Abstract
Prostate cancer (PCa) is the most commonly diagnosed cancer in men in the Western world. The transition of androgen-dependent PCa to castration-resistant (CRPC) is a major clinical manifestation during disease progression and presents a therapeutic challenge. Our studies have shown that genetic ablation of inhibitor of differentiation 4 (Id4), a dominant-negative helix loop helix protein, in mice results in prostatic intraepithelial neoplasia lesions and decreased Nkx3.1 expression without the loss of androgen receptor (Ar) expression. ID4 is also epigenetically silenced in the majority of PCa. However, the clinical relevance and molecular pathways altered by ID4 inactivation in PCa are not known. This study investigates the effect of loss of ID4 in PCa cell lines on tumorigenicity and addresses the underlying mechanism. Stable silencing of ID4 in LNCaP cells (L-ID4) resulted in increased proliferation, migration, invasion, and anchorage-independent growth. An increase in the rate of tumor growth, weight, and volume was observed in L-ID4 xenografts compared with that in the LNCaP cells transfected with nonspecific short hairpin RNA (L+ns) in noncastrated mice. Interestingly, tumors were also observed in castrated mice, suggesting that loss of ID4 promotes CRPC. RNA sequence analysis revealed a gene signature mimicking that of constitutively active AR in L-ID4, which was consistent with gain of de novo steroidogenesis. Prostate-specific antigen expression as a result of persistent AR activation was observed in L-ID4 cells but not in L+ns cells. The results demonstrate that ID4 acts as a tumor suppressor in PCa, and its loss, frequently observed in PCa, promotes CRPC through constitutive AR activation.
Androgen receptor (AR) acts as a tumor suppressor in the normal prostate but transitions to an oncogene in prostate cancer (PCa) which, following initial antiandrogen therapies, often recurs as castration-resistant prostate cancer (CRPC) (1). AR activation in response to circulating androgens in the early stages of the disease and later to intratumoral androgens as a result of de novo steroidogenesis appears to be a key transitional event (2). The mechanisms leading to gain of de novo steroidogenesis are not well established, but the activity of steroidogenic enzymes such as CYP17A1, which is currently targeted by the new generation of drugs such abiraterone acetate (3), is increased in CRPC.
Inhibitor of differentiation 4 (ID4), a dominant-negative helix-loop-helix protein, belongs to the basic helix-loop-helix (HLH) family of transcription factors (4). Similar to its other family members (ID1, ID2, and ID3), ID4 also lacks the DNA binding basic domain and acts as a dominant-negative transcriptional regulator of basic HLH transcription factors (4). ID1, ID2, and ID3, are generally considered to be as tumor promoters/supporting oncogenes. Conversely, ID4 has been proposed as a potential tumor suppressor based on the evidence that it is epigenetically silenced in many cancers (5–8).
ID4 is highly expressed in normal ductal epithelial cells of the prostate but rarely in stromal cells (9, 10). In PCa, ID4 expression is progressively lost with increasing stage of the disease (9). We and others have shown that loss of ID4 in PCa is primarily due to promoter hypermethylation (9, 11, 12). Our previous studies on prostate glands from Id4−/− mice demonstrated prostatic intraepithelial neoplasia (PIN) lesions as early as 6 weeks. These results suggested that loss of ID4 expression could be an early event in PCa (10). AR expression and cellular localization within Id4−/− and wild-type prostate are similar, but AR transcriptional activity in Id4−/− mice is attenuated, resulting in decreased expression of Nkx3.1, an androgen-regulated gene required for normal prostate development (10).
We previously reported high levels of ID4 expression in the androgen-sensitive PCa cell line LNCaP, which has low tumorigenic potential (9, 12). Conversely, ID4 is epigenetically silenced in the highly tumorigenic C81 PCa cell line (9). C81 cells are AR-positive but androgen-independent derivatives of LNCaP cells cultured progressively under androgen-deprived conditions (13). These observations collectively support the role of ID4 as a putative tumor suppressor, possibly by regulating normal AR activity in the prostate. Whether the loss of ID4, commonly seen in PCa, also results in deregulated AR activity leading to CRPC remains to be investigated. The present study was therefore conceived to determine whether loss of ID4 leads to PCa and, more specifically, to CRPC and the possible underlying mechanism. Our results suggest that knockdown of ID4 in LNCaP cells promotes tumorigenicity with a gene expression signature that resembles constitutive AR activity in castrated mice. We propose that epigenetic inactivation of ID4 results in CRPC, a condition of significant clinical interest.
Materials and Methods
Cell lines and ID4 silencing
The PCa cell line LNCaP was used to stably silence ID4 using gene-specific short hairpin RNA retroviral vectors (L-ID4 cells) as described in our previous study (14). C81 cells were kindly provided by Professor Ming-Fong Lin (University of Nebraska Medical Center, Omaha, Nebraska) (15). The LNCaP cells transfected with nonsilencing short hairpin RNA (L+ns cells) were used as controls. For measuring androgen responses, the cells were cultured in charcoal-stripped fetal bovine serum (cFBS) for 24 hours and subsequently treated with vehicle or 10 nM synthetic androgen R1881 (metribolone) and/or 25 μM bicalutamide (Casodex; a gift from AstraZeneca) for 24 hours.
Proliferation assay
Cell proliferation was determined using a CellTiter 96 nonradioactive cell proliferation assay (Promega) according to the manufacturer's protocol.
Scratch wound, transwell migration, and soft agar colony-forming assay
Scratch wound and transwell migration assays were performed as described previously (16). For the soft agar colony-forming assay, L+ns and L-ID4 cells (104 cells/well) in 1.5 mL of 0.3% agar (Difco) supplemented with complete RPMI 1640 medium plus 10% FBS were layered over the 0.5% agar base layer in 6-well plates. The plates were incubated at 37°C with 5% CO2 in a humidified incubator for 14 days; fresh medium was overlaid every 3 days. The plates stained in crystal violet solution were air dried, and the colony numbers were counted using a microscope.
Immunocytochemistry (ICC) and immunohistochemistry (IHC)
ICC and IHC protein localization studies on cells grown in glass chamber slides or paraffin-embedded 5-μm tissue sections, respectively, were performed as described previously (17) using protein specific antibodies (see Supplemental Table 1). The respective antigens were not detected with nonimmune IgG, which was used as a control for all immune localization studies (data not shown).
Immunoblot analysis
Cellular, nuclear, and cytoplasmic proteins were prepared from cultured PCa cell lines using M-PER and N-PER kits (Thermo Scientific). Immunoblot analyses using protein-specific antibodies were performed as described earlier (10) (Supplemental Table 1). The LAS 3000 imager (Fuji) and ImageQuant software were used to capture and quantify the images.
Real-time quantitative RT-PCR (qRT-PCR)
RNA (2 μg) isolated from fresh-frozen xenografts or cell lines was reverse transcribed in a final volume of 25 μL as per standard protocols (10, 18). Reverse-transcribed RNA was used for qRT-PCR with gene-specific primers (10) (Supplemental Table 2). Total RNA from xenografts was isolated by an E.Z.N.A. DNA/RNA kit (Omega Bio-Tek).
Animal studies
L+ns and L-ID4 cells (2 × 106) suspended in 100 μL of serum-free RPMI 1640 medium containing Matrigel (1:1 [v/v]; BD Biosciences) were injected subcutaneously into both flanks of 4-week-old castrated (C) and noncastrated (NC) male nu/nu athymic nude mice (Charles River) using a 27-gauge syringe. The nu/nu mice were maintained at the Mercer University Vivarium. All studies were approved by the Clark Atlanta and Mercer University committee for the use and care of animals. Tumors were harvested as soon as their volume reached around 800 mm3 (range, 6–8 weeks). The growth of the tumors was measured each week using the equation[L (length) × W (width)2]/2. At the end of the experiments, the mice were killed by asphyxiation, the tumors were surgically removed and weighed, and the volume was measured. Harvested tumors were fixed in 10% buffered formalin. The fixed tumors were paraffin embedded, sectioned (5 μm), and either stained with hematoxylin and eosin or used for IHC. Images were captured using a Zeiss microscope with an Axiom Cam version 4.5 imaging system. For tumor imaging, the mice were anesthetized with 2% isoflurane throughout the procedure. The IRDye 800CW epidermal growth factor targeting agent (1 nm) was injected via the tail vein into mice bearing subcutaneous tumors and evaluated for tumor-specific retention of the fluorophore by near-infrared fluorescence imaging (Odyssey CLx; LI-COR Biosciences).
