Abstract
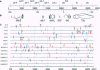
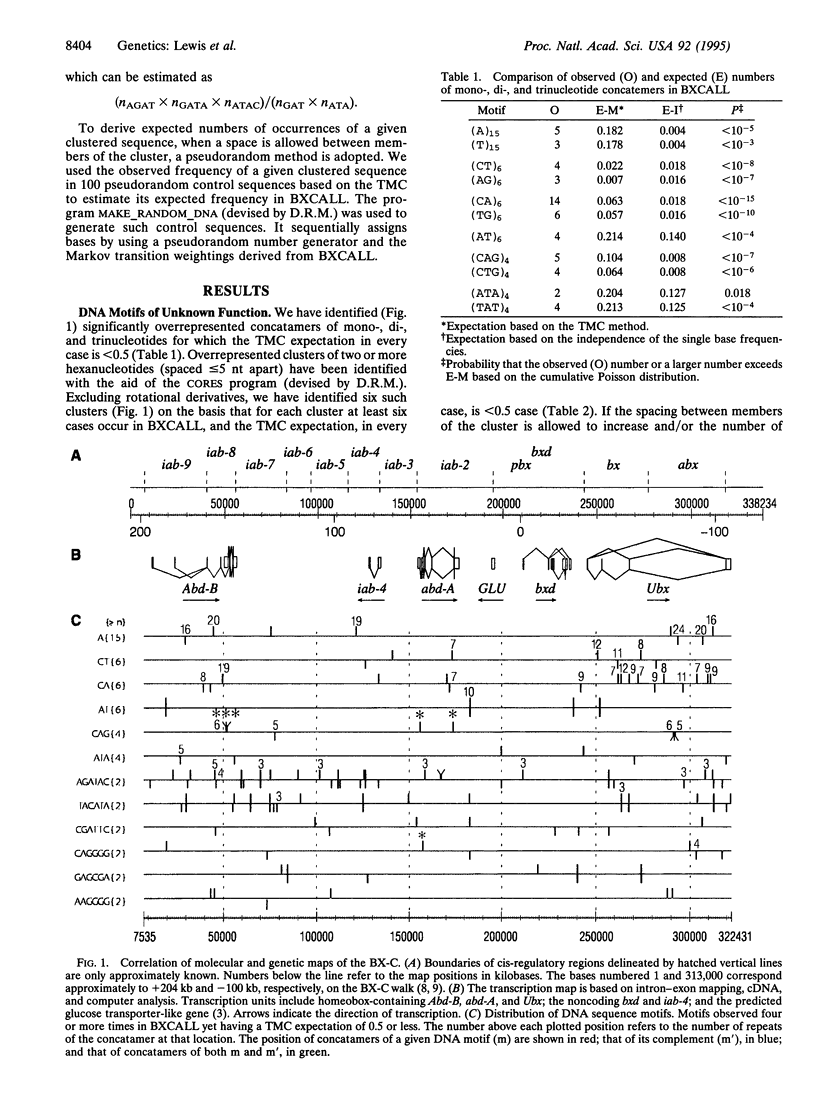
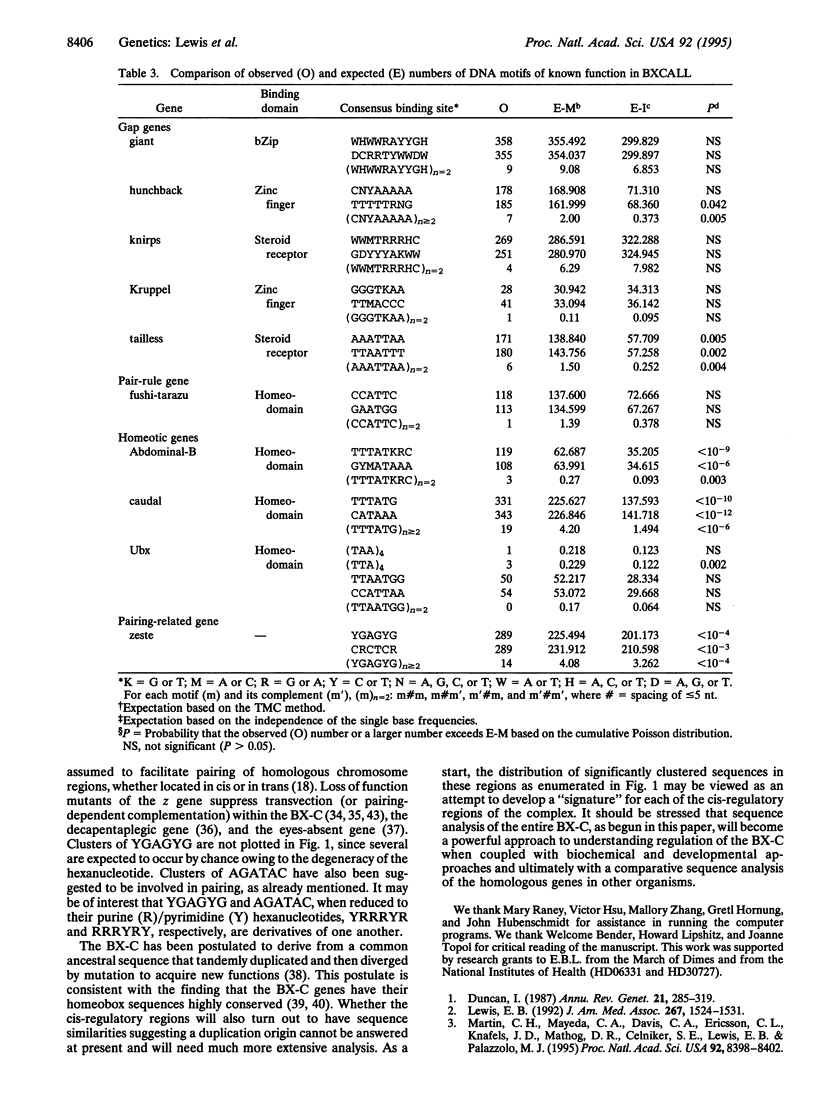
The bithorax complex (BX-C) of Drosophila, one of two complexes that act as master regulators of the body plan of the fly, has now been entirely sequenced and comprises approximately 315,000 bp, only 1.4% of which codes for protein. Analysis of this sequence reveals significantly overrepresented DNA motifs of unknown, as well as known, functions in the non-protein-coding portion of the sequence. The following types of motifs in that portion are analyzed: (i) concatamers of mono-, di-, and trinucleotides; (ii) tightly clustered hexanucleotides (spaced < or = 5 bases apart); (iii) direct and reverse repeats longer than 20 bp; and (iv) a number of motifs known from biochemical studies to play a role in the regulation of the BX-C. The hexanucleotide AGATAC is remarkably overrepresented and is surmised to play a role in chromosome pairing. The positions of sites of highly overrepresented motifs are plotted for those that occur at more than five sites in the sequence, when < 0.5 case is expected. Expected values are based on a third-order Markov chain, which is the optimal order for representing the BXCALL sequence.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Benson M., Pirrotta V. The Drosophila zeste protein binds cooperatively to sites in many gene regulatory regions: implications for transvection and gene regulation. EMBO J. 1988 Dec 1;7(12):3907–3915. doi: 10.1002/j.1460-2075.1988.tb03277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988 Jun 3;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Capovilla M., Eldon E. D., Pirrotta V. The giant gene of Drosophila encodes a b-ZIP DNA-binding protein that regulates the expression of other segmentation gap genes. Development. 1992 Jan;114(1):99–112. doi: 10.1242/dev.114.1.99. [DOI] [PubMed] [Google Scholar]
- Dearolf C. R., Topol J., Parker C. S. The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature. 1989 Sep 28;341(6240):340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- Duncan I. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- Ekker S. C., Jackson D. G., von Kessler D. P., Sun B. I., Young K. E., Beachy P. A. