Abstract
Erdheim-Chester disease (ECD) is a rare histiocytosis with a high prevalence of BRAF V600E mutation (>50% of patients). Patients with BRAF-mutant ECD can respond to BRAF inhibitors. Unfortunately, the lack of adequate archival tissue often precludes BRAF testing. We hypothesized that cell-free DNA (cfDNA) from plasma or urine can offer an alternative source of biologic material for testing. We tested for BRAF V600E mutation in cfDNA from the plasma and urine of 6 ECD patients. In patients with available archival tissue, the result of BRAF mutation analysis was concordant with plasma and urine cfDNA results in all 3 patients (100% agreement, kappa 1.00). In all 6 patients, BRAF mutation analysis of plasma and urine cfDNA was concordant in 5 of 6 patients (83% agreement, kappa 0.67). Testing for BRAF V600E mutation in plasma and urine cfDNA should be further investigated as an alternative to archival tissue mutation analysis.
Keywords: BRAF, cell-free DNA, plasma, urine, Erdheim-Chester disease
INTRODUCTION
Erdheim-Chester disease (ECD) is a rare form of non-Langerhans cell histiocytosis affecting adults, which is associated with xanthogranulomatous infiltration of foamy macrophages.[1-3] ECD is deemed to be driven by increased signaling within the mitogen-activated-protein kinase pathway. Advances in genome sequencing technologies led to the identification of BRAF V600E mutations in at least 50% of patients with ECD.[4] Furthermore, preliminary results suggest that patients with ECD and BRAF mutations can benefit from targeted inhibition of BRAF protein with BRAF inhibitors.[5] Unfortunately, archival tissue often does not provide an adequate amount of DNA for molecular testing. Therefore, novel technologies allowing mutation analysis to be performed using alternative sources of biologic material are needed.[6]
Cell-free DNA (cfDNA) is released to the circulation from cells undergoing apoptosis, necroptosis and active secretion and has been identified in the plasma and urine of patients with cancer.[7, 8] Detecting and quantifying the amount of mutant cfDNA fragments harboring specific mutations can be used as an alternative to tissue testing. Some data suggest that the amount of mutant DNA correlates with tumor burden and can be used to identify the emergence of resistant mutations.[9-14] The concept of mutation testing from urine cfDNA has been assessed in a pilot study in patients with advanced colorectal cancer and other colorectal diseases in which KRAS mutations in urine cfDNA were concordant in 95% of cases with KRAS mutation status in tumor tissue.[15] We examine in our study whether urine and plasma cfDNA can be used as an alternative to tissue biopsies for BRAF V600E mutation testing in patients with ECD.
RESULTS AND DISCUSSION
A total of 6 patients with ECD were enrolled. Their median age at diagnosis was 46 years (range, 26 to 71 years) and most patients were white 4 (67%) and male 4 (67%). Tumor tissue BRAF V600E mutation testing with targeted next-generation sequencing and/or allele-specific PCR in the CLIA-certified laboratory was requested for all 6 patients, but 3 patients had insufficient tissue samples, precluding mutation analysis (Table 1). BRAF V600E mutations were detected in 2 (67%) of 3 tested tumor tissue samples, 3 (50%) of 6 plasma cfDNA samples and 4 (67%) of urine cfDNA samples (Figure 1). Observed agreements were 100% (3 of 3, kappa 1.00) for tumor tissue and plasma cfDNA, 100% (3 of 3, kappa 1.00) for tumor tissue and urine cfDNA, and 83% (5 of 6, kappa 0.67) for plasma cfDNA and urine cfDNA (Table 1). In addition, there were no BRAF V600E mutations in plasma and urine cfDNA from 14 patients with metastatic cancer with confirmed wt BRAF in their tumor tissue (data not shown). Finally, only one patient (patient 1) was treated with a BRAF inhibitor; however, the treatment outcomes were not available at the time of analysis.
Table 1. Urine and plasma cell-free DNA BRAF VGOOE mutations.