Next-generation sequencing and network analysis
RNA was isolated from L+ns and L-ID4 cells (pooled from 2 sets of experiments) and sent to Otogenetics Corporation for RNA sequencing (RNA-seq). Paired-end FASTQ files received from Otogenetics were mapped using the Tophat version 2.0.3 pipeline to the University of California, Santa Cruz (UCSC) hg19.fa and hg19_genes.gtf references downloaded from Illumina iGenomes (19). The resulting *.bam files were used to calculate fragments per kilobases of exons per million mapped reads and gene expression differences using the cuffdiff program from within the Cufflinks package and hg19_genes.gtf as the reference file (20). The 41 significantly differentially expressed genes were submitted to Ingenuity Systems for comprehensive pathway and network analysis.
Testosterone ELISA
L+ns, L-ID4, and C81 cells were cultured in 96-well plates (10 000 cells in triplicate). Twenty-four hours after plating, the complete medium with 10% FBS was replaced with 10% charcoal-stripped media. The medium was collected after 72 hours, and cells were counted again. The medium was used for quantitating testosterone by an ELISA kit (R&D System) as per the manufacturer's recommendation.
Data and statistical analysis
The National Institutes of Health ImageJ (21) was used for counting cells stained positive for their respective antigens in IHC studies (10). qRT-PCR data were analyzed using the ΔΔCt method (9). The within-group Student t test was used for evaluating the statistical differences between groups.
Results
ID4 expression in PCa and disease-free survival
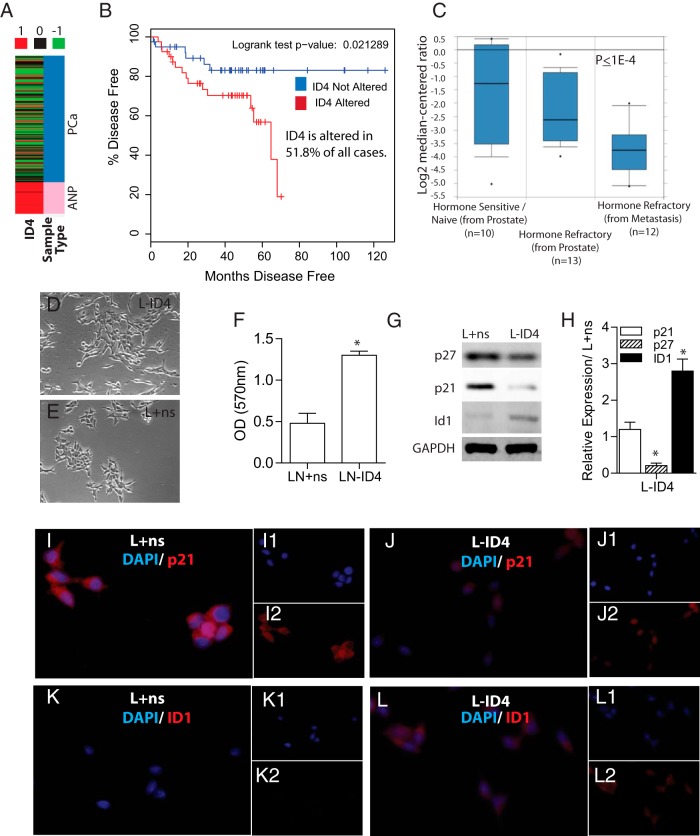
Decreased ID4 expression in prostate adenocarcinoma compared with that in adjacent normal prostate (Figure 1A) was observed in The Cancer Genome Atlas (TCGA) (22) prostate cancer adenocarcinoma (PRAD) gene expression (Illumina HiSeq) database. ID4 expression was also down-regulated in 51.8% of cases (mRNA expression, all complete tumors, Z-score threshold ±2.0) in the Memorial Sloan Kettering Cancer Center Prostate Adenocarcinoma (23) dataset (cBioPortal for Cancer Genomics) (24, 25), with a significant reduction in disease-free survival compared with that of cases expressing ID4 (P = .02) (Figure 1B). These results are consistent with decreased ID4 expression in PCa reported in our earlier studies (9, 12). ID4 expression was also decreased in CRPC compared with that in hormone-naive (P < .0004, ≥2-fold change, default) in the prostate dataset of Tamura et al (26) in Oncomine (Figure 1C).
Figure 1.
Expression of ID4 correlates inversely with prostate cancer. A, ID4 expression in prostate adenocarcinoma (PCa, blue) compared with that in adjacent normal prostate (ANP, pink) in The Cancer Genome Atlas (TCGA) prostate cancer adenocarcinoma (PRAD) gene expression (Illumina HiSeq) database. B, Of cases in the Memorial Sloan Kettering Cancer Center Prostate Adenocarcinoma dataset (cBioPortal for Cancer Genomics, mRNA expression, all complete tumors, Z-score threshold ±2.0), 58.1% of those with altered ID4 expression demonstrated a significant reduction in disease-free survival compared with those in which ID4 expression was not altered (P = .02). C, Expression of ID4 is decreased in CTPC compared with that in hormone-naive prostate cancer. The box and whisker plots from Oncomine were obtained from the prostate dataset of Tamura et al (26) and grouped by “Prostate hormone therapy (HT) response status (detailed).” The default thresholds were used (2-fold change, P < .00004). D and E, Morphology of L-ID4 (C) and L+ns cells (D) (phase-contrast images ×200). F, Proliferation rate of L+ns and L-ID4 cells expressed as absorption at 570 nm due to conversion of tetrazolium salt to formazan (means ± SEM, n = 5; *, P < .001) G, Immunoblot analysis of p27, p21, and ID1 protein levels in L+ns and L-ID4 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. A representative blot of at least 3 experiments is shown. H, Expression of p27, p21, and ID1 mRNA by real-time qRT-PCR using gene-specific primers (data were normalized to β-actin followed by relative expression compared with the respective genes in L+ns [set to 1]) (means ± SEM, n = 3; *, P < .001). I–L, Immunocytochemical localization of p21 (I and J) and ID1 (K and L) in L+ns (I and K) and L-ID4 (J and L) cells. Red staining is gene specific and blue is 4′,6-diamidino-2-phenylindole (DAPI) (see respective insets). A representative image of 3 experiments is shown.
ID4 knockdown promotes proliferation of LNCaP cells
LNCaP cells were used as the primary model to investigate the effect of ID4 on the PCa phenotype. LNCaP cells express ID4 as opposed to DU145 cells that lack ID4 expression because of hypermethylation of CpG dinucleotides within its promoter (9). LNCaP cells are also AR positive, androgen sensitive, less proliferative, and less tumorigenic than the highly tumorigenic DU145 cells (27).
L-ID4 cells established in our laboratory (14) appeared mesenchymal, elongated, and dispersed (Figure 1D), whereas L+ns cells clumped together with typical LNCaP morphology (Figure 1E). The proliferation rate of L-ID4 cells was approximately 2.5-fold higher than that of L+ns cells cultured in FBS (Figure 1F). Concomitant with increased proliferation, the expression (protein [Figure 1G] and mRNA [Figure 1H]) of cyclin-dependent kinase inhibitor 1B (CDKN1B, p27) decreased in L-ID4 compared with that in L+ns cells. Interestingly, cyclin-dependent kinase inhibitor 1A (CDKN1A, p21) protein declined (Figure 1G), but the mRNA remained unchanged in L-ID4 compared with that in L+ns cells (Figure 1H). The immunocytochemical localization also suggested a decline in CDKN1A protein in L-ID4 (Figure 1H). The discrepancy between CDKN1A mRNA and protein expression could be due to increased protein degradation (28). The expression of ID1 is also associated with increased proliferation (29). As expected, ID1 expression increased in L-ID4 compared with that in L+ns cells (Figure 1, F, G, and K).