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994 Aug 1;13(15):3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., Young K. E., von Kessler D. P., Beachy P. A. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991 May;10(5):1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart W. M. Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2636–2640. doi: 10.1073/pnas.79.8.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M., Gerwin N., Taubert H., Jäckle H. Competition for overlapping sites in the regulatory region of the Drosophila gene Krüppel. Science. 1992 Apr 3;256(5053):94–97. doi: 10.1126/science.1348871. [DOI] [PubMed] [Google Scholar]
- Hogness D. S., Lipshitz H. D., Beachy P. A., Peattie D. A., Saint R. B., Goldschmidt-Clermont M., Harte P. J., Gavis E. R., Helfand S. L. Regulation and products of the Ubx domain of the bithorax complex. Cold Spring Harb Symp Quant Biol. 1985;50:181–194. doi: 10.1101/sqb.1985.050.01.024. [DOI] [PubMed] [Google Scholar]
- Jiang J., Hoey T., Levine M. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 1991 Feb;5(2):265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- Karch F., Bender W., Weiffenbach B. abdA expression in Drosophila embryos. Genes Dev. 1990 Sep;4(9):1573–1587. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- Karch F., Weiffenbach B., Peifer M., Bender W., Duncan I., Celniker S., Crosby M., Lewis E. B. The abdominal region of the bithorax complex. Cell. 1985 Nov;43(1):81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- Kennison J. A. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993 Mar;9(3):75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- LEWIS E. B. Pseudoallelism and gene evolution. Cold Spring Harb Symp Quant Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- Leiserson W. M., Bonini N. M., Benzer S. Transvection at the eyes absent gene of Drosophila. Genetics. 1994 Dec;138(4):1171–1179. doi: 10.1093/genetics/138.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. Regulation of the genes of the bithorax complex in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:155–164. doi: 10.1101/sqb.1985.050.01.021. