Abstract
The hypothalamic arcuate nucleus controls many critical homeostatic functions including energy homeostasis, reproduction, and motivated behavior. Although G protein–coupled receptors (GPCRs) are involved in the regulation of these functions, relatively few of the GPCRs have been identified specifically within the arcuate nucleus. Here, using TaqMan low-density arrays we quantified the mRNA expression of nonolfactory GPCRs in mouse arcuate nucleus. An unprecedented number of GPCRs (total of 292) were found to be expressed, of which 183 were known and 109 were orphan GPCRs. The known GPCR genes expressed were classified into several functional clusters including hormone/neurotransmitter, growth factor, angiogenesis and vasoactivity, inflammation and immune system, and lipid messenger receptors. The plethora of orphan genes expressed in the arcuate nucleus were classified into 5 structure-related classes including class A (rhodopsin-like), class B (adhesion), class C (other GPCRs), nonsignaling 7-transmembrane chemokine-binding proteins, and other 7-transmembrane proteins. Therefore, for the first time, we provide a quantitative estimate of the numerous GPCRs expressed in the hypothalamic arcuate nucleus. Finally, as proof of principle, we documented the expression and function of one of these receptor genes, the glucagon-like peptide 1 receptor (Glp1r), which was highly expressed in the arcuate nucleus. Single-cell RT-PCR revealed that Glp1r mRNA was localized in proopiomelanocortin neurons, and using whole-cell recording we found that the glucagon-like peptide 1-selective agonist exendin-4 robustly excited proopiomelanocortin neurons. Thus, the quantitative GPCR data emphasize the complexity of the hypothalamic arcuate nucleus and furthermore provide a valuable resource for future neuroendocrine/endocrine-related experiments.
The hypothalamic arcuate nucleus is a small but complex center that controls many critical physiological functions including energy homeostasis, reproduction, locomotor activity, and motivated behaviors (1–9). Many of these functions are controlled or modulated via G protein–coupled receptors (GPCRs), such as opioid, melanocortin, neuropeptide Y (NPY), GPR54, γ-aminobutyric acid (GABA) B, serotonin, and adrenergic receptors (10–18). Although much is known about the anatomical distribution and physiological functions of many of the GPCRs that are expressed in the hypothalamus (19–22), there is still an information gap concerning the quantitative expression of GPCR subtypes and the expression of orphan GPCRs within the arcuate nucleus.
GPCRs comprise the largest family of cell surface signaling molecules and are encoded by approximately 800 genes in the human genome (23). Approximately half of these are olfactory or taste receptors, and the remaining (∼375) are nonolfactory (23). In addition, there are many orphan GPCRs, so called because the endogenous ligand and functions are not known (21, 23).
To elucidate the expression of nonolfactory GPCRs, we performed TaqMan low-density arrays (384-well setting) and investigated nonolfactory GPCR mRNA expression in the arcuate nucleus of female mice. Interestingly, we found that 292 of the 353 GPCRs in the array, of which 109 were orphans, were expressed in the arcuate nucleus. The expression of these orphans within the arcuate nucleus argues for a role in homeostatic functions, which may help to deorphanize the receptors because many neuropeptides and other ligands for GPCRs for this brain region are known (8, 21).
The array revealed that GABAB R1 and R2 are highly expressed, which confirms our previous in vitro electrophysiology findings dating back over 20 years (18, 24–27). The array also revealed new candidates such as glucagon-like peptide (GLP) 1 receptor (Glp1r), which was highly expressed in the arcuate nucleus. As proof of principle, the GLP1-selective agonist exendin-4 induced an inward current via activation of a nonselective cationic conductance along with inhibition of an inwardly rectifying potassium conductance in proopiomelanocortin (POMC) neurons. Therefore, this quantitative analysis provides insight into the complexity of GPCR signaling in the hypothalamic arcuate nucleus and is a valuable resource for directing future experiments.
Materials and Methods
Animals and treatments
All animal treatments were in accordance with institutional guidelines based on National Institutes of Health standards and were performed with institutional animal care and use committee approval at the Oregon Health and Science University. Adult female and male wild type (C57BL/6) and POMC-enhanced green fluorescent protein (EGFP) mice (C57BL/6; originally obtained from Dr Malcolm Low) (12) were used for a GPCR low-density array and/or verification experiments. The mice were selectively bred in-house and were maintained under controlled temperature (25°C) and photoperiod (12:12 hours light/dark cycle) conditions with food and water ad libitum. To limit the influence of reproductive hormones on gene expression, adult females were ovariectomized (OVX) under ketamine-xylazine (1 and 0.1 mg/10 g, respectively) anesthesia, treated once with carprofen (4 mg/kg) for analgesia, and allowed to recover for 7 to 13 days. Gonadally intact male and female POMC-EGFP mice were used for whole-cell recording and for POMC cell harvesting.