ID4 knockdown promotes cell migration, invasion, and anchorage-independent cell growth
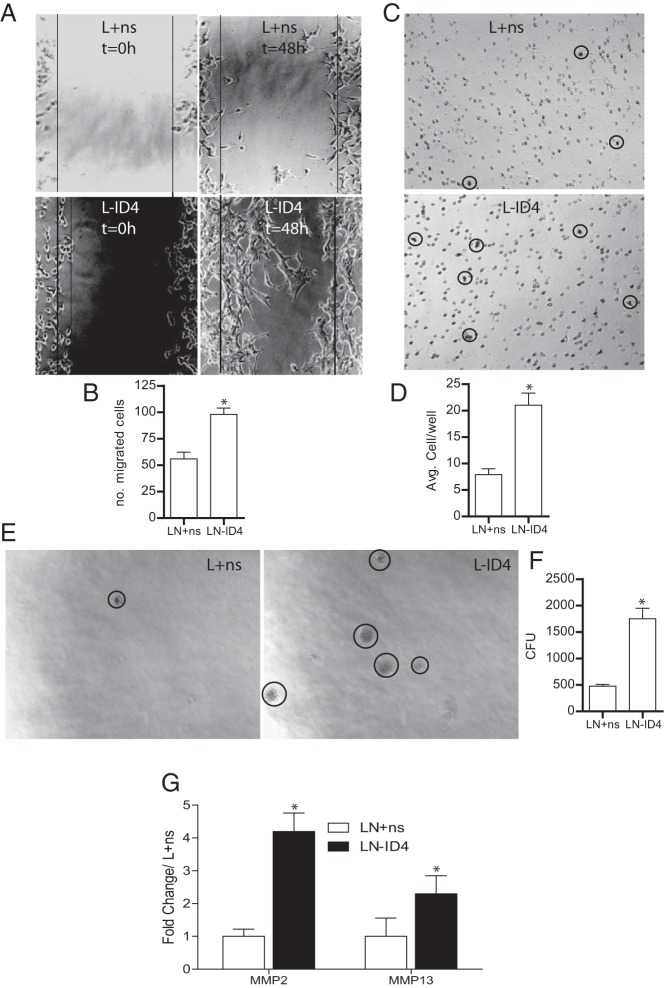
Forty-eight hours after the scratch wound, a larger fraction of L-ID4 than L+ns cells migrated into the wound space (Figure 2, A and B). At least 3 times higher invasion through Matrigel was observed with L-ID4 than with L+ns cells (Figure 2, C and D). The anchorage-independent growth of L-ID4 cells in soft agar demonstrated an approximately 4-fold increase in the number of colonies of L-ID4 cells compared with that of the L+ns controls (Figure 2, E and F). Increased invasion of L-ID4 cells was associated with 4- and 2-fold higher expression of the matrix-degrading enzymes matrix metallopeptidase (MMP) 2 and MMP13, respectively (Figure 2G). These results suggested that loss of ID4 promotes cell migration, invasion, and anchorage-independent growth, all hallmarks of an aggressive cancer.
Figure 2.
Motility, invasion, and anchorage-independent growth of L-ID4 cells. A, Motility of L+ns and L-ID4 cells was assessed by a wound-healing scratch assay. The boundaries of the scratch are indicated by the solid lines. A representative image of 3 different experiments is shown. B, Numbers of cells within the scratch (shown in panel A) were counted after 48 hours and are plotted as means ± SEM (n = 3; *, P < .001). C, Invasive capacity of L+ns and L-ID4 cells across Matrigel. Cells migrated across the Matrigel membrane (circled, representative image shown) were stained (HEMA2 stain set, Fisher Scientific). D, Numbers of cells migrated across the Matrigel membrane (shown in panel C) were counted and are expressed as means ± SEM (n = 3; *, P < .001). E, Anchorage-independent growth of L+ns and L-ID4 cells was assessed in a soft agar assay (representative image). The visible colonies are circled. F, Numbers of independent colonies were counted (panel E) and are expressed as means ± SEM (n = 3; *, P < .001). CFU, colony-forming units. G, Expression of MMP2 and MMP13 by real-time qRT-PCR. Data are expressed as means ± SEM (*, P < .001) from 3 experiments and are normalized to the expression of the respective genes in L+ns set to 1.
Knockdown of ID4 promotes increased tumor growth of subcutaneous xenografts in vivo
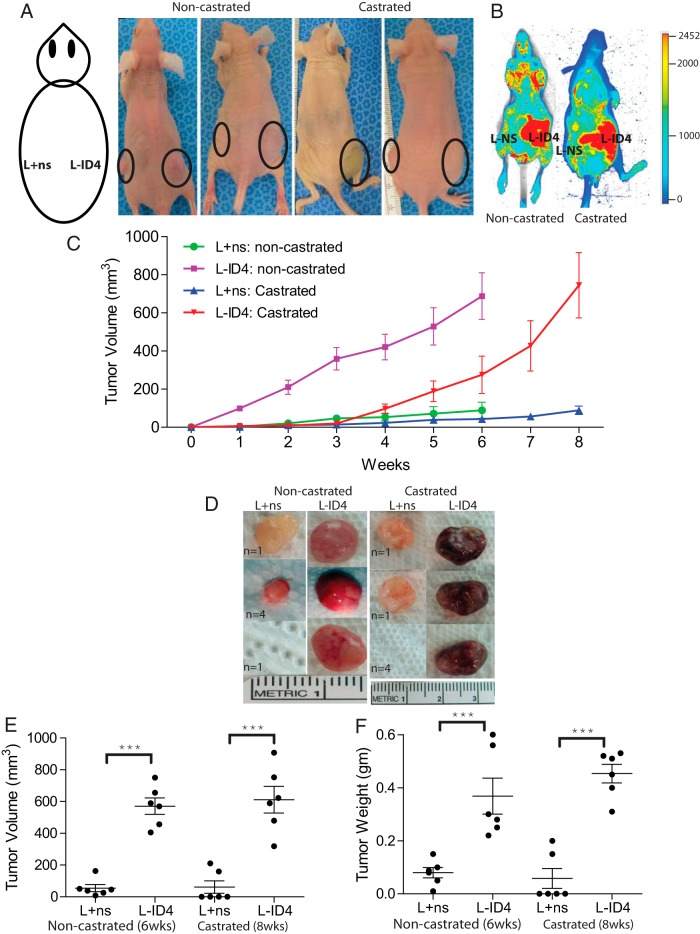
The L+ns and L-ID4 cells were subcutaneously injected into the flanks of NC and C nude male mice (Figure 3A) to investigate the effect of ID4 on androgen-dependent and -independent tumor formation, respectively. L-ID4 tumors in NC mice were observed within 1 week of injection and grew progressively over the period of 6 weeks by which time the experiment was terminated (Figure 3C). In contrast, a latent period of approximately 3 weeks of no L-ID4 tumor growth followed by accelerated tumor growth was observed in C mice (Figure 3C). At the end of the experiments (6 weeks for NC mice and 8 weeks for C mice), the tumors were excised and volume and weights were measured. In NC mice, the L+ns cells formed smaller tumors in 5 of 8 mice (62.5%), whereas no tumor was detected in 3 of 8 mice (37.5%) (Figure 3D). In contrast, 8 of 8 (100%) tumors were observed at the right flank with L-ID4 cells in NC mice. In the C group, only 2 of 8 mice (25%) showed measurable tumor growth on the left flank injected with L+ns cells, whereas tumors were detected at the right flank of all 8 mice injected with L-ID4 cells (100%), suggesting that L-ID4 cells are capable of forming tumors even at castration levels of androgens (Figure 3D). The tumor weights and volume were significantly higher than those of their respective controls in both NC and C mice (Figure 3, E and F). The weight and volume of L-ID4 tumors between NC and C mice were not statistically different (Figure 3, E and F). Interestingly, the tumors formed by L-ID4 cells in C mice appeared bloodier than their counterparts in NC mice (Figure 3D). The L-ID4 tumors in C mice required a shorter time (5 weeks) to achieve tumor volume and weight similar to those of the NC mice (Figure 3C). These observations might suggest increased angiogenesis (30) and a more aggressive phenotype that is usually associated with CRPC. The lag time in C mice also suggests that L-ID4 may require time to adapt to castrated levels of androgens before initiating in vivo growth. Collectively, these results indicated that knockdown of ID4 enhances tumorigenicity of PCa cells and promotes the CRPC phenotype.
