SUMMARY
Centrioles are 9-fold symmetric structures duplicating once per cell cycle. Duplication involves self-oligomerization of the centriolar protein SAS-6, but how the 9-fold symmetry is invariantly established remains unclear. Here, we found that SAS-6 assembly can be shaped by preexisting (or mother) centrioles. During S phase, SAS-6 molecules are first recruited to the proximal lumen of the mother centriole, adopting a cartwheel-like organization through interactions with the luminal wall, rather than via their self-oligomerization activity. The removal or release of luminal SAS-6 requires Plk4 and the cartwheel protein STIL. Abolishing either the recruitment or the removal of luminal SAS-6 hinders SAS-6 (or centriole) assembly at the outside wall of mother centrioles. After duplication, the lumen of engaged mother centrioles becomes inaccessible to SAS-6, correlating with a block for re-duplication. These results lead to a proposed model that centrioles may duplicate via a template-based process to preserve their geometry and copy number.
INTRODUCTION
Centrioles are composed of microtubules invariably organized in a radial 9-fold symmetry. The 9-fold symmetry is widely thought to derive from a geometric scaffold known as the cartwheel, which is characterized by a central hub from which nine spokes emanate (Anderson and Brenner, 1971). The cartwheel is present at the proximal lumen of centrioles, coincident with several centriolar proteins including SAS-6 (Nakazawa et al., 2007), STIL/SAS-5 (Stevens et al., 2010), CPAP (Kleylein-Sohn et al., 2007) and CEP135 (Kleylein-Sohn et al., 2007). SAS-6, in particular, has been shown to form the primary backbone of the cartwheel (Kitagawa et al., 2011; van Breugel et al., 2011); it exists as a dimer, and can self-oligomerize via an N-terminal head domain forming a ring resembling the central hub, and C-terminal tails pointing outwards as spokes (Kitagawa et al., 2011; van Breugel et al., 2011). Biochemical and structural studies, however, revealed some flexibility in the dimer structure and a relatively weak interaction interface between the N-terminal head domains (Kitagawa et al., 2011; van Breugel et al., 2011), allowing SAS-6 dimers to adopt variable oligomeric conformations in addition to nine dimers (Cottee et al., 2011; Kitagawa et al., 2011; van Breugel et al., 2011). As such, it is unclear how invariant 9-fold symmetry is achieved (Cottee et al., 2011).
“Self-assembly” as the prevailing model for centriole biogenesis is also supported by the observation that centrioles can form in the absence of preexisting centrioles, in a process known as “de novo assembly” (Azimzadeh et al., 2012; Khodjakov et al., 2002; Szollosi et al., 1972; Vladar and Stearns, 2007). The number of centrioles formed through the de novo pathway is highly variable, posing a grave risk for dividing cells that require strict control over centriole numbers to maintain genomic stability (Ganem et al., 2009) and proper cilia function (Mahjoub and Stearns, 2012). Thus, de novo assembly is normally inhibited in cycling cells (La Terra et al., 2005), where canonical duplication dominates. It is unclear whether or not canonical duplication and de novo assembly initiate centriole assembly through the same mechanism, but in either case, “symmetry-ensuring” activities are required to guide proper self-oligomerization of SAS-6 into a precisely 9-fold symmetrical structure (Cottee et al., 2011). The nature of such symmetry-ensuring activities for both cases, however, is unknown.
Unlike de novo assembly, in cycling cells, new centrioles are born in close proximity to a preexisting (mother) centriole, where the accumulation of SAS-6 (oligomers) at the side of mother centrioles is thought to mark the beginning of centriole assembly (Strnad et al., 2007). Interestingly, in vertebrate cycling cells, before newborn centrioles are transformed to mother centrioles, their cartwheel structures are lost from the proximal lumen (Vorobjev and Chentsov, 1980; Vorobjev and Chentsov Yu, 1982). Cartwheel removal occurs during mitosis (Arquint and Nigg, 2014), and SAS-6 and STIL are further eliminated by the proteasome-mediated degradation (Arquint and Nigg, 2014; Strnad et al., 2007). These “cartwheel-less” centrioles have an empty proximal lumen, but retain their 9-fold symmetry, and are active in supporting duplication, suggesting that they may contain the “symmetry-ensuring” activity for SAS-6 assembly.