Experimental design
RNA preparation and TaqMan GPCR low-density array analysis
OVX female POMC-EGFP mice (3–6 months old) were used for arcuate nucleus microdissection and RNA extraction. To obtain sufficient RNA for the GPCR array determinations, RNA from 6 to 7 mice was used to form 1 pool, and 4 different pools of RNA (Arc1, Arc2, Arc3, and Arc4) were prepared for the experiments. Each pool of RNA was DNase treated and reverse transcribed as described below. Initially, each pool of cDNA was subjected to quality control, which included performing real-time quantitative PCR (qPCR) analysis for the μ-opioid GPCR and the orphan GPCR12 using SYBR Green real-time PCR assays (see below). The housekeeping gene, β-actin mRNA, was also measured as an overall control gene. GPCR12, in addition to the μ-opioid receptor (28), was used for test purposes because we had found previously that this orphan receptor was expressed in the arcuate nucleus.
Samples from each pool were run as a separate GPCR array, ie, 4 different cDNA pools for 4 different arrays as outlined below. The TaqMan-type quantitative GPCR low-density array (384-well; Applied Biosystems [ABI]) was preformatted, and targets included retinal receptors, small molecule receptors, chemokine receptors, and others including classic endogenous control targets from already known and orphan GPCRs for a total of 353 GPCRs.
Tissue dissection, RNA extraction, and cDNA synthesis
The arcuate nucleus was microdissected from OVX POMC-EGFP mice, and RNA was extracted as described previously (29). In brief, a brain slicer (EM Corporation) was used to produce in each animal a 1-mm coronal block, which extended from the rostral to the caudal borders of the arcuate nucleus. This tissue block was placed in RNAlater (Ambion; Thermo Fisher Scientific Inc) for tissue RNA preservation. After 2 to 3 hours in the RNAlater solution, the arcuate nucleus was dissected with the aid of a dissecting microscope. Total RNA was extracted under RNase-free conditions using an Ambion RNAqueous Micro kit and quantified with a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies). Total RNA was DNAse-1 treated (DNA-free kit; Ambion) at 37°C for 30 minutes to eliminate genomic DNA contamination. All RNA samples produced purity values with an OD260/280 of 1.9 to 2.1. We have established the method of RNA extraction using RNAlater to protect the RNA before extraction, followed by use of an RNAqueous Micro kit for extraction to produce RNA for both the microarray and qPCR. The RNA integrity was analyzed by running 250 ng of RNA on a 1.2% denaturing agarose gel in 1× 3-(N-morpholino)propanesulfonic acid (MOPS) buffer and ribosomal RNAs 28S and 18S were visualized with ethidium bromide (Figure 1A). Reverse transcription was performed for first-strand cDNA synthesis with 400 ng of total RNA, random primers (100 ng/reaction; Promega), and SuperScript III RT (200 U/reaction; Invitrogen) in a total volume of 20 μL. Reverse transcription was conducted with the following protocol: 25°C for 5 minutes, 50°C for 60 minutes, and 70°C for 15 minutes, followed by cooling to 12°C. The cDNAs from several reactions were combined to generate each pool, and the pools were stored at −20°C. Aliquots of the respective arcuate tissue RNA were used as negative controls (no RT) and processed simultaneously with the experimental samples.
Figure 1.