Figure 3.
Inactivation of ID4 in LNCaP cells promotes tumor growth in vivo. A, L+ns and L-ID4 cells in Matrigel were injected into the left and right flanks of nude mice (male nu/nu), respectively, as shown in the schematic diagram. Tumor growth is shown by solid circles. B, At the end of the experiments, mice were injected with IRDye 800CW endothelial growth factor receptor targeting agent and evaluated for tumor-specific retention of the fluorophore by near-infrared fluorescence imaging (red). C, Volumes of the tumors were measured weekly (expressed as cubic millimeters, means ± SEM, n = 6/group). The noncastrated mice were killed at 6 weeks, whereas castrated mice were killed at 8 weeks. D, Representative xenograft images with numbers of mice with similar tumor (n) are shown. E and F, Respective volumes and weights (means ± SEM, n = 6) of the tumors after excision from the mice (***, P < .001, between L+ns and L-ID4 tumors in noncastrated and castrated mice, respectively).
Xenograft morphology and ID4, Ki67, CD31, ID1, and lamin expression
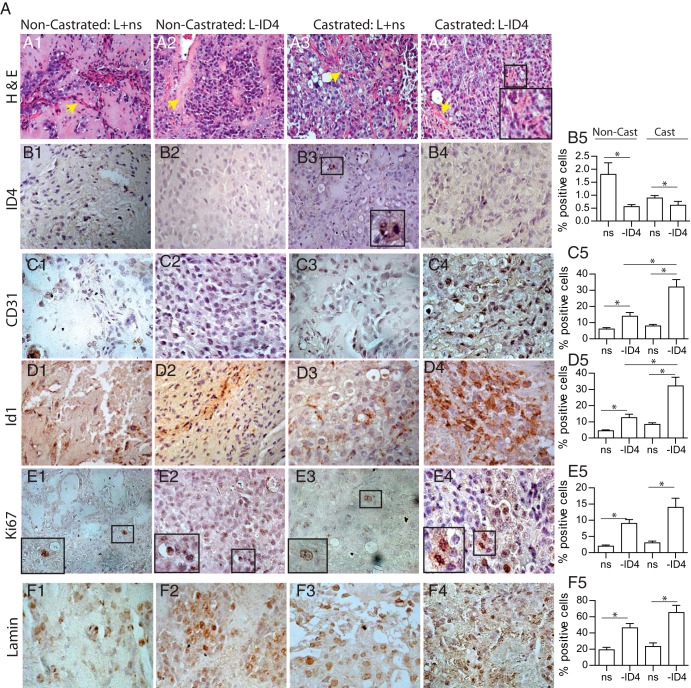
The histological examination of the tumors demonstrated an abundance of supporting fibromuscular tissue tracts (Figure 4, A1, A2, and A3, yellow arrows). Infiltrating red blood cells were observed more frequently in C L-ID4 xenografts, suggesting increased vascularization (Figure 4A4).
Figure 4.
Histological and immunohistological analysis of L+ns and L-ID4 xenografts from noncastrated and castrated nude mice. A1–A4, hematoxylin and eosin staining of tumor xenografts. The yellow arrows are indicative of fibromuscular tracts. The inset in A4 (castrated L-ID4) shows the infiltrating red blood cells. B1–F5, Immunohistochemical localization (brown staining) and quantitation of ID4 (B1–B5), CD31 (C1–C5), ID1 (D1–D5), Ki67 (E1–E5), and human-specific lamin (F1–F5) in tumor xenografts. The insets in the panels are enlarged versions of the respective boxed regions. All images are representative and at ×400 magnification. B5, C5, D5, E5, and F5 show means ± SEM (*, P < .001) of ID4-, CD31-, ID1-, Ki67-, and lamin-positive cells, respectively, in 5 random fields. Note the very small number of ID4-positive cells in all xenografts. Representative data are shown.
As expected, ID4 immunoreactivity was undetectable in L-ID4 xenografts but was detected in L+ns xenografts (Figure 4, B1–B4, and B5, ID4-positive cells). We have previously shown that ID4 is regulated by androgens in LNCaP cells (18). Thus, the unexpectedly low ID4 expression in C L+ns xenografts compared with that in NC L+ns cells (Figure 4B5) could possibly be due to lack of androgens in C mice.
The number of CD31-positive cells (a measure of neovascularization) (Figure 4, C1–C5) in C L-ID4 tumors was significantly higher than that in C L+ns tumors and NC L-ID4 tumors (Figure 4C5). High ID1 expression is also associated with increased proliferation (31), angiogenesis, and neovascularization (32) in PCa (33). In our studies, the expression of ID1 was also higher in L-ID4 xenografts than in L+ns xenografts (Figure 4, D1–D5). These results suggested that loss of ID4 increased tumor vasculature and a lack of circulating androgens potentiated vascularization of the tumors that was evident in the bloodier tumors observed in this group.
Ki-67 expression was also higher in NC and C L-ID4 tumors (Figure 4, E1–E5). Thus, increased tumor growth resulted from an increase in proliferation of L-ID4 cells than of L+ns cells in vivo. Predominant human-specific lamin A (using a human-specific antibody) expression observed in L-ID4 tumors obtained from NC and C mice (Figure 4, F1–F5) further suggested that increased tumor volume is due to the expansion of LNCaP cells and not due to mouse-derived stromal cells.
RNA-seq revealed a gene expression profile consistent with activation of androgens in L-ID4 cells
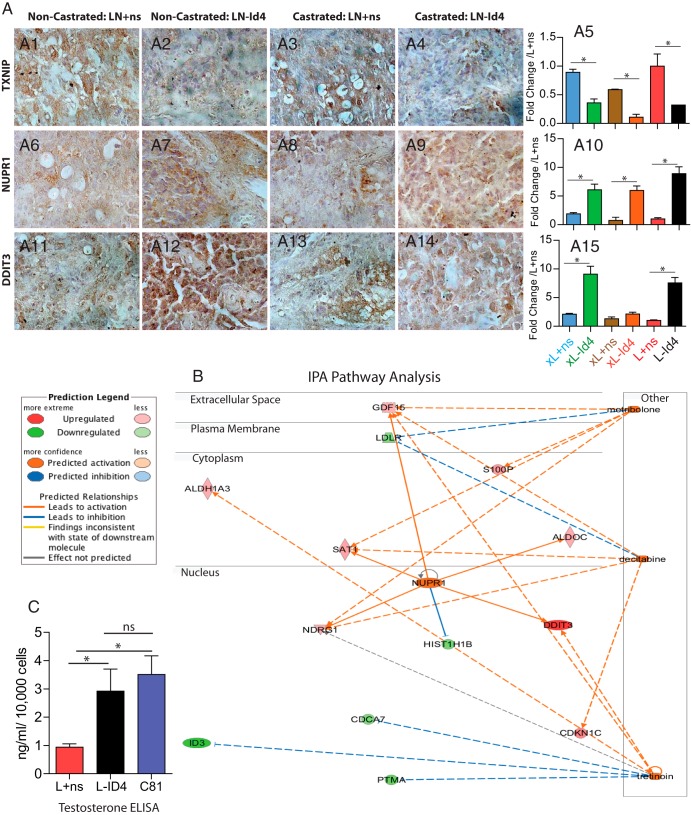
RNA-seq was performed to further evaluate the molecular basis of the CRPC phenotype in L-ID4 cell lines compared with that in L+ns cell lines. A pooled poly(dT)-primed cDNA library for each cell line was subjected to paired-end sequencing. The L+ns library yielded 22 914 987 mapped reads, whereas the L-ID4 library yielded 24 387 809 mapped reads. Default cuffdiff settings yielded 41 genes with significant gene expression differences between the 2 cell lines. There were 18 genes up-regulated and 23 genes down-regulated in L-ID4 relative to L+ns cell lines (Table 1). Both reads of a single paired-end fragment mapped to ID4 in the L+ns cells, whereas no reads mapped to ID4 in L-ID4 cells, consistent with down-regulation of ID4 in the L-ID4 cell line. The down-regulation of thioredoxin-interacting protein (TXNIP) (Figure 5, A1–A4) and up-regulation of nuclear protein, transcriptional regulator, 1 (NUPR1) (Figure 5, A6–A9), and DNA damage inducible transcript 3 (DDIT3) (Figure 5, A11–A14) in tumor xenografts (NC and C, IHC) and in the L-ID4 cell line (qRT-PCR) (Figure 5, A5, A10, and A15), respectively, supported the RNA-seq data (Table 1). The .bam files have been deposited into the Sequence Read Archive (SRA) resource at National Center for Biotechnology Information (NCBI) under BioProject identification number PRJNA229873.