RESULTS
SAS-6 is transiently recruited to the proximal lumen of mother centrioles in early S phase
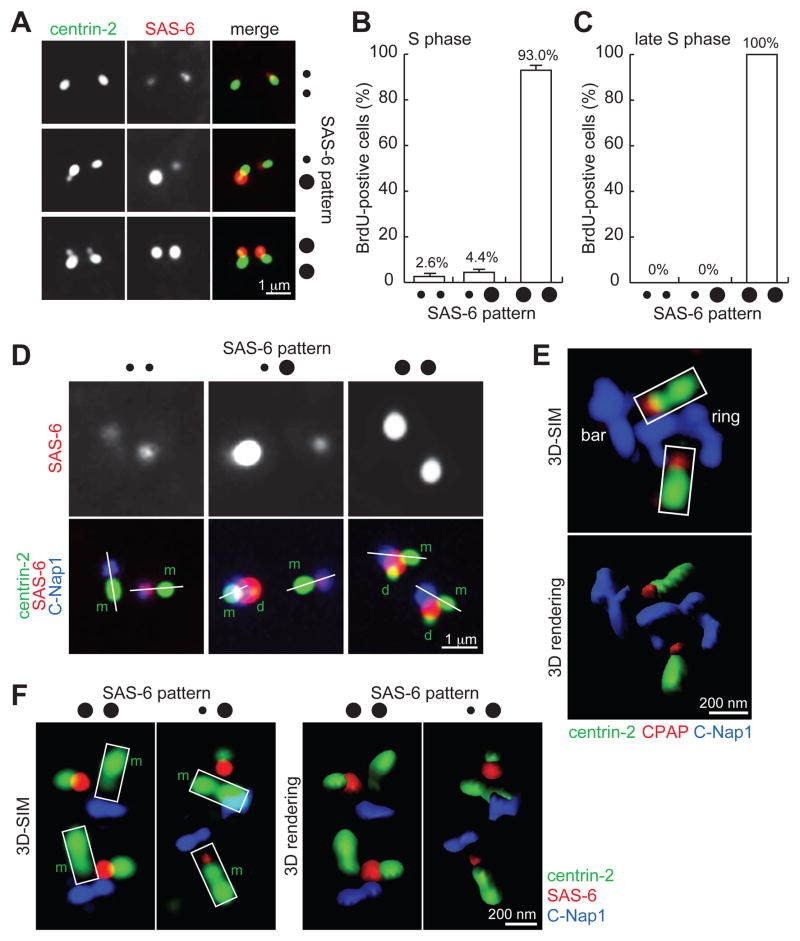
To understand how a mother centriole supports the assembly of a new centriole, we revisited SAS-6 recruitment during centriole duplication. In unsynchronized cells transiently labeled with BrdU, we noticed three distinct localization patterns of SAS-6 during S phase (Figure 1A). In addition to the previously documented pattern of two bright SAS-6 foci in most of cells (Strnad et al., 2007) (••; 93.0%), we also found in a small fraction of S-phase cells displaying one bright and one weak SAS6 foci (••; 4.4%), and even less frequently, those with two weak SAS-6 foci (••; 2.6%) (Figure 1B). The bright SAS6 foci were detected only on duplicated centrioles (doublets), while the weak foci were found exclusively on unduplicated centrioles (singlets) (Figure 1A). Due to the low representation of the weak SAS-6 foci in the population, we asked whether they were a stochastic anomaly or represented a transient, early stage of centriole duplication. SAS-6 localization patterns were analyzed in cells that had entered S phase for at least 4 hours, as revealed by pulse/chase BrdU labeling. All late S-phase cells were found to carry two bright SAS-6 foci (••; 100%) (Figure 1C), strongly suggesting that a small amount of SAS-6 is recruited to mother centrioles in early S-phase, prior to the formation of new centrioles.
Figure 1. SAS-6 is transiently recruited to the proximal lumen of mother centrioles in early S-phase.
(A) U2OS cells in S phase were processed for immunofluorescence to visualize centriole (centrin-2) and SAS-6.
(B) Quantification of cells with weak SAS-6 foci in S phase. Error bars, standard deviation. n>80, N=3.
(C) Quantification of weak SAS-6 foci in late S phase. n>100, N=3.
(D) Weak SAS-6 localizes to the proximal end of mother centrioles. Linear alignment of centrin-2 (distal-end component) and C-Nap1 (proximal end) is marked by a white line. Here, rabbit antibodies recognizing the C-terminus of C-Nap1 were used with a mouse anti-SAS-6 antibody. m, mother; d, daughter.
(E) 3D-SIM image of luminal components CPAP and centrin-2 in comparison to non-luminal component C-Nap1. Here, a mouse antibody recognizing the N-terminus of C-Nap1 was used with a rabbit anti-CPAP antibody. Imaris 3D rendering is presented as indicated.
(F) 3D-SIM images of SAS-6, C-Nap1, and centrin in S-phase centrioles stained with the antibodies indicated. m, mother.
All S-phase cells in (A), (B), (C), (D) and (F) were identified by BrdU labeling (see Experimental Procedures), not shown.
