c2X::bcsE D418A from E. coli | This study |
| pMAL::bcsE(GIL) | pMAL-c2X::bcsE (152-523aa) | This study |
| pMAL::KBbcsE | pMAL-c2X::bcsE from K. pneumoniae ATCC 700721 | This study |
| pMAL::KBbcsE(R393D) | pMAL-c2X::bcsE R393D from K. pneumoniae | This study |
| pMAL::KBbcsE(D396A) | pMAL-c2X::bcsE D396A from K. pneumoniae | This study |
| pMAL::STMbcsE | pMAL-c2X::bcsE from S. Typhimurium | This study |
| pMAL::vpsT | pMAL-c5x::vpsT from V. cholerae | Krasteva et al., 2010 |
| pSRKTc | Tetr vector | Khan et al., 2008 |
| pSUB11 | 3xFLAG expressing vector | Uzzau et al., 2001 |
, American Type Culture Collection.
DRaCALA
The E. coli AG1 strains of the ASKA library were inoculated into 96-well plates in fresh LB medium supplemented with chloramphenicol and grown on a rotation shaker (180 rpm) at 25 °C for 10 h. Protein overexpression was induced for 4 h with isopropyl-β-D-thiogalactopyranoside (IPTG, final concentration, 1 mM). Cells were collected by centrifugation, resuspended in a c-di-GMP binding buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM EDTA, 10% glycerol, pH 7.5) containing 100 μg/mL lysozyme (Qiagen), incubated for 1 h and subjected to two freeze-thaw cycles. Cell lysates were subsequently stored at -80 °C.
After thawing, cell lysates (20 μl) were mixed with fluorescently or radioisotope-labeled c-di-GMP, incubated for 20 min, and a fraction (2 μl) of these mixtures were spotted on nitrocellulose membrane (BioRad) using an eight-channel pipette. 2′-fluo-aminohexylcarbamoyl-c-di-GMP (2′-fluo-AHC-cdiGMP) manufactured by Biolog (Germany) was purchased from Axxora, Inc.
33P-c-di-GMP was prepared in house using α33P-GTP (Perkin Elmer) as substrate and a DGC from Synechocystis sp. PCC 6803, Slr1143, purified as described earlier from E. coli DH5α (pMslr) (Ryjenkov et al., 2005). The reaction mixture (Slr1143 [2 μM] and α33P-GTP [25 μM]) was incubated for 6 h at 37°C, boiled at 100 °C for 5 min, and denatured proteins were removed by centrifugation. This mixture containing approximately 90% 33P-c-di-GMP was used without further purification.
After spotting, membranes were allowed to dry. Subsequently, they were either scanned for fluorescence (2′-fluo-AHC-cdiGMP) using a GE Typhoon FLA 9500 scanner (λexc, 488 nm, λemi, 520 nm) or exposed to X-ray film (33P-c-di-GMP) for 2 days prior to film development. Fluorescence intensities of the spots were quantified by ImageQuant 5.2 (Molecular Dynamics), where necessary.
Protein overexpression and purification
The bcsE genes were amplified from genomic DNA of E. coli MG1655, S. Typhimurium LT2 and Klebsiella pneumoniae ATCC 700721. The fragments were subsequently cloned into the pET23a (Novagen) and pMAL-c2x (NEB) vectors. The BcsE::His6 fusion protein was expressed and purified as previously reported (Ryjenkov et al., 2006). Briefly, IPTG (0.3 mM, final concentration) was added to exponentially growing (A600, 0.6) strain BL21[DE3] (Novagen) containing the pET::bcsE plasmid. After 10 h of induction at 18 °C, cells were pelleted and resuspended in a buffer containing 300 mM NaCl, 50 mM NaH2PO4 (pH 7.4) and 5% glycerol as well as protease inhibitors (phenylmethylsulfonyl fluoride and P8465; Sigma). Cells were disrupted using a French pressure minicell (Spectronic Instruments), followed by brief ultrasonification (Sonifier 250; Branson). Crude cell extracts were centrifuged at 35,000 x g for 25 min, and the supernatant was loaded onto the Co2+ resin (Pierce) for affinity purification.
The MBP-BcsE fusion proteins were expressed in XL-Blue (Stratagene) containing appropriate pMAL::bcsE plasmids. Cells were grown to A600, 0.6, after which IPTG (final concentration, 0.5 mM) was added. Cells were collected after an overnight cultivation at room temperature. The cell pellet was resuspended in buffer containing 200 mM NaCl, 25 mM Tris (pH 8.0), 0.5 mM EDTA and 5% glycerol and disrupted as described above. MBP-BcsE was purified by affinity chromatography on amylose resin (NEB).