Table 1.
List of Significantly Up-Regulated and Down-Regulated Genes in L-ID4 Compared With Those in L+ns Control Cells
| Gene Symbol | Entrez Gene Name | Location | Type | Fold Regulation |
|---|---|---|---|---|
| DDIT3 | DNA-damage-inducible transcript 3 | Nucleus | Transcription regulator | 15.6465 |
| AMACR | α-Methylacyl-CoA racemase | Cytoplasm | Enzyme | 6.4582 |
| PRSS1 | Protease, serine,1 (trypsin 1) | Extracellular space | Peptidase | 6.1290 |
| SLC26A3 | Solute carrier family 26, member 3 | Plasma membrane | Transporter | 6.0425 |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | Nucleus | Other | 5.1054 |
| RGS2 | Regulator of G-protein signaling 2, 24 kDa | Nucleus | Other | 4.4968 |
| DIO1 | Deiodinase, iodothyronine, type I | Cytoplasm | Enzyme | 4.1950 |
| S100P | S100 calcium binding protein P | Cytoplasm | Other | 3.7255 |
| SYCE3 | Synaptonemal complex central element protein 3 | Nucleus | Other | 3.6100 |
| SAT1 | Spermidine/spermine N1-acetyltransferase 1 | Cytoplasm | Enzyme | 3.3894 |
| CRISP3 | Cysteine-rich secretory protein 3 | Extracellular space | Other | 3.3368 |
| FXYD3 | FXYD domain containing ion transport regulator 3 | Plasma membrane | Other | 3.0587 |
| SYT4 | Synaptotagmin IV | Cytoplasm | Transporter | 3.0510 |
| ALDOC | Aldolase C, fructose-bisphosphate | Cytoplasm | Enzyme | 2.9990 |
| NUPR1 | Nuclear protein, transcriptional regulator, 1 | Nucleus | Transcription regulator | 2.9928 |
| ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | Cytoplasm | Enzyme | 2.9411 |
| NDRG1 | N-myc downstream regulated 1 | Nucleus | Kinase | 2.9277 |
| GDF15 | Growth differentiation factor 15 | Extracellular space | Growth factor | 2.8433 |
| MCM5 | Minichromosome maintenance complex component 5 | Nucleus | Enzyme | −2.7266 |
| TMEM179B | Transmembrane protein 179B | Other | Other | −2.7503 |
| TUBA1A | Tubulin, α 1a | Cytoplasm | Other | −2.8099 |
| RPN1 | Ribophorin I | Cytoplasm | Enzyme | −2.8183 |
| GPX4 | Glutathione peroxidase 4 | Cytoplasm | Enzyme | −2.8186 |
| NUP188 | Nucleoporin 188 kDa | Nucleus | Other | −2.8307 |
| ATP6V0B | ATPase, H+ transporting, lysosomal 21 kDa, V0 subunit b | Cytoplasm | Transporter | −2.8322 |
| YEATS4 | YEATS domain containing 4 | Nucleus | Transcription regulator | −2.8511 |
| CDCA5 | Cell division cycle associated 5 | Cytoplasm | Other | −2.9944 |
| FH | Fumarate hydratase | Cytoplasm | Enzyme | −3.0990 |
| RNF26 | Ring finger protein 26 | Other | Other | −3.2874 |
| RNF114 | Ring finger protein 114 | Extracellular space | Other | −3.3081 |
| LDLR | Low-density lipoprotein receptor | Plasma membrane | Transporter | −3.3277 |
| MRPL16 | Mitochondrial ribosomal protein L16 | Cytoplasm | Other | −3.5228 |
| CDCA7 | Cell division cycle associated 7 | Nucleus | Other | −3.5235 |
| RPS15 | Ribosomal protein S15 | Cytoplasm | Other | −3.5653 |
| TUBB4B | Tubulin, β 4B class IVb | Cytoplasm | Other | −3.6613 |
| HIST1H1B | Histone cluster 1, H1b | Nucleus | Other | −3.7330 |
| PTMA | Prothymosin, α | Nucleus | Other | −4.3649 |
| LSM7 | LSM7 homolog, U6 small nuclear RNA associated (Saccharomyces cerevisiae) | Nucleus | Other | −4.5048 |
| NDUFS7 | NADH dehydrogenase (ubiquinone) | Cytoplasm | Enzyme | −5.7413 |
| ID3 | Inhibitor of DNA binding 3, helix-loop-helix protein | Nucleus | Transcription regulator | −9.0171 |
| TXNIP | Thioredoxin interacting protein | Cytoplasm | Other | −11.0709 |
Figure 5.
Comprehensive pathway, network analysis, and expression of significantly regulated genes in L+ns and L-ID4 cells and xenografts. A, Immunohistological analysis of the 3 most highly regulated genes in L+ns and L-ID4 detected by next-generation sequencing (RNA-seq) (Table 1). A1–A4, TXNIP. A6–A9, NUPR1, A11–A14, DDIT3. Representative images at ×400 are shown. A5, A10, and A15, expression of TXNIP (A5), NUPR1 (A10), and DDIT3 (A15) by real-time qRT-PCR in RNA isolated from tumor xenografts (indicated by prefix x) and L+ns and L-ID4 cell lines. The expression is normalized to the respective mRNA expression in L+ns cell lines (means ± SEM; *, P < .001). B, Pathway and network analysis of the 41 most regulated genes (shown in Table 1). Three major pathways were detected (metribolone (R1881), decitabine (5-azacitidine), and tretinoin (retinol). See the legend on the left for additional descriptions. C, Testosterone ELISA on L+ns, L-ID4, and C81 conditioned media. Testosterone levels (means ± SEM of 3 experiments in triplicate) are represented as nanograms per milliliter per 10,000 cells. C81 cells were used as positive controls. ns, not significant.
The 41 significantly differentially expressed genes were analyzed through Ingenuity Pathway Analysis Upstream Regulator Analysis (IPA-URA) (Ingenuity Systems, www.ingenuity.com). The goal of IPA-URA is to identify upstream regulators and predict whether they are activated or inhibited, given the observed gene expression changes. The analysis revealed that L-ID4 cells had gene expression profiles similar to those observed after activation with metribolone (R1881, a synthetic androgen analog), NUPR1, decitabine (5-aza-2′-deoxycytidine), and tretinoin (all-trans retinoic acid) (Figure 5B and Supplemental Table 3). Consistent with these results, NUPR1 is 1 of the 18 genes that is significantly up-regulated in our data and is predicted to be activated by Ingenuity analysis. In addition, 5 target genes (GDF15, LDLR, NDRG1, S100P, and SAT1) responded in L-ID4 cells as if the cells had been treated with metribolone, indicating activation of the AR. These results prompted us to investigate whether L-ID4 cells gained the capacity to synthesize androgens, the natural analog of metribolone.