To define the specific localization of SAS-6 within centrioles, we examined the localization of SAS-6 relative to the proximal-end marker C-Nap1 (Fry et al., 1998; Mayor et al., 2000), and distal-end marker centrin. Consistent with previous reports (Strnad et al., 2007), bright SAS-6 foci were always tethered to the side of mother centrioles, as revealed by the non-linear alignment of the centrin, SAS-6, and C-Nap1 signals (Figure 1D). Strikingly, the majority of weak SAS-6 foci were present at the proximal end of mother centrioles, situated in between centrin and C-Nap1 foci in a linear fashion (Figure 1D). To determine whether the weak SAS6 foci reside in the proximal lumen of mother centrioles, we examined their localization by super-resolution 3D structured illumination (3D-SIM) microscopy. Unlike conventional fluorescence microscopy, where C-Nap1 was presented as a solid point (Figure 1D), C-Nap1 was resolved into a ring (top view) or a bar (side view) when analyzed with 3D-SIM (Figure 1E and 1F). Note that using an antibody against the N-terminal domain (Figure 1E), C-Nap1 was visualized as a larger ring/bar compared to that by C-terminal antibodies (Figure 1F). This is in agreement with previous immuno-electron microscopy studies showing that C-Nap1 is a non-luminal proximal-end component of centrioles (Fry et al., 1998; Mayor et al., 2000). Importantly, luminal components such as CPAP and centrin were found to localize above the void zone of the C-Nap1 ring or in the middle region atop the C-Nap1 bar (Figure 1E). Moreover, unlike the strong SAS-6 focus present clearly outside of the mother centriole (98%, n=58, Figure 1F), nearly all weak SAS-6 foci were located at the space between centrin and C-Nap1 in the mother centriole lumen (97%, n=58, Figure 1F), a pattern similar to that of CPAP. Together, we conclude that SAS-6 is first recruited to the proximal lumen of the mother centriole during early S phase.
SAS-6 adopts a cartwheel-like organization at mother centrioles independently of its self-oligomerization activity
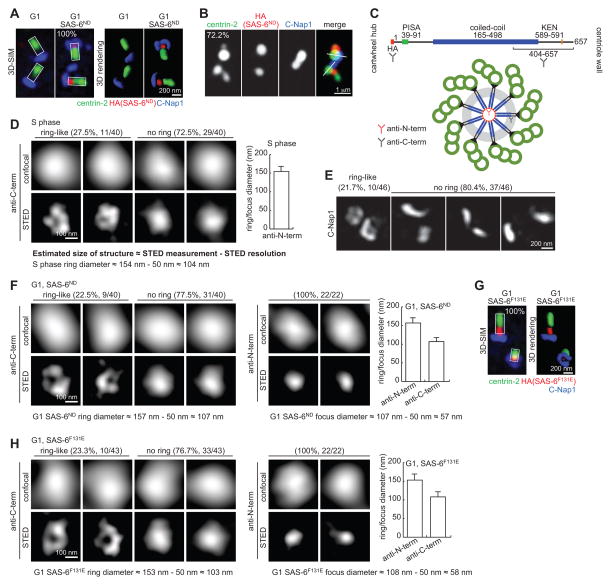
Luminal SAS-6 accumulates only transiently in S phase, suggesting that an activity in S phase efficiently promotes SAS-6 release from the lumen. To investigate the luminal recruitment of SAS-6 independent of other S-phase activities, an inducible cell line was created in which SAS-6 could be ectopically expressed in G1 phase. SAS-6 is normally degraded in G1 by APC/Ccdh1, but a mutation of the degron (KEN box) renders it non-degradable (Strnad et al., 2007), referred to here as SAS-6ND. To assess whether SAS-6ND could localize to G1 centrioles, retinal pigment epithelial (RPE-1) cells that had been arrested in G0/G1 phase by serum starvation were induced to mildly express SAS-6ND through an inducible promoter, and subjected to 3D-SIM analysis. Intriguingly, newly synthesized SAS-6ND stably localized to the proximal lumen of all G1-arrested centrioles (Figure 2A; 100%). Moreover, when cells were released into S phase, SAS-6ND was efficiently removed from the lumen and became externally attached (Figure 2B), confirming the presence of S-phase activities that release luminal SAS-6. This assay recapitulates and temporally separates the two SAS-6 localization steps seen in wild-type centrioles during early S phase.
Figure 2. G1 centrioles can recruit and organize SAS-6 into an ordered structure similar to the cartwheel.
(A) RPE-1 cells arrested in G1 for 48 hours followed by expression of HA-tagged SAS-6ND were imaged by 3D-SIM. n>45, N=3.
(B) G1-arrested RPE-1 cells expressing SAS-6ND as described in (A) were released into S phase by serum reintroduction and stained with the indicated antibodies. S-phase cells were identified by BrdU labeling, not shown. n>21, N=3.
(C) Schematic diagram of domain organization of SAS-6 and the epitopes recognized by the antibodies used in STED analyses.
(D) Representative STED images of SAS-6 staining with anti-C-term antibody on normal S-phase centrioles were shown. Graph presents STED measurements of ring/focus diameter by averaging the long and short axes. Actual size of the structure is estimated by subtracting STED resolution from STED measurements as indicated. Error bar, standard deviation. n=40.
(E) Frequency of observing C-Nap1 ring under 3D-SIM. n=46.
(F) STED imaging of SAS-6ND stained with anti-C-term and anti-N-term antibodies. Actual size of the structure is estimated as indicated. Error bars, standard deviation. n as indicated.
(G) SAS-6F131E localizes to the proximal lumen of G1 centrioles. 3D-SIM image of G1-arrested RPE-1 cells inducibly expressing HA-tagged SAS-6F131E as described in (A). n>39, N=3.