Purified proteins were desalted using Zeba spin desalting columns (7-kD cut-off, Pierce), which were pre-equilibrated with a c-di-GMP-binding buffer. Protein purity was assessed by SDS-PAGE. Protein concentration was measured using a Bradford protein assay kit (Biorad).
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange II site-directed mutagenesis kit according to the manufacturer's instructions (Agilent Technologies). All mutations were confirmed by DNA sequencing.
Equilibrium dialysis
Protein-c-di-GMP binding was examined by equilibrium dialysis in Dispo-Biodialyzer cassettes (The Nest Group) as previously reported (Ryjenkov et al., 2006). Nucleotide concentrations were quantified by HPLC as described earlier (Ryjenkov et al., 2006). Dissociation constants, Kd, were calculated by the GraphPad Prism software, version 5.0 (GraphPad Software) using a nonlinear regression model.
Construction of a BcsA-3xFlag strain
One-step gene inactivation (Datsenko and Wanner, 2000) and the tetRA cassette (Karlinsey, 2007) was used to create a C-terminal BcsA-3xFLAG fusion protein by scar-less insertion of the sequence encoding 3xFLAG upstream of the stop codon of the bcsA gene leading to a non-polar construct. In the first step, the stop codon of bcsA was replaced with the tetRA element which provides tetracycline resistance (TetR). TetR clones carrying a correct insertion were purified and transformed with DNA fragments encoding 3xFLAG amplified from pSUB11 (Table 1) flanked by sequences up- and downstream of the bcsA stop codon. TetS agar (Karlinsey, 2007) was used to select for clones where tetRA was replaced by the 3xFLAG fragment and candidate clones were purified at least twice at 42°C. Correct construction of the bcsA-3xFLAG strain was verified with PCR and sequencing for different clones and the cellulose expression phenotype was tested.
Construction of a bcsE deletion mutant
Construction of chromosomal deletions of bcsE was performed by replacement of the gene with a tetRA cassette using one-step gene inactivation (Datsenko and Wanner, 2000). Tn10dTc (Karlinsey, 2007) was used as a template. In brief, bcsE was deleted by replacing its ORF, except 40 nucleotides in the beginning and the end, with the tetRA cassette in the tetAR direction. All constructed mutants were verified with PCR. To restore expression of the genes downstream of bcsE in the bcsE101 mutant, 15 μg/mL tetracycline was added to the medium.
Assessment of cellulose levels
To assess cellulose levels qualitatively, S. Typhimurium UMR1, MAE97 and their derivatives (Table 1) were cultivated on LB agar lacking NaCl supplemented with Congo red dye (final concentrations, 40 μg/mL Congo red and 20 μg/mL Coomassie Brilliant Blue) or on Calcofluor (final concentration, 50 μg/mL) agar plates. Colony morphotypes were assessed after growth at 37°C for 24-72 h for MAE97 derivatives, and after growth at 28°C for 24-48 h for UMR1 derivatives.
Cellulose production was quantified as described (Pultz et al., 2012) with minor modifications. In brief, a black 96 well microtiter plate with glass bottom (Greiner) was inoculated with 200 μL LB without salt agar containing 50 μg/mL Calcofluor and supplemented with 0.01% L-arabinose, 15 μg/mL tetracycline and 100 μg/mL ampicillin. Eight μL of a bacterial suspension (A600, 0.1) taken from an overnight culture on a LB without salt agar plate was added into each well and the plate was incubated for 24 h at 37°C. The emission intensity at 460 nm was read after excitation at 355 nm in a multilabel reader (VICTORTM X3, Perkin Elmer). Statistical analysis was performed via an unpaired t-test with two-tailed P-value (*** is p<0.0001) using Prism 5 (GraphPad Software).
BcsA-3xFLAG detection
Five mg of cell biomass grown at 28 °C overnight on LB agar lacking NaCl with appropriate antibiotics was mixed with a 200 μl volume of urea buffer (8 M urea, 2% SDS, 11% glycerol, 62.5 mM Tris-HCl, pH 6.8), and sonicated 4 times (10 sec, 3.5 amplitude-microns) on ice. Samples were stored at -20°C. Cell lysates were subjected to SDS-PAGE separation, and proteins were subsequently transferred onto a PVDF membrane (Millipore). BcsA-3xFLAG was detected by using an anti-FLAG-tag antibody (1:1500; Sigma) and a horseradish peroxidase-conjugated goat-anti-mouse antibody (1:7500; Jackson Immunoresearch).