| Patient# | Age at diagnosis | Gender | Involvement | Urine BRAF VGOOE/WT | Plasma BRAF VGOOE/WT | Patient Tissue BRAFstatus |
|---|---|---|---|---|---|---|
| 1 | 59 | Male | CNS, cardiac, bones, renal | V600E (22.590%) | V600E (8.598%) | V600E |
| 2* | 43 | Male | CNS, bones, renal | V600E (0.311%) | V600E (1.522%) | V600E |
| 3 | 26 | Male | Skin | Wild-type (0.010%) | Wild-type (0.063%) | Wild-type |
| 4 | 71 | Female | Bones, lymph nodes | V600E (0.159%) | Wild-type (0.047%) | Unknown** |
| 5 | 43 | Female | CNS, bones | V600E (4.940%) | V600E (0.261%) | Unknown** |
| 6 | 49 | Male | CNS, cardiac, omentum, retroperitoneum | Wild-type indeterminate (0.079%) | Wild-type (0.048%) | Unknown** |
CNS; central nervous system
Urine and plasma collected on different dates
lnsuficient tissue for molecular analysis
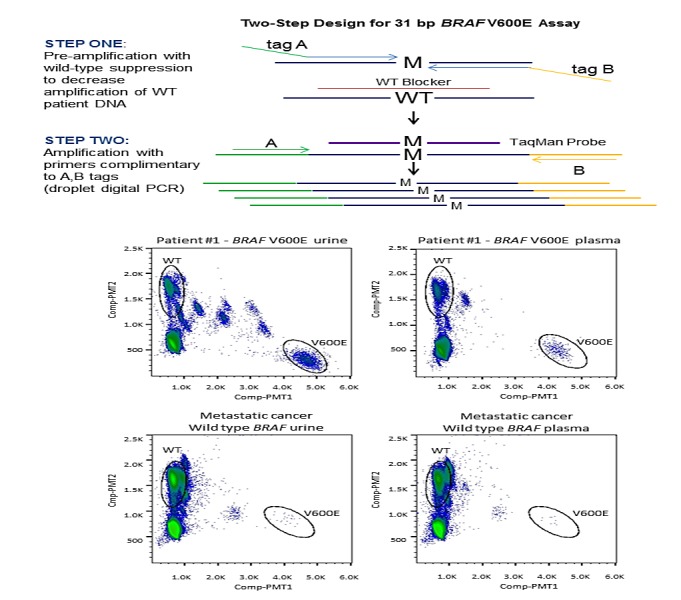
BRAF V600E mutations have been reported in more than 50% of patients with ECD.[4] In addition, preliminary data reveal encouraging activity of BRAF inhibitors such as vemurafenib in ECD patients with BRAF V600E mutations.[5] Mutation analysis of tumor tissue remains a gold standard for molecular analysis; however, in some disorders such as ECD, available tumor tissue often does not provide enough DNA for molecular analysis. In our experience, archival tissue testing for BRAF mutations is not feasible in up to 60% of patients.[6] This creates a major hurdle for further implementation of personalized therapies into the ECD therapeutic armamentarium since BRAF inhibitors in general can be effective in patients with BRAF mutations but detrimental in patients without them.[16] Therefore, there is a clear need for a new and easily obtainable source of material that can be used to analyze tumor molecular aberrations. [7, 8] cfDNA is released to the circulation from cells undergoing apoptosis, necroptosis and active secretion and has been identified in the plasma or urine of patients with cancer.[9-14] Arguably, cfDNA can originate from multiple tumor sites and its molecular analysis may perhaps better reflect prevailing molecular aberrations.[9, 10]
Our study suggests that mutation analysis of plasma and/or urine cfDNA from patients with ECD can be concordant with archival tissue and should be investigated as its alternative in furthering personalized therapy for patients whose tumor tissue is in short supply.
METHODS
Patients with ECD were referred to the Clinical Center for Targeted Therapy at The University of Texas MD Anderson Cancer Center (MD Anderson) and prospectively enrolled starting in January 2013. In addition, 14 patients with metastatic cancers with known wild-type (wt) BRAF in tumor tissue were used as a negative control group. Database registration of patients and pathology assessment were performed at MD Anderson. The study and all treatments were conducted in accordance with MD Anderson Institutional Review Board guidelines. A total of 10mL blood samples and approximately 60-120mL of urine from each consented patient were used for DNA isolation.
Urine cfDNA was isolated by adding urine to an ion-exchange resin (GE Healthcare; Pittsburgh, PA). Nucleic acid was eluted with a chaotropic agent and subsequently purified by a silica-based resin (QIAGEN; Germantown, MD). The eluate was further purified using a specific molecular cutoff filter concentrator (Millipore; Billerica, MA), followed by a size exclusion column (Bio-Rad; Hercules, CA). Plasma cfDNA was isolated using the QIAamp Circulating Nucleic Acid Kit (QIAGEN; Germantown, MD) according to the manufacturer's instructions.