(H) STED imaging of SAS-6F131E stained with anti-C-term and anti-N-term antibodies. Actual size of the structure is estimated as indicated. Error bars, standard deviation. n as indicated.
To examine the organization of SAS-6ND at the proximal lumen of G1 centrioles, super-resolution stimulated emission depletion (STED) microscopy at a resolution of ~50 nm was used (Donnert et al., 2007; Hell and Wichmann, 1994; Klar et al., 2000). We first examined the cartwheel structure of S-phase centrioles using a monoclonal antibody against the C-terminal tail of SAS-6 (Figure 2C). In these experiments, and depending on the orientation of the centrioles imaged, the C-termini of SAS-6 should occasionally be resolved into a ring under STED imaging. Indeed, 27.5% of SAS-6 signals in S-phase centrioles were visualized as a ring-like structure (Figure 2D), comparable to the frequency of observing a C-Nap1 ring by 3D-SIM (Figure 2E). The average diameter of the ring was ~100 nm, which is consistent with previous electron microscopy analyses of the cartwheel (Guichard et al., 2012; Kitagawa et al., 2011). Interestingly, the size of the solid foci was also measured at ~100 nm on average, revealing potentially the thickness of the SAS-6 stack. The same C-terminal antibody was then used to analyze SAS-6ND at G1 centrioles. Strikingly, ~22.5% of SAS-6ND signals in G1 centrioles displayed a ~100 nm ring (Figure 2F). In contrast, when antibodies recognizing the N-terminal hemagglutinin (HA) tag of SAS-6ND were used, tight foci of ~55 nm in size were consistently seen (Figure 2F, right panel), much smaller than the foci visualized with the C-terminal antibody (~100 nm). These results suggest that the luminal SAS-6 in G1 centrioles adopts an organized structure, with the N-termini of SAS-6ND clustered at the center of the lumen, resembling the central hub, and the C-terminal tails spread out radially from the center, resembling the spokes of a cartwheel.
SAS-6 dimers are able to self-oligomerize through the N-terminal globular domain to form a cartwheel-like ring (Kitagawa et al., 2011; van Breugel et al., 2011). It is therefore possible that pre-assembled cartwheels localize to the centriolar lumen. Alternatively, SAS-6 dimers may first localize to the centriole lumen followed by oligomerization. To differentiate between these two scenarios, SAS-6ND harboring the F131E mutation that has been previously shown to disrupt the oligomerization property of SAS-6 (Kitagawa et al., 2011; van Breugel et al., 2011) was inducibly expressed in G1-arrested cells. SAS-6F131E, which contains an intact C-terminal tail, localized efficiently to the proximal lumen of G1 centrioles (Figure 2G), indicating that SAS-6 dimers can be individually recruited. More surprisingly, STED imaging analyses revealed that in the absence of self-oligomerization, luminal SAS-6F131E was organized into a cartwheel-like configuration, showing a ring-like shape or large focus of ~100 nm when probed with the C-terminal antibody, and compact foci of ~55 nm when the N-terminus was labeled (Figure 2H). This result suggests that mother centrioles recruit and then organize SAS-6 molecules into a highly ordered structure, and that this is largely independent of the self-oligomerization activity of SAS-6.
SAS-6 recruitment to the proximal lumen is mediated by the C-terminal tail, and requires luminal protein CPAP
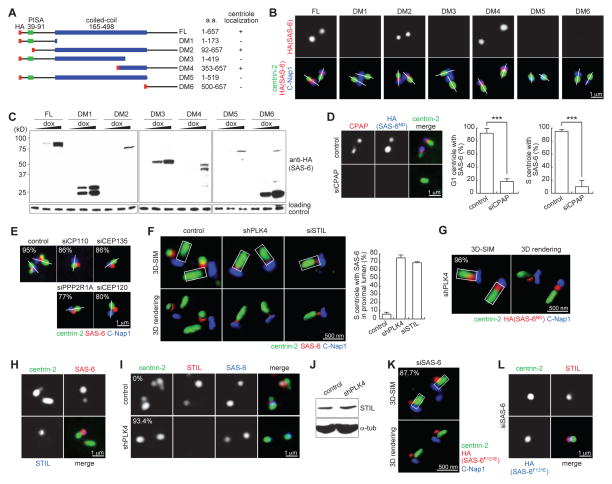
To determine the mechanism by which SAS-6 is recruited to the proximal lumen of a preexisting centriole, different domains of SAS-6ND were inducibly expressed in G1-arrested cells, and their localizations examined (Figure 3A and B). All SAS-6 fragments lacking the C-terminal domain (DM1, DM3 and DM5) failed to localize to G1 centrioles (Figure 3A-C). Interestingly, a fragment of the C-terminal domain (DM6) alone was insufficient for centriolar targeting (Figure 3A and B). Only when a small portion of coiled-coil domain was also present (DM4), could it effectively localize to centrioles (Figure 3A and B), suggesting that both the C-terminal tail and the dimerization of SAS-6 through its coiled-coil domain are important for recruitment to centriole lumen. To further test if centriolar components present at the wall of the proximal lumen are required for SAS-6 recruitment, we focused on the luminal protein CPAP and CEP135 that have been shown to form a complex with SAS-6 (Lin et al., 2013). In agreement with the previous report (Lin et al., 2013), incomplete but severe reduction of CEP135, which efficiently blocked centriole duplication, had no effect on SAS-6 recruitment (not shown; see below Figure 3E). In contrast, SAS-6 recruitment was disrupted in centrioles depleted of CPAP in both G1 and S-phase cells (Figure 3D), supporting the idea that the luminal protein CPAP mediates the recruitment of SAS-6. The role of CEP135 in luminal SAS-6 recruitment is uncertain at this point, as our RNAi could not completely remove centriole-bound CEP135 (Lin et al., 2013). These results together indicate that SAS-6 dimers are individually recruited to the centriole lumen, and subsequently aligned by the radial, 9-fold symmetrical body frame of a preexisting centriole.