Bioinformatic analyses
The sequences of the N- and C-terminal fragments of E. coli BcsE and its central GIL (DUF2819) domain were compared against the nonredundant protein database at the NCBI and UniProtKB using PSI-BLAST (Altschul et al., 1997) and JackHMMer (Finn et al., 2011), respectively. The secondary structure of the GIL domain was predicted using Jpred3 (Cole et al., 2008). Comparisons of the GIL domain to the libraries of known protein domains and 3D structures were performed using CD-Search (Marchler-Bauer and Bryant, 2004), HHPred (Söding et al., 2005) and GenThreader (McGuffin and Jones, 2003) with default parameters.
Supplementary Material
Acknowledgments
We are thankful to Romy Reimann and Uwe Remminghorst for strain construction, Fitnat Yildiz and Gary Roberts for the VpsT and Clp overexpression plasmids, respectively, Hermann Schätzl for access to Typhoon, Galina Selivanova for the use of the multilabel reader, and Kurt Miller for manuscript proofreading. This study was supported by the United States National Science Foundation (MCB1052575 to MG), NIH Intramural Research Program at the National Library of Medicine (MYG) and the Swedish Research Council Natural Sciences (621-2010-5755), Petrus and Augusta Hedlund Foundation and Carl Tryggers Foundation CTS 10:324 (to UR). Andrea Blanka was supported by the International Research Training Group 1273 funded by the German Research Foundation (DFG).
Footnotes
The authors have no conflict of interest to declare.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Barends TR, Hartmann E, Griese JJ, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Baratto A, Dorel C, Landini P. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol. 2006;188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel J, Bertinetti D, Möller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jänsch L, Herberg FW, Häussler S. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Meth. 2012;88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang B, Fang X, Means WJ, McCormick RJ, Gomelsky M, Zhu MJ. c-di-GMP signaling regulates E. coli O157:H7 adhesion to colonic epithelium. Vet Microbiol. 2013;164:344–351. doi: 10.1016/j.vetmic.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Jahn CE, Selimi DA, Barak JD, Charkowski AO. The Dickeya dadantii biofilm matrix consists of cellulose nanofibres, and is an emergent property dependent upon the type III secretion system and the cellulose synthesis operon. Microbiology. 2011;157:2733–2744. doi: 10.1099/mic.0.051003-0. [DOI] [PubMed] [Google Scholar]
- Karlinsey JE. Lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Meth Enzymol. 2007;421:199–209. doi: 10.1016/S0076-6879(06)21016-4. [DOI] [PubMed] [Google Scholar]
- Khan SR, Gaines J, Roop RM, 2nd, Farrand SK. Broad-host-range expressionvectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol. 2008;74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey MM, Partridge JD, Green J. Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiol. 2010;156:2873–2886. doi: 10.1099/mic.0.037887-0. [DOI] [PubMed] [Google Scholar]
- Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quere B, Ghigo JM. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol Microbiol. 2009;72:724–740. doi: 10.1111/j.1365-2958.2009.06678.x. [DOI] [PubMed] [Google Scholar]
- Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013;32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Zhang G, Wood TK. Escherichia coli BdcA controls biofilm dispersal in Pseudomonas aeruginosa and Rhizobium meliloti. BMC Res Notes. 2011;4:447. doi: 10.1186/1756-0500-4-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteom. 2012;75:4874–4878. doi: 10.1016/j.jprot.2012.05.033. [DOI] [PubMed] [Google Scholar]
- Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povolotsky TL, Hengge R. ‘Life-style’ control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J Biotechnol. 2012;160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol. 2012;86:1424–40. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucl Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- Römling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cel Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Rohde M, Olsen A, Normark S, Reinköster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Sierralta WD, Eriksson K, Normark S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol. 1998;28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Russell MH, Bible AN, Fang X, Gooding J, Campagna S, Gomelsky M, Alexandre G. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway promotes sensory adaptation. mBio. 2013;4:e00001–13. doi: 10.1128/mBio.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Römling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual. 2nd. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saxena IM, Lin FC, Brown RM., Jr Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990;15:673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013;195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Remminghorst U, Ahmad I, Zakikhany K, Römling U. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol. 2009;191:3928–3937. doi: 10.1128/JB.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Standal R, Iversen TG, Coucheron DH, Fjaervik E, Blatny JM, Valla S. A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J Bacteriol. 1994;176:665–672. doi: 10.1128/jb.176.3.665-672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Lori C, Boehm A, Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2013;32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschowri N, Busse S, Hengge R. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 2009;23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol. 2011;407:633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica Serovar Typhimurium. J Bacteriol. 2011;193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Hatamoto Y, Kutsukake K. Functional and expressional analyses of the anti-FlhD4C2 factor gene ydiV in Escherichia coli. Microbiol. 2012;158:1533–1542. doi: 10.1099/mic.0.056036-0. [DOI] [PubMed] [Google Scholar]
- Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, et al. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