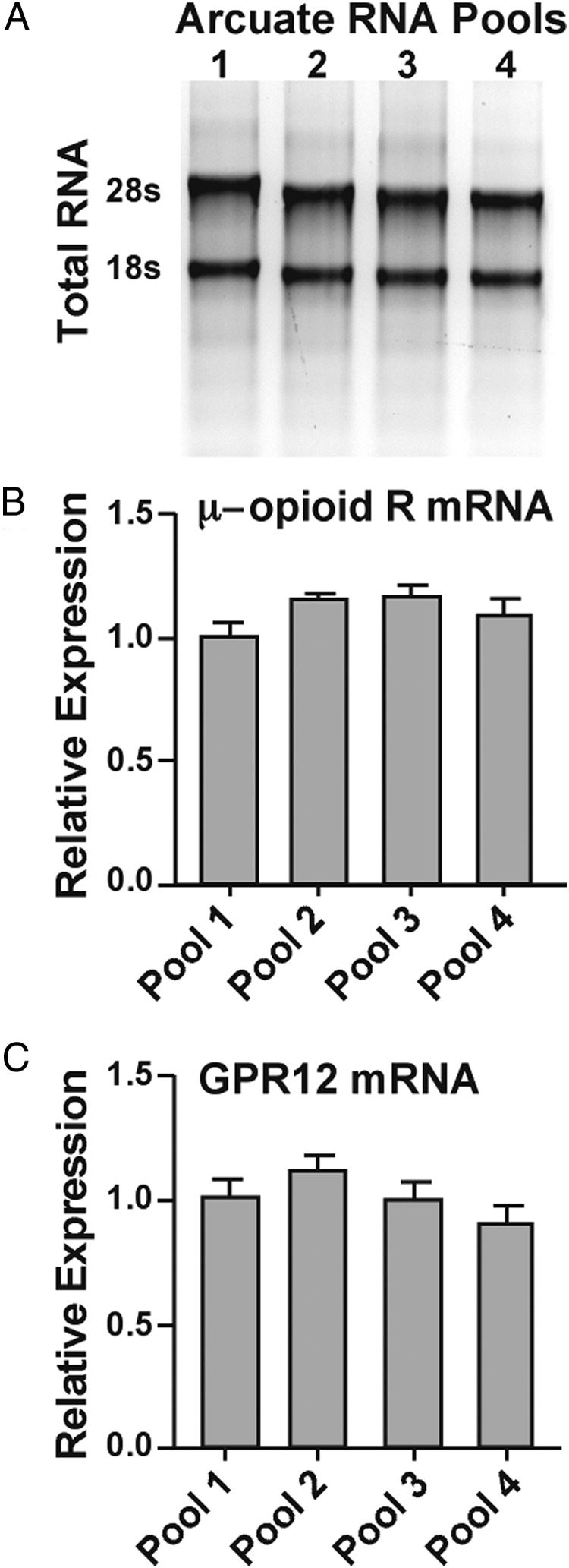
Assessment of RNA quality in the samples used for PCR array analysis. A, Samples containing 250 ng of RNA from arcuate pools 1 to 4 were run on a 1.2% denaturing agarose gel in 1× MOPS buffer and visualized with ethidium bromide. The gel depicts 2 distinct ribosomal RNA bands (28S and 18S) for each sample. B and C, μ-Opioid receptor (μ-opioid R) and GPR12 mRNA levels were measured in the 4 independent RNA samples from arcuate nuclei using reverse transcription and real-time qPCR. This was done to ascertain that there were no significant differences in RNA expression measured in the 4 independent samples to be used for PCR array determinations.
qPCR of arcuate tissue cDNA using mouse GPCR low-density array
The cDNA from each arcuate pool was first evaluated using individual real-time qPCR assays for the μ-opioid receptor and GPR12. Primers were designed for GPR12 using accession number NM_00101094 (127-bp product, forward primer 445–466 nt, and reverse primer 550–571 nt). Primers for the μ-opioid receptor and the control gene (β-actin) were as described previously (30). Next, the respective cDNA pools were analyzed using the mouse TaqMan GPCR low-density array (384-well setting) according to the manufacturer's instruction on an ABI HT 7900 real-time PCR system. In brief, the samples were loaded into 8 reservoirs, each with 100 μl for a total of 800 μl for each array. A single array was run per animal pool. Four arrays were run for 4 distinct animal pools with the following reaction conditions: 50°C for 2 minutes and 94.5°C for 10 minutes, followed by 40 cycles of 97°C for 30 seconds and 59.7°C for 1 minute. In addition to the 353 GPCR genes, the GPCR array had 25 non-GPCR genes and 3 control genes: 18S, β-actin (Actb), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Verification of GPCR low-density array results using individual GPCR TaqMan assays
The results of the low-density arrays were verified using individual TaqMan qPCR assays with the same primers and probes as those used in the array and purchased individually from ABI: GABAB receptor 2 (Gabbr2), μ-opioid receptor (Oprm1), 5-hydroxytryptamine (serotonin) (5HT) receptor 2C (Htr2c), CRH receptor 2 (Crhr2), and the control housekeeping gene β-actin (Actb). For these test experiments, we used the same arcuate tissue samples as used in the low-density arrays. The individual real-time qPCR assays were run under the following conditions: 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute using TaqMan Gene Expression Master Mix (ABI) in 96-well plates in a 7500 fast real-time PCR system (ABI). The results were compared with the low-density array results for the same genes. In addition, the expression of selected GPCRs (GLP1R and GLP2R, μ-opioid receptor, and κ-opioid receptor) was confirmed using Power SYBR Green Master Mix (ABI) on an ABI fast real-time PCR instrument as described previously (29, 31). Primers were designed to span introns using Clone Manager software (Professional Suite version 8; Scientific and Educational Software) and synthesized by Invitrogen (Thermo Fisher Scientific Inc). Real-time qPCR primers for μ-opioid receptor and κ-opioid receptors were as described previously (30). qPCR primers for GLP1R and GLP2R, respectively, were as follows: accession number NM_021332 (109-bp product, forward primer 379–400 nt, and reverse primer 468–487 nt) and accession number NM_175681 (94-bp product, forward primer 1379–1399 nt, and reverse primer 1452–1472 nt.