Figure 3. Molecular requirements for SAS-6 recruitment to and release from centriole lumen.
(A) Collections of various SAS-6 deletion mutants tagged with HA (red). Centriolar localizations of these mutants are summarized.
(B and C) Localization and expression of SAS-6 mutants. Indicated SAS-6 mutants were inducibly expressed in G1-arrested RPE-1 cells, and their luminal localizations were examined.
(D) HeLa cells depleted of CPAP (two sequential RNAi for 6 days) were induced to express SAS-6ND. Centrioles in G1 or S-phase cells were examined for SAS-6 localization as indicated. Similar results were obtained with three independent siRNA oligos (see Experimental Procedures). Error bars, standard deviation. n>38, N=3. The significance (two-tailed t-test) is indicated, *** P < 0.0001.
(E) U2OS cells depleted of the indicated protein were examined for the localization of the endogenous SAS-6 during S phase. n>31, N=2.
(F) U2OS cells depleted of PLK4 or STIL were examined for the localization of the endogenous SAS-6 during S phase by 3D-SIM. Error bars, standard deviation. n>26, N=3.
(G) U2OS cells depleted of PLK4 were induced to express SAS-6ND. The duplication status and localization of the elevated SAS-6ND were examined and quantified in S-phase cells by 3D-SIM with indicated antibodies. Note that duplication was not rescued, and S-phase cells containing unduplicated centrioles (centrin singlet) were counted. n>25, N=3.
(H) Wild-type, early S-phase U2OS cells containing one bright and one weak SAS-6 foci were examined for the localization of STIL. Note that STIL only associates with the bright SAS-6 focus.
(I) STIL localization in control or PLK4-depleted U2OS cells expressing SAS-6ND was examined. n>29, N=3.
(J) Immunoblots showing STIL level is unaffected by PLK4 knockdown.
(K and L) SAS-6F131E was inducibly expressed in U2OS cells depleted of the endogenous SAS-6. Localizations of SAS-6F131E (K) and STIL (L) were then examined in S-phase cells by 3D-SIM and regular microscopy, respectively as indicated. n>24, N=3.
S-phase cells in (D), (E), (F), (G), (I), (K) and (L) were identified by BrdU labeling, not shown.
See also Figure S1.
Release of luminal SAS-6 from mother centrioles requires PLK4 and STIL, and is a pre-requisite for daughter centriole formation
To explore the process by which luminal SAS-6 is released from the centriolar lumen, centriolar proteins required for duplication were systematically depleted. Centriole duplication failed in all cases, but resulted in distinct patterns of SAS-6 localization. In cells where CP110, CEP135, PPP2R1A or CEP120 were depleted or severely reduced, SAS-6 was consistently seen at the side of mother centrioles (Figure 3E), suggesting that these proteins are not critically involved in SAS-6 recruitment, but regulate later stages of centriole assembly. In contrast, in cells depleted of PLK4 or STIL, centriole duplication was arrested at a step where the endogenous SAS-6 was persistently trapped at the proximal lumen of mother centrioles (Figure 3F). Consistent with the reported role of PLK4 in stabilizing SAS-6 (Puklowski et al., 2011), PLK4-depleted centrioles containing weak or no SAS-6 signals were noticed, with 43% of centrioles marked with weak (luminal) SAS-6, and 57% with no detectable SAS-6 (Suppl. Figure S1A), raising a possibility that the duplication defect is due to SAS-6 insufficiency. Intriguingly, overexpression of SAS-6ND in PLK4-depleted cells failed to rescue duplication, and the elevated SAS-6 remained trapped at the proximal lumen of nearly all PLK4-depleted centrioles (Figure 3G), revealing a new role of PLK4 in the release of luminal SAS-6. Next, we examined the involvement of STIL, a critical centriole biogenesis factor known to interact with CPAP (Arquint et al., 2012; Cottee et al., 2013; Tang et al., 2011; Vulprecht et al., 2012) and Plk4 (Firat-Karalar et al., 2014). Strikingly, in early S-phase cells containing one bright and one weak SAS-6 foci as described above in Figure 1, STIL was always associated with bright SAS-6 foci, but rarely seen at centrioles with weak SAS-6 signals (Figure 3H), suggesting that STIL specifically marks the population of SAS-6 that is tethered at the side, but not in the lumen, of mother centrioles. Consistently, no STIL was detected in PLK4-depleted centrioles where high level of SAS-6 was trapped in the lumen (Figure 3I), although the cellular level of STIL was not changed (Figure 3J), suggesting that STIL functions downstream of PLK4 to promote SAS-6 release. This pathway may add new insights into the relationship of SAS-6 with Ana2/SAS-5/STIL and PLK4/ZYG-1 reported in Drosophila and C. elegans (Lettman et al., 2013; Qiao et al., 2012; Stevens et al., 2010). Together, we conclude that the disassociation of luminal SAS-6 from mother centrioles is mediated by PLK4 and STIL, a pre-requisite for subsequent formation of daughter centrioles at the side of mother centrioles.