Loss of ID4 expression promotes de novo steroidogenesis
Three-fold higher levels of testosterone were observed in L-ID4 than in L+ns conditioned media. The levels of testosterone in L-ID4 cells were similar to those observed in C81 cells, which are known to synthesize testosterone (34) (Figure 5C). These results indicated that knockdown of ID4 in LNCaP cells phenocopies AR activation in part due to de novo steroidogenesis.
Persistent AR expression and activity in ID4 knockdown LNCaP cells
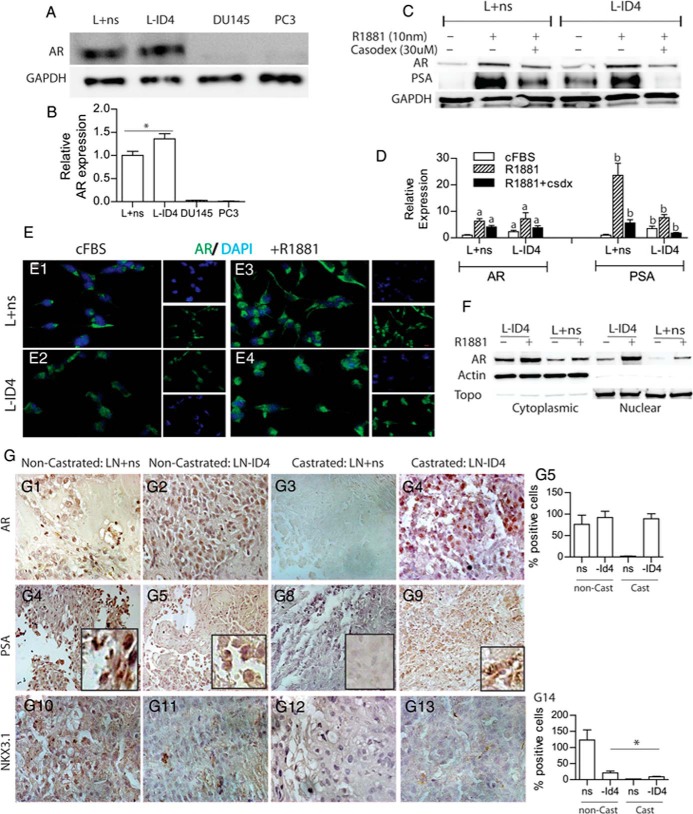
AR expression (Figure 6, A and B) was significantly higher in L-ID4 than in L+ns cells in FBS. AR in L-ID4 cells cultured for 24 hours in cFBS was also significantly higher than that in L+ns cells (Figure 6, C and D, and E1 and E2, ICC, green staining). The immunoblots of cytoplasmic and nuclear extracts and ICC further demonstrated that AR is cytoplasmic in L+ns cells and cytoplasmic and nuclear in L-ID4 cells in cFBS (Figure 6, E1, E2, and F). A similar increase in AR protein was observed between L+ns and L-ID4 cells after treatment with R1881 (Figure 6, C and D). These results were also recapitulated in ICC studies, which demonstrated that AR is translocated to the nucleus in L+ns cells (Figure 6, E3 and E4) upon R1881 treatment. The effect of ID4 knockdown on AR localization by immunoblot analysis also confirmed that a larger fraction of AR is nuclear in L-ID4 than in L+ns cells in cFBS (Figure 6F).
Figure 6.
AR expression and activity in L+ns and L-ID4 cell lines and xenografts. A, AR protein levels in L+ns and L-ID4 cultured in 10% FBS for 24 hours. DU145 and PC3 cells were used as negative controls for AR expression. B, Semiquantitative AR protein levels normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and then to AR levels in L+ns (set to 1) from 3 different experiments (means ± SEM, n = 3; *, P < .001). C, Protein levels of AR and PSA (a marker for AR activity) in L+ns and L-ID4 cells in response to R1881. The cells were cultured for 24 hours in cFBS before treatment with R1881 (10 nM) or R1881 + Casodex (30 μM, antiandrogen) for 24 hours. D, Semiquantitative AR and PSA protein levels normalized to GAPDH and then to AR or PSA levels in L+ns (set to 1) from 3 different experiments (means ± SEM, n = 3; a and b, P < .001 for AR and PSA in L+ns set to 1, respectively). GAPDH was used as a loading control. A representative immunoblot of 3 different experiments is shown. E, Immunofluorescence localization of AR (green) in L+ns (E1 and E3) and L-ID4 (E3 and E4) in the presence (E3 and E4) or absence (E1 and E2) of 10 nM R1881 for 24 hours. Representative images from 3 different experiments are shown. DAPI, 4′,6-diamidino-2-phenylindole. F, Cytoplasmic vs nuclear immunolocalization of AR in L+ns and L-ID4 cells in response to R1881 (10 nM). Topoisomerase (Topo) and actin were used to determine the purity and loading of nuclear and cytoplasmic extracts, respectively. G, Immunohistological localization of AR (G1–G4), PSA, (G6–G9) and NKX3.1 (G10–G13) (markers of AR transcriptional activity) protein levels in L+ns and L-ID4 xenografts from noncastrated (non-Cast) and castrated (Cast) mice. All images are representative at ×400 magnification. G5 and G14 show means ± SEM of AR- and NKX3.1-positive nuclei counted in 5 random fields (*, P < .001). PSA protein expression was not quantitated because of the diffuse nature of the staining.
The expression of prostate-specific antigen (PSA), an androgen response marker, was significantly higher in L-ID4 cells, but was undetectable in L+ns cells in cFBS (Figure 6, C and D). In the presence of the R1881, PSA increased significantly in L+ns and L-ID4 cells compared with that in their respective untreated controls (Figure 6, C and D). In both cell lines, R1881-dependent expression of AR and PSA was reversed by the antiandrogen Casodex, suggesting androgen-dependent regulation. These results led us to conclude that AR is constitutively expressed and active in L-ID4 compared with that in L+ns cells, possibly due to the increased steroidogenic capacity of L-ID4 cells (Figure 5B).
AR expression was observed in all xenografts except in L+ns tumors obtained from C mice (Figure 6, G1–G5). Because androgens are also required to maintain AR expression (35), we speculate that long-term deprivation of androgens in C mice could result in undetectable AR in L+ns xenografts (Figure 6G3). The expression of PSA also supported AR expression in the tumors. Whereas no PSA was observed in L+ns tumors in C mice (Figure 6G8), PSA immunoreactivity was observed in L+ns (Figure 6G6) and L-ID4 (Figure 6G7) tumors in NC mice and L-ID4 tumors in C mice (Figure 6G8). These results are also in accordance with PSA expression in the absence or presence of R1881 in the in vitro cell line data discussed above. The expression of NKX3.1, an androgen-regulated gene, was similar to PSA and AR expression in NC L+ns xenografts but was significantly attenuated in NC L-ID4 xenografts and undetectable in C mice xenografts. NKX3.1 is a PCa tumor suppressor (36); hence, its loss of expression is consistent with tumor growth in L-ID4 xenografts. Furthermore, previous studies have also shown that ID4 knockdown (in LNCaP cells and in Id4−/− mice) resulted in attenuated NKX3.1 expression (10).
Discussion
Studies from our laboratory, including meta-analyses, have demonstrated that ID4 expression is decreased in PCa (9, 12). In particular, a strong association between decreased ID4 expression and hormone-refractory metastatic PCa and shorter disease-free survival shows direct clinical relevance of ID4. In this study, we demonstrate that knockdown of ID4, which mirrors ID4 expression in PCa increases tumorigenicity both in vitro and in vivo and promotes CRPC. We focused primarily on the LNCaP cell line model because it closely resembles the androgen-sensitive PCa model and under appropriate conditions can transition to an androgen-independent cell line in vitro and in vivo.