POMC-EGFP cell harvest, cDNA synthesis, and single-cell PCR
These procedures were conducted as described previously (32–34). In brief, four 250-μm basal hypothalamic slices were cut on a vibratome and placed in an auxiliary chamber containing oxygenated artificial cerebrospinal fluid (aCSF). The slices were allowed to recover for 1 to 2 hours in the auxiliary chamber. Thereafter, the arcuate nucleus was microdissected and digested with protease (1 mg/ml aCSF) for approximately 15 minutes at 37°C. The cells were dispersed onto a 60-mm glass-bottom Petri dish and were perfused continually with aCSF at a rate of 2.0 ml/min. The dispersed fluorescent cells were visualized using a Leica inverted fluorescent microscope, patched, and then harvested with gentle suction to the pipette and expelled into siliconized microcentrifuge tubes containing 5 μl of RNasin solution. Individual POMC-EGFP cells were harvested from 6 animals (3 males and 3 females). Perfused aCSF was regularly collected along with the harvested cells and processed together with the cells and other control samples.
Single POMC cells were reverse transcribed for the first-strand cDNA synthesis using both random primers (100 ng/reaction) and anchored oligo(dT) primers (400 ng/reaction; Invitrogen) and SuperScript III RT (100 U/reaction) in a total volume of 20 μl. One cell from each animal was processed without the RT enzyme for a negative control. Basal medial hypothalamic tissue RNA used as positive and negative controls was processed similarly with and without RT, respectively. Initially, all GFP harvested cells were identified as POMC cells using single-cell PCR as described previously (33).
In addition, primers were designed for the GLP1R and GLP2R for single-cell PCR analysis using Clone Manager software and synthesized by Invitrogen. Single-cell GLP1R and GLP2R primers were, respectively, as follows: accession number NM_021332 (149-bp product, forward primer 474–494 nt, and reverse primer 602–622 nt) and accession number NM_175681 (171-bp product, forward primer 665–676 nt, and reverse primer 816–835 nt). POMC primers for single-cell analysis were as described previously (33).
GPCR array data analysis
The array data analysis was conducted using ABI RQ Manager 1.2 software. The cycle threshold (CT) was set at 0.3 ΔRn (according to the manufacturer's specification) for all 4 plates, and the upper limit of the PCR cycles was 35 for a real product. Then the raw data were exported into the Microsoft Excel program for further analysis. Relative quantification analysis was done using the comparative ΔΔCT method (35, 36). The data are expressed as an n-fold change in gene expression normalized to a reference gene, β-actin (GAPDH gave similar results, whereas 18S rRNA was expressed in quantities that were too high to provide a useful control) and relative to a calibrator (the mean ΔCT of all GPCR genes with CT ≤35 cycles). Means ± SEM from the 4 pools were used to determine the relative quantitative expression of GPCR mRNAs in the arcuate nucleus.
Whole-cell recording
Based on our findings that Glp1r mRNA was equally expressed in male and female POMC neurons and that the GLP1R agonist exendin-4 (200 nM; Tocris Bioscience) induced similar inward currents in male and female POMC neurons, adult male POMC-EGFP mice were used for the quantitative analysis of the effects of exendin-4 (33). Coronal slices containing the arcuate nucleus were prepared for electrophysiological recording. The electrophysiological instrumentation, recording methods, and solutions were as we have reported previously (33). Exendin-4 (200 nM) was applied to POMC neurons after the establishment of the whole-cell patch-clamp configuration. Depolarization and neuronal firing were measured under current clamp conditions. In addition, exendin-4–induced currents were measured under voltage-clamp conditions. The current-voltage relationship of the exendin-4–activated current was constructed from the currents induced by a family of voltage steps from −120 to −40 mV with a duration of 1 second (Vhold = −60 mV).