To examine if SAS-6 release depends on its self-oligomerization activity, SAS-6F131E was expressed in cells depleted of the endogenous SAS-6. As reported previously (Kitagawa et al., 2011; van Breugel et al., 2011), centriole duplication failed in this case, but strikingly, SAS-6F131E was seen attaching to the outside wall of mother centrioles during S phase (Figure 3K) and co-localizing with STIL (Figure 3L). Thus consistent with the previous study done in worms (Lettman et al., 2013), we suggest that self-oligomerization activity of SAS-6 is critically involved in later stages of centriole duplication, downstream of the initial recruitment (Figure 2G), organization (Figure 2H), and release (Figure 3K and L) of luminal SAS-6. Moreover, we found that SAS-6 deletion mutants, DM2 and DM4, both of which localize to the lumen of centrioles independent of STIL during G1 phase, can tether to the side of mother centrioles in S phase (Suppl. Figure S1B), co-localizing with STIL (Suppl. Figure S1C). In contrast, mutants that cannot localize to the lumen of G1 centrioles (DM1, 3, 5 or 6) also failed to associate with mother centrioles during S phase (Suppl Figure S1D; not shown), even for DM5, which contains the intact coiled-coil domain known to interact with STIL (Qiao et al., 2012). These results, together with those seen in CPAP depletion, support the idea that luminal targeting of SAS-6 plays a critical role in the formation of the SAS-6/STIL structure at the side of mother centrioles, consistent with the chronology of SAS-6 recruitment during early S phase.
Recruitment of luminal SAS-6 to mother centrioles is inhibited by centriole engagement
Centriole engagement is known to block centriole duplication, while disengagement releases the block and licenses duplication (Tsou and Stearns, 2006a, b). However, the step at which duplication is specifically regulated by engagement/disengagement is unknown. Interestingly, the luminal recruitment of SAS-6, which marks one of the earliest steps of centriole duplication, can occur at disengaged centrioles in G1 cells (Figure 2A), but not engaged mother centrioles in S phase (Figure 1F), suggesting that luminal recruitment of SAS-6 may be regulated by centriole engagement/disengagement. In agreement with this, the earlier observation that disengaged centrioles in S phase are capable of recruiting luminal SAS-6 (Figure 3F and G) indicates that recruitment is not simply regulated by cell cycle cues. To test whether the opposite condition also holds, we generated engaged centrioles in G1 cells expressing SAS-6ND by blocking centriole disengagement in mitosis with Plk1 inhibitors as described previously (Figure 4A) (Tsou et al., 2009; Wang et al., 2011). Intriguingly, while SAS-6ND was recruited to all disengaged centrioles in control G1 cells, no SAS-6ND was detected in mother centrioles that remained engaged to their daughters (Figure 4B; 90%). These data suggest that centriole engagement and disengagement critically regulate the ability of mother centrioles to assemble luminal SAS-6, consistent with their roles in blocking and licensing centriole duplication respectively.
Figure 4. Engaged centrioles block luminal recruitment of SAS-6.
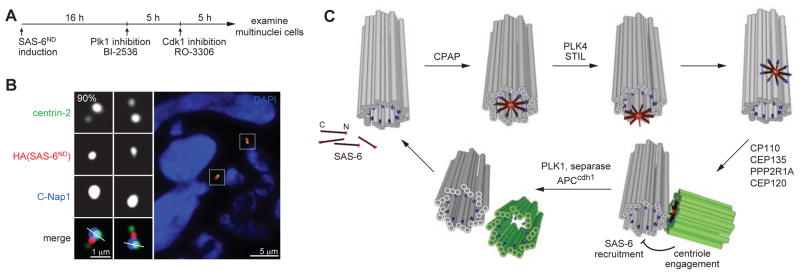
(A) Schematic outlining the generation of G1 cells with engaged centrioles as described previously (Tsou et al., 2009; Wang et al., 2011). SAS-6ND expression was induced in proliferating cells for 16 hours before adding BI-2536 to inhibit Plk1. Mitotic arrested cells were forced to exit mitosis 3 hours after Plk1 inhibition by Cdk1 inhibitor RO-3306 for 5 hours. Multinuclei cells, which were cells that had gone through mitosis in the absence of Plk1 activity, were examined for the centriolar localization of SAS-6 with indicated antibodies. n>26, N=3.