Enhanced tumor formation by cells with ID4 knockdown (L-ID4) unequivocally supports the tumor suppressor activity of ID4 that correlates with the epigenetic inactivation of ID4 in PCa (9, 11). The cancer phenotypes including proliferation, migration, invasion, and anchorage-independent colony formation are significantly increased in L-ID4 cells. Elevated MMP2 and MMP13 (37, 38) expression, which is often associated with more aggressive PCa and promotes migration through extracellular matrix barriers, is also found in L-ID4 cells. Reduced expression of the cyclin-dependent kinase inhibitors CDKN1A (p21) and CDKN1B (p27) supports the higher proliferative index of L-ID4 cells. Conversely, ectopic expression of ID4 in DU145 and PC3 cells leads to cell cycle arrest, which is associated with increased CDKN1A expression (12).
ID1 expression is elevated in PCa (17). Ectopic ID1 expression in LNCaP cells also promotes androgen independence, proliferation (39), and migration (40). Increased migration of LNCaP cells overexpressing ID1, a phenotype that is essentially similar to L-ID4 cells, is in part due to increased MMP2 activity (41). It is possible that the elevated ID1 expression in L-ID4 cells could lead to androgen independence, increased tumorigenicity, and invasiveness. ID1 promotes proliferation by down-regulating CDKN1A (42), whereas ectopic ID4 expression attenuates proliferation by up-regulating CDKN1A. An inverse correlation between ID1 and ID4 expression in L+ns and L-ID4 cells, PCa (41), and ID4 knockout mice that develop PIN-like lesions (10) is therefore noteworthy. These results suggest that ID1 may actually compensate for the loss of ID4 in terms of expression, but fails to compensate functionally and instead promotes a PCa phenotype.
The major observation made in this study is the acquisition of a castration-resistant phenotype of L-ID4 cells both in vitro and in vivo. The bloody tumors formed by LNCaP ID4 cells in castrated nude mice are similar to the bloody LNCaP xenografts observed in castrated SCID mice (43). The authors attributed this tumor phenotype to CRPC (43). C81 cells, the androgen-independent derivative of LNCaP cells, which lack ID4 expression due to promoter hypermethylation (9), also form dark tumors in castrated nude mice (44).
At the molecular level, we show a strong association between known markers of PCa progression (Ki67, PSA, ID1, NKX3.1, and CD31) with L-ID4 xenografts. The gene expression signature (RNA-seq) revealed in L-ID4 cells was predicted by IPA-URA to be consistent with that seen by constitutive activation of AR by metribolone (methyltreienolone, R1881), a synthetic androgen. These analyses are also supported by increased AR expression and activity in terms of PSA expression that was insensitive to R1881 treatment in L-ID4 cells and xenografts, a hallmark of androgen independence. One of the mechanisms that could explain the L-ID4 cells as a phenocopy of R1881 treatment is the gain in de novo steroidogenesis by L-ID4 cells. Studies have suggested that PCa cells acquire the ability to synthesize intratumoral androgens (2) as a compensatory mechanism to castration (chemical and physical), triggering AR activation and disease progression. Both in vitro and in vivo studies have also shown increased testosterone and dihydrotestosterone levels in castration-resistant LNCaP tumors (45) and C81 cells (34), a derivative of LNCaP cells cultured progressively under androgen-deprived conditions (13). Incidentally, androgen-insensitive C81 cells also lack ID4 expression (9), suggesting that loss of ID4 could promote CRPC. Thus, deficiency of ID4 could potentiate intratumoral androgen synthesis, leading to constitutive AR expression and activity. The down-regulation of PSA expression by the androgen antagonist Casodex suggested that AR activation in L-ID4 was still dependent on ligand binding in vitro. These results suggest that CRPC could retain sensitivity to antiandrogens as shown recently by the new generation of nonsteroidal antiandrogens (eg, MDV3100) (46).
In this study, we did not investigate the expression of enzymes that are involved in androgen biosynthesis in L-ID4 cells; hence, the de novo steroidogenesis mechanism in L-ID4 is speculative. Moreover, sensitive and accurate quantitative analysis of testosterone and other intermediate steroid precursors/metabolites may provide a better insight into the mechanisms involved in the gain of the CRPC phenotype of L-ID4 cells in vitro and in vivo.
The RNA-seq–based gene expression profile also supported increased tumorigenicity (AMACR [47], PRSS1, CDKN1C (48), NUPR1 [49], and NDRG1 [50]) and the CRPC phenotype of L-ID4 (CRISP3 [51], and S100P [52]). The decitabine-regulated gene signature in the IPA-UPA also reflects the extensive promoter hypomethylation in L-ID4 cells, consistent with hypomethylation of S100P in PCa (52).
ID4 lacks DNA binding activity and hence is essentially incapable of regulating gene expression directly. This is perhaps reflected by the small number of genes (41) that were found to be differentially expressed between LNCaP and LNCaP-ID4 cells. An important question that remains to be addressed is the structural basis of the unique function of ID4 compared with that of ID1, ID2, and ID3. Outside of the conserved HLH domain, the N- and C- terminal ends of ID4 are unique (alanine and proline rich, respectively) and could provide some insight into the structural properties of ID4, resulting in its distinct biological activity. We anticipate that ID4 could regulate the activity of as yet unknown, but distinct, transcription factors or a host of other proteins that could indirectly result in its tumor suppressor activity.
To fully understand the role of ID4 in PCa and CRPC, a cohort study that can establish a relationship between ID4 expression, AR status (androgen sensitive or insensitive), and disease stage needs to be performed. The xenograft studies also need to be reevaluated to explore the effect of ID4 on PCa and CRPC by using an inducible ID4 knockdown in LNCaP and other ID4-expressing and androgen-sensitive cell lines such as LAPC4. Nevertheless, we expect that the studies reported herein are the first steps to unraveling the complex role of ID4 in PCa. The knowledge gained from characterizing the prostate phenotype from Id4−/− mice (10) (PIN lesions, PTEN−, pAkt+, Id1+, Myc+, NKX3.1−, and AR+) could be integrated with these results to develop a working model by which knockdown of ID4 promotes the PCa and CRPC phenotype.
Additional material
Supplementary data supplied by authors.
Acknowledgments
The work was supported by the National Cancer Institute, National Institutes of Health (Grant CA128914 to J.C.) and in part by the National Center for Research Resources, Research Center in Minority Institutions, National Institutes of Health (Grant G12RR03062 (to the Center for Cancer Research and Therapeutic Development).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AR
- androgen receptor
- C
- castrated
- CRPC
- castration-resistant prostate cancer
- DDIT3
- DNA damage inducible transcript 3
- HLH
- helix-loop-helix
- ICC
- immunocytochemistry
- ID4
- inhibitor of differentiation 4
- IHC
- immunohistochemistry
- IPA-URA
- Ingenuity Pathway Analysis Upstream Regulator Analysis
- MMP
- matrix metalloproteinase
- NC
- noncastrated
- NUPR1
- nuclear protein, transcriptional regulator, 1
- PCa
- prostate cancer
- PIN
- prostatic intraepithelial neoplasia
- PSA
- prostate-specific antigen
- qRT-PCR
- quantitative RT-PCR
- RNA-seq
- RNA sequencing
- TXNIP
- thioredoxin-interacting protein.