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. The 1991 Albert Lasker Medical Awards. Clusters of master control genes regulate the development of higher organisms. JAMA. 1992 Mar 18;267(11):1524–1531. [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Wallrath L. L., Granok H., Elgin S. C. (CT)n (GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol Cell Biol. 1993 May;13(5):2802–2814. doi: 10.1128/mcb.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. H., Mayeda C. A., Davis C. A., Ericsson C. L., Knafels J. D., Mathog D. R., Celniker S. E., Lewis E. B., Palazzolo M. J. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci U S A. 1995 Aug 29;92(18):8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Regulski M., Dessain S., McGinnis N., McGinnis W. High-affinity binding sites for the Deformed protein are required for the function of an autoregulatory enhancer of the Deformed gene. Genes Dev. 1991 Feb;5(2):278–286. doi: 10.1101/gad.5.2.278. [DOI] [PubMed] [Google Scholar]
- Sauer F., Jäckle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991 Oct 10;353(6344):563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Schier A. F., Gehring W. J. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992 Apr 30;356(6372):804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimell M. J., Simon J., Bender W., O'Connor M. B. Enhancer point mutation results in a homeotic transformation in Drosophila. Science. 1994 May 13;264(5161):968–971. doi: 10.1126/science.7909957. [DOI] [PubMed] [Google Scholar]
- Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995 Jun;7(3):376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Stanojević D., Hoey T., Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. 1989 Sep 28;341(6240):331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Thali M., Müller M. M., DeLorenzi M., Matthias P., Bienz M. Drosophila homoeotic genes encode transcriptional activators similar to mammalian OTF-2. Nature. 1988 Dec 8;336(6199):598–601. doi: 10.1038/336598a0. [DOI] [PubMed] [Google Scholar]
- Treisman J., Desplan C. The products of the Drosophila gap genes hunchback and Krüppel bind to the hunchback promoters. Nature. 1989 Sep 28;341(6240):335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Tremml G., Bienz M. Induction of labial expression in the Drosophila endoderm: response elements for dpp signalling and for autoregulation. Development. 1992 Oct;116(2):447–456. doi: 10.1242/dev.116.2.447. [DOI] [PubMed] [Google Scholar]