(B) SAS-6 recruitment to mother centriole lumen is inhibited by engaged centrioles. Immunofluorescence image of a G1 cell containing engaged centrioles with indicated antibodies.
(C) Model for a template-based mechanism of centriole duplication. SAS-6 dimers are individually recruited to the proximal lumen of mother centrioles, where they are organized by the surrounding geometry to undergo 9-fold symmetrical assembly. The assembled SAS-6 oligomer is then released from the lumen and forms the cartwheel that drives daughter centriole formation. Centriole engagement blocks re-recruitment of SAS-6 to the mother centriole lumen and thus prevents centriole re-duplication. Centriole disengagement and cartwheel removal at the end of mitosis allow the centrioles to be used as a template for their own biogenesis. Mother centriole (grey), daughter centriole (green), CPAP (blue), SAS-6 (red).
DISCUSSION
Our results demonstrate that SAS-6 is transiently recruited to the proximal lumen of disengaged mother centrioles in early S phase, before it is seen co-localizing with STIL at the external site for new centriole formation. Luminal targeting of SAS-6 is mediated through the C-terminal tail of SAS-6 and the luminal wall protein CPAP, shaping SAS-6 into a cartwheel-like organization independent of its self-oligomerization activity. The removal or release of luminal SAS-6 requires STIL and the kinase Plk4. Abolishing either the recruitment or the removal of luminal SAS-6 leads to a block of SAS-6 assembly (or centriole duplication) at the outside wall of mother centrioles. After duplication, the lumen of the engaged mother centriole is prevented from recruiting new SAS-6 until disengagement occurs at the end of mitosis, correlating with the block and license for centriole duplication. Notably, due to technical limitation and the transient nature of the process, we have not been able to determine if the SAS-6 structure formed in the lumen “directly re-localizes” to the outside wall of the mother centriole. These results, however, prompted us to reevaluate centriole duplication in cycling cells, and consider a template-based mechanism by which the geometric shape and copy number of the centriole can be faithfully preserved (see Figure 4C for the proposed model).
A recent report showed that high levels of the exogenous SAS-6 can accumulate into a torus-like pattern around the proximal end of mother centrioles, co-localizing with the pericentriolar marker (Keller et al., 2014). In our study, both the endogenous and the mildly expressed exogenous SAS-6 were seen to localize at the lumen of mother centrioles, without forming a torus-like pattern.
It is known that centrioles can form through de novo assembly (Azimzadeh et al., 2012; Khodjakov et al., 2002; Szollosi et al., 1972; Vladar and Stearns, 2007), suggesting that the template-based replication proposed here may be specifically used in cycling cells where centriole number is strictly controlled. In this case, mother centrioles act as the template to catalyze and guide the assembly of the SAS-6 ring, which can otherwise be formed through other mechanisms such as self-assembly but is at lower rate and without numerical controls. We note that loss of cartwheel, which empties the proximal lumen of a centriole, appears to be species specific, raising a question as to whether the template-based replication proposed here can apply to centrioles constantly occupied with the cartwheel as seen in other species (Guichard et al., 2012). Interestingly, the cartwheel is known to contain a stack of SAS-6 oligomers that varies in height (Guichard et al., 2012), suggesting that centrioles may serve as a SAS-6 reservoir that grows and shrinks constantly. It is thus tempting to speculate that mother centrioles, acting as a reservoir, can inducibly release a fraction of the SAS-6 stack for new centriole assembly, a model that has been proposed previously (Mignot, 1996). In vertebrate cycling cells, however, the program may be modified such that the SAS-6/STIL-based cartwheel at the daughter centriole is “non-releasable”, and must be eliminated before the next cell cycle to allow the assembly of “releasable SAS-6”, ensuring a tight control of centriole duplication.
EXPERIMENTAL PROCEDURES
Details of experimental procedures regarding cell culture, RNAi and stable cell lines, microscopy, and antibodies are provided in Supplemental Experimental Procedures.
Super-resolution microscopy
DeltaVision OMX (Applied Precision) or a home-built continuous-wave (CW) STED (Liao Lab, Columbia University) was employed. DeltaVision OMX was equipped with Olympus 100×, 1.4 NA oil objective; 405 nm, 488 nm, and 593 nm laser illumination; and standard excitation and emission filter sets. Images were acquired using a 125-nm z-step size and then deconvolved and aligned in three dimensions with softWoRx 5.0.0 (Applied Precision). The STED imaging was performed as previously decribed (Yang et al., 2013). Briefly, a 491-nm solid-state laser (Calypso 25, Cobolt AB) and a 592-nm 1W CW laser (VFL-P-1000-592, MPB Communications) were used to supply the excitation beam and the depletion beam, respectively. Two laser beams were coupled and focused onto the sample through a 100× oil immersion objective (Olympus UPLSAPO100x-1.4 NA). An avalanche photodiode (APD) module (SPCM-AQR-15, PerkinElmer) was used for signal collection. For quantitative measurement, STED images were first cleaned by a mean filter of 0.5 pixels and background subtracted, and then were fitted with a two-dimensional Gaussian function,
to identify the principal axes and to find the FWHMs, where c1 and c2 were used to determine the FWHM by multiplying the factor of . For image presentation, confocal and STED images were smoothed and contrast-enhanced with generalized Tikhonov regularization with ImageJ (Tikhonov and Arsenin, 1977).