References
- 1. Wang J, Shang ZQ, Niu YJ. Androgen receptor roles in benign and malignant prostate disease. Clin Oncol Cancer Res. 2011;8:85–91. [Google Scholar]
- 2. Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab. 2010;21:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kobayashi T, Inoue T, Kamba T, Ogawa O. Experimental evidence of persistent androgen-receptor-dependency in castration-resistant prostate cancer. Int J Mol Sci. 2013;14:15615–15635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riechmann V, van Crüchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22:749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu L, Liu C, Vandeusen J, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–274. [DOI] [PubMed] [Google Scholar]
- 6. Chen SS, Claus R, Lucas DM, et al. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood. 2011;117:862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan AS, Tsui WY, Chen X, et al. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene. 2003;22:6946–6953. [DOI] [PubMed] [Google Scholar]
- 8. Castro M, Grau L, Puerta P, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharma P, Chinaranagari S, Patel D, Carey J, Chaudhary J. Epigenetic inactivation of inhibitor of differentiation 4 (Id4) correlates with prostate cancer. Cancer Med. 2012;1:176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma P, Knowell AE, Chinaranagari S, et al. Id4 deficiency attenuates prostate development and promotes PIN-like lesions by regulating androgen receptor activity and expression of NKX3.1 and PTEN. Mol Cancer. 2013;12:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vinarskaja A, Goering W, Ingenwerth M, Schulz WA. ID4 is frequently downregulated and partially hypermethylated in prostate cancer. World J Urol. 2012;30:319–325. [DOI] [PubMed] [Google Scholar]
- 12. Carey JP, Asirvatham AJ, Galm O, Ghogomu TA, Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Igawa T, Lin FF, Lee MS, Karan D, Batra SK, Lin MF. Establishment and characterization of androgen-independent human prostate cancer LNCaP cell model. Prostate. 2002;50:222–235. [DOI] [PubMed] [Google Scholar]
- 14. Knowell A, Patel D, Morton D, Sharma P, Glymph S, Chaudhary J. Id4 dependent acetylation restores mutant-p53 transcriptional activity. Mol Cancer. 2013;12:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin MF, Meng TC, Rao PS, Chang C, Schonthal AH, Lin FF. Expression of human prostatic acid phosphatase correlates with androgen-stimulated cell proliferation in prostate cancer cell lines. J Biol Chem. 1998;273:5939–5947. [DOI] [PubMed] [Google Scholar]
- 16. Walker L, Millena AC, Strong N, Khan SA. Expression of TGFβ3 and its effects on migratory and invasive behavior of prostate cancer cells: involvement of PI3-kinase/AKT signaling pathway. Clin Exp Metastasis. 2013;30:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sharma P, Patel D, Chaudhary J. Id1 and Id3 expression is associated with increasing grade of prostate cancer: Id3 preferentially regulates CDKN1B. Cancer Med. 2012;1:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Asirvatham AJ, Schmidt MA, Chaudhary J. Non-redundant inhibitor of differentiation (Id) gene expression and function in human prostate epithelial cells. Prostate. 2006;66:921–935. [DOI] [PubMed] [Google Scholar]
- 19. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldman M, Craft B, Swatloski T, et al. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 2013;41(Database issue):D949–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, Furihata M, Tsunoda T, et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. [DOI] [PubMed] [Google Scholar]
- 27. Venanzoni MC, Giunta S, Muraro GB, et al. Apolipoprotein E expression in localized prostate cancers. Int J Oncol. 2003;22:779–786. [PubMed] [Google Scholar]
- 28. Lu Z, Hunter T. Ubiquitylation and proteasomal degradation of the p21Cip1, p27Kip1 and p57Kip2 CDK inhibitors. Cell Cycle. 2010;9:2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. [DOI] [PubMed] [Google Scholar]
- 30. Huss WJ, Hanrahan CF, Barrios RJ, Simons JW, Greenberg NM. Angiogenesis and prostate cancer: identification of a molecular progression switch. Cancer Res. 2001;61:2736–2743. [PubMed] [Google Scholar]
- 31. Ouyang XS, Wang X, Ling MT, Wong HL, Tsao SW, Wong YC. Id-1 stimulates serum independent prostate cancer cell proliferation through inactivation of p16INK4a/pRB pathway. Carcinogenesis. 2002;23:721–725. [DOI] [PubMed] [Google Scholar]
- 32. Lyden D, Young AZ, Zagzag D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. [DOI] [PubMed] [Google Scholar]
- 33. Ling MT, Lau TC, Zhou C, et al. Overexpression of Id-1 in prostate cancer cells promotes angiogenesis through the activation of vascular endothelial growth factor (VEGF). Carcinogenesis. 2005;26:1668–1676. [DOI] [PubMed] [Google Scholar]
- 34. Dillard PR, Lin MF, Khan SA. Androgen-independent prostate cancer cells acquire the complete steroidogenic potential of synthesizing testosterone from cholesterol. Mol Cell Endocrinol. 2008;295:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takeda H, Nakamoto T, Kokontis J, Chodak GW, Chang C. Autoregulation of androgen receptor expression in rodent prostate: Immunohistochemical and in situ hybridization analysis. Biochem Biophys Res Commun. 1991;177:488–496. [DOI] [PubMed] [Google Scholar]
- 36. Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao X, Thrasher JB, Pelling J, Holzbeierlein J, Sang QX, Li B. Androgen stimulates matrix metalloproteinase-2 expression in human prostate cancer. Endocrinology. 2003;144:1656–1663. [DOI] [PubMed] [Google Scholar]
- 38. Pang ST, Flores-Morales A, Skoog L, Chuan YC, Nordstedt G, Pousette A. Regulation of matrix metalloproteinase 13 expression by androgen in prostate cancer. Oncol Rep. 2004;11:1187–1192. [PubMed] [Google Scholar]
- 39. Ling MT, Wang X, Lee DT, Tam PC, Tsao SW, Wong YC. Id-1 expression induces androgen-independent prostate cancer cell growth through activation of epidermal growth factor receptor (EGF-R). Carcinogenesis. 2004;25:517–525. [DOI] [PubMed] [Google Scholar]
- 40. Ling MT, Wang X, Ouyang XS, et al. Activation of MAPK signaling pathway is essential for Id-1 induced serum independent prostate cancer cell growth. Oncogene. 2002;21:8498–8505. [DOI] [PubMed] [Google Scholar]
- 41. Coppe JP, Itahana Y, Moore DH, Bennington JL, Desprez PY. Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin Cancer Res. 2004;10:2044–2051. [DOI] [PubMed] [Google Scholar]
- 42. Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andersen RJ, Mawji NR, Wang J, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–546. [DOI] [PubMed] [Google Scholar]
- 44. Lai KP, Huang CK, Chang YJ, Chung CY, et al. New therapeutic approach to suppress castration-resistant prostate cancer using ASC-J9 via targeting androgen receptor in selective prostate cells. Am J Pathol. 2013;182:460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Locke JA, Nelson CC, Adomat HH, Hendy SC, Gleave ME, Guns ES. Steroidogenesis inhibitors alter but do not eliminate androgen synthesis mechanisms during progression to castration-resistance in LNCaP prostate xenografts. J Steroid Biochem Mol Biol. 2009;115:126–136. [DOI] [PubMed] [Google Scholar]
- 46. Tran C, Ouk S, Clegg NJ, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carnell AJ, Kirk R, Smith M, McKenna S, Lian LY, Gibson R. Inhibition of human α-methylacyl CoA racemase (AMACR): a target for prostate cancer [published online ahead of print August 8, 2013]. ChemMedChem. 2013. doi: 10.1002/cmdc.201300179. [DOI] [PubMed] [Google Scholar]
- 48. Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. [DOI] [PubMed] [Google Scholar]
- 49. Chowdhury UR, Samant RS, Fodstad O, Shevde LA. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 2009;28:225–232. [DOI] [PubMed] [Google Scholar]
- 50. Song Y, Oda Y, Hori M, et al. N-myc downstream regulated gene-1/Cap43 may play an important role in malignant progression of prostate cancer, in its close association with E-cadherin. Hum Pathol. 2010;41:214–222. [DOI] [PubMed] [Google Scholar]
- 51. Dahlman A, Edsjö A, Halldén C, et al. Effect of androgen deprivation therapy on the expression of prostate cancer biomarkers MSMB and MSMB-binding protein CRISP3. Prostate Cancer Prostatic Dis. 2010;13:369–375. [DOI] [PubMed] [Google Scholar]
- 52. Wang Q, Williamson M, Bott S, Brookman-Amissah N, et al. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–6565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.