Urine and plasma cfDNA were quantified by a droplet digital PCR (ddPCR; QX-100, BioRad; Hercules, CA) assay to a 44bp amplicon of RNase P, a single-copy gene. Quantified DNA (12.4ng to 60ng) was used for a two-step PCR assay for rare mutant allele detection of a 31bp region containing BRAF V600E (Figure 1). The first step involved pre-amplification with two primers flanking the BRAF V600E locus, where both primers contain non-complementary 5' tags which hybridize to second round primers. A complementary blocking oligonucleotide suppressed wt BRAF amplification, achieving enrichment of the mutant BRAF V600E sequence within the pre-amplification step. The second step entailed a duplex ddPCR reaction using FAM (V600E BRAF) and VIC (wt BRAF) TaqMan probes to enable differentiation of mutant versus wild-type quantification, respectively. The RainDrop ddPCR instrument (RainDance; Billerica, MA) was used for PCR droplet separation, fluorescent reading, and counting droplets containing mutant sequence, wt sequence, or unreacted probe. For a given patient sample, the assay reported BRAF V600E mutation fragments detected as a percentage of detected wt BRAF. Previous to this study, accuracy of the urine-based ddPCR BRAF V600E assay was verified in 89 urine specimens from 50 healthy control samples (Precision Med; Solana Beach, CA) and 39 samples from 20 patients with known positive BRAF V600E mutation tissue biopsies as determined in a CLIA laboratory.[17] Thresholds for mutation detection in urine were determined by assessing these data using a classification tree. Minimizing the percentage of false negatives was given a higher importance than minimizing false positives. Thresholds were defined as no detection – wt (<0.05%), indeterminate (0.05% - 0.107%), and detected – V600E (>0.107%). For plasma detection, plasma from 13 patients with wt BRAF metastatic cancer was used to determine a threshold for detection of BRAF V600E mutations. For plasma, >0.094% mutant, equivalent to three standard deviations (0.021%) above the mean of wt BRAF controls (0.031%), was considered positive for BRAF V600E mutation.
Acknowledgments
Adriana Muniz-Fernandez and Mary Barry are acknowledged for their technical expertise. We also thank Joann Aaron, MA, for editorial assistance.
REFERENCES
- 1.Janku F, Amin HM, Yang D, Garrido-Laguna I, Trent JC, Kurzrock R. Response of histiocytoses to imatinib mesylate: fire to ashes. J Clin Oncol. 2010;28(31):e633–636. doi: 10.1200/JCO.2010.29.9073. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud L, Hervier B, Neel A, Hamidou MA, Kahn JE, Wechsler B, Perez-Pastor G, Blomberg B, Fuzibet JG, Dubourguet F, Marinho A, Magnette C, Noel V, Pavic M, Casper J, Beucher AB, et al. CNS involvement and treatment with interferon-alpha are independent prognostic factors in Erdheim-Chester disease: a multicenter survival analysis of 53 patients. Blood. 2011;117(10):2778–2782. doi: 10.1182/blood-2010-06-294108. [DOI] [PubMed] [Google Scholar]
- 3.Janku F, Munoz J, Subbiah V, Kurzrock R. A tale of two histiocytic disorders. Oncologist. 2013;18(1):2–4. doi: 10.1634/theoncologist.2012-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haroche J, Charlotte F, Arnaud L, von Deimling A, Helias-Rodzewicz Z, Hervier B, Cohen-Aubart F, Launay D, Lesot A, Mokhtari K, Canioni D, Galmiche L, Rose C, Schmalzing M, Croockewit S, Kambouchner M, et al. High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120(13):2700–2703. doi: 10.1182/blood-2012-05-430140. [DOI] [PubMed] [Google Scholar]
- 5.Haroche J, Cohen-Aubart F, Emile JF, Arnaud L, Maksud P, Charlotte F, Cluzel P, Drier A, Hervier B, Benameur N, Besnard S, Donadieu J, Amoura Z. Dramatic efficacy of vemurafenib in both multisystemic and refractory Erdheim-Chester disease and Langerhans cell histiocytosis harboring the BRAF V600E mutation. Blood. 2013;121(9):1495–1500. doi: 10.1182/blood-2012-07-446286. [DOI] [PubMed] [Google Scholar]
- 6.Janku F, Munoz J, Fayad LE, Lin PP, Cohen PR, Esmaeli B, Hymes SR, Jackson TL, Barnes TG, Kurzrock R. Outcomes of patients with Erdheim-Chester disease treated at MD Anderson Cancer Center. Erdheim-Chester disease International Medical Symposium; San Diego, CA. 2013. [Google Scholar]
- 7.De Mattos-Arruda L, Cortes J, Santarpia L, Vivancos A, Tabernero J, Reis-Filho JS, Seoane J. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol. 2013;10(7):377–389. doi: 10.1038/nrclinonc.2013.80. [DOI] [PubMed] [Google Scholar]
- 8.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 9.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, Hadfield J, May AP, Caldas C, Brenton JD, Rosenfeld N. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science translational medicine. 2012;4(136):136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 10.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 11.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 12.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C, Sartore-Bianchi A, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su YH, Wang M, Brenner DE, Norton PA, Block TM. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Annals of the New York Academy of Sciences. 2008;1137:197–206. doi: 10.1196/annals.1448.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 17.Janku F, Falchook GS, Piha-Paul SA, Naing A, Tsimberidou AM, Holley VR, Karp DD, Zinner RG, Fu S, Wheler JJ, Hong DS, Meric-Bernstam F, Stepanek VM, Luthra R, Leppin L, Hassaine L, et al. Detection and monitoring of BRAF and KRAS mutations in cell-free urinary DNA of metastatic cancer patients by digital droplet PCR. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics Proceedings. 2013. Abstract B175.