Supplementary Material
HIGHLIGHTS.
SAS-6 is recruited to the proximal lumen of mother centrioles in early S phase
SAS-6 is organized into a cartwheel-like configuration by the centriole lumen
Release of luminal SAS-6 from mother centrioles requires PLK4 and STIL
Centriole engagement/disengagement regulates luminal recruitment of SAS-6
Acknowledgments
We thank E. Nigg (U. of Basel) and M. Mahjoub (Washington U) for antibodies; A. Hall and C. Haynes at MSKCC for comments on the manuscript. We are grateful for A. North (Rockefeller University) for the use of superresolution microscopy (OMX, DeltaVision), supported by the award S10RR031855 from the National Center for Research Resources. This work was supported by the National Institutes of Health grants GM088253 to M.-F. B. Tsou.
Footnotes
AUTHOR CONTRIBUTIONS
M.K., C.F. and M-F.B.T. designed experiments. M.K. and C.F. performed most of the experiments. T-T.Y. and J-C.L. performed and analyzed STED imagining. C.F., M.K. and M-F.B.T. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson RG, Brenner RM. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. The Journal of cell biology. 1971;50:10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Current biology : CB. 2014;24:351–360. doi: 10.1016/j.cub.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. Journal of cell science. 2012;125:1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF. Centrosome loss in the evolution of planarians. Science. 2012;335:461–463. doi: 10.1126/science.1214457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, Andreeva A, Oegema K, Lea SM, Raff JW, van Breugel M. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife. 2013;2:e01071. doi: 10.7554/eLife.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Raff JW, Lea SM, Roque H. SAS-6 oligomerization: the key to the centriole? Nature chemical biology. 2011;7:650–653. doi: 10.1038/nchembio.660. [DOI] [PubMed] [Google Scholar]
- Donnert G, Keller J, Wurm CA, Rizzoli SO, Westphal V, Schönle A, Jahn R, Jakobs S, Eggeling C, Hell SW. Two-color far-field fluorescence nanoscopy. Biophysical Journal. 2007;92:L67–L69. doi: 10.1529/biophysj.107.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity Interactions among Centrosome Components Identify Regulators of Centriole Duplication. Current biology : CB. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. The Journal of cell biology. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gonczy P. Cartwheel architecture of Trichonympha basal body. Science. 2012;337:553. doi: 10.1126/science.1222789. [DOI] [PubMed] [Google Scholar]
- Hell S, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Optics Letters. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Keller D, Orpinell M, Olivier N, Wachsmuth M, Mahen R, Wyss R, Hachet V, Ellenberg J, Manley S, Gonczy P. Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. The Journal of cell biology. 2014;204:697–712. doi: 10.1083/jcb.201307049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. The Journal of cell biology. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci U S A. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Developmental cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. The Journal of cell biology. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Developmental cell. 2013;25:284–298. doi: 10.1016/j.devcel.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, Chou EJ, Wu CT, Tang TK. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. The EMBO journal. 2013;32:1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Current biology : CB. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. The Journal of cell biology. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot JP. New hypothesis on the replication of centrioles and basal bodies. Comptes rendus de l’Academie des sciences Serie III, Sciences de la vie. 1996;319:1093–1099. [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Current biology : CB. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, Grunwald V, Kubicka S, Pich A, Manns MP, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nature cell biology. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G. SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly. The EMBO journal. 2012;31:4334–4347. doi: 10.1038/emboj.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Developmental cell. 2010;19:913–919. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Developmental cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. Journal of cell science. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. The EMBO journal. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov A, Arsenin V. Solutions of ill-posed problems. Winston & Sons; 1977. [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Current opinion in cell biology. 2006a;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006b;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Developmental cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. The Journal of cell biology. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. The ultrastructure of centriole in mammalian tissue culture cells. Cell Biol Int Rep. 1980;4:1037–1044. doi: 10.1016/0309-1651(80)90177-0. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. The Journal of cell biology. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S, et al. STIL is required for centriole duplication in human cells. Journal of cell science. 2012;125:1353–1362. doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. The Journal of cell biology. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Hampilos PJ, Nathwani B, Miller CH, Sutaria ND, Liao JC. Superresolution STED microscopy reveals differential localization in primary cilia. Cytoskeleton (Hoboken) 2013;70:54–65. doi: 10.1002/cm.21090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.