Results
Quality control of the experimental procedure
To assess the purity of the RNA samples, we measured the OD260/280 and found this to be 2.07 ± 0.01 (n = 4), which indicates excellent purity of the extracted RNA (37). In addition, we analyzed the 28S/18S rRNA bands and found 2 distinct bands with 28S being more robust than 18S in all 4 of the pooled arcuate samples, an indication that the RNA quality was good (Figure 1A). Further quality control assays for RNA extraction and cDNA synthesis were performed through measurements of the μ-opioid receptor and GPR12 as target genes and β-actin as internal control using RT-PCR in the four different arcuate samples. Each arcuate sample was analyzed at least 6 times. This analysis revealed that the mRNA expression of the μ-opioid receptor and GPR12 were consistent across the four arcuate samples (Figure 1, B and C), suggesting that the RNA and cDNA amount and quality were similar in the 4 different arcuate samples (P > .05; one-way ANOVA).
GPCR array analysis
With use of the GPCR low-density array approach, transcript levels for 353 target GPCRs were profiled in the adult mouse arcuate nucleus on 4 different plates. Based on this analysis, we identified the expression levels of 183 of the already known GPCRs (51.84%) and 109 putative orphan GPCRs (30.88%) (Figure 2 and Tables 1–9). The remaining 61 GPCR genes in the array (17.28%), including 36 known and 25 orphan GPCRs, were undetectable (CT >35 cycles) in the arcuate nucleus. The relative mRNA expression levels of the various GPCRs were quantified and arbitrarily classified as high-expressing (>5-fold), moderate-expressing (1- to 5-fold), or low-expressing (<1-fold) genes. The expression profiles for the 4 independent arcuate tissue pools run on the 4 different plates were consistent. Therefore, most of the 353 GPCR genes were expressed in the hypothalamic arcuate nucleus.
Figure 2.
Pie graph of classified GPCR genes (292) expressed in mouse hypothalamic arcuate nucleus. Thirteen classified groups including 8 known functional groups and 5 orphan groups of GPCR genes are represented by a different color in each group. The known functional GPCR groups contain 183 genes including neurotransmitter receptors (44; 15%), neuropeptide receptors (26; 9%), hormone receptors (31; 11%), receptors having angiogenesis and vasoactivity (10; 3%), receptors with inflammation and immune system regulation (16; 5%), lipid messenger receptors (20; 7%), growth factor receptors (12; 4%), and other known G protein-coupled receptors (24; 8%). The orphan GPCR groups contain 109 genes including class A orphans (66; 23%), class B orphans (23; 8%), class C orphans (4; 1%), nonsignaling 7TM chemokine-binding proteins (2; 1%), and other 7TM protein genes (14; 5%). The number on each slice of pie indicates the number of the genes in each classified group (see the names of the genes in each group in Tables 1–9).
Table 1.
Classification and Relative Quantity of Neurotransmitter Receptors in the Arcuate Nucleus (44)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Acetylcholine receptors (muscarinic) (5) | ||
| Chrm1-Mm00432509_s1 | 10.0196 ± 1.0778 | Cholinergic receptor, muscarinic 1 |
| Chrm2-Mm01701855_s1 | 0.9426 ± 0.0615 | Cholinergic receptor, muscarinic 2, cardiac |
| Chrm3-Mm00446300_s1 | 4.4877 ± 0.3574 | Cholinergic receptor, muscarinic 3, cardiac |
| Chrm4-Mm00432514_s1 | 0.9044 ± 0.0365 | Cholinergic receptor, muscarinic 4 |
| Chrm5-Mm01701883_s1 | 7.4446 ± 0.3187 | Cholinergic receptor, muscarinic 5 |
| Adenosine receptors (4) | ||
| Adora1-Mm01308023_m1 | 14.2690 ± 4.5901 | Adenosine A1 receptor |
| Adora2a-Mm00802075_m1 | 0.2879 ± 0.0255 | Adenosine A2a receptor |
| Adora2b-Mm00839292_m1 | 1.7697 ± 0.0568 | Adenosine A2b receptor |
| Adora3-Mm00802076_m1 | 0.2163 ± 0.0206 | Adenosine A3 receptor |
| Adrenergic receptors (9) | ||
| Adra1a-Mm00442668_m1 | 6.6485 ± 0.3875 | Adrenergic receptor, α1a |
| Adra1b-Mm00431685_m1 | 2.5781 ± 0.3029 | Adrenergic receptor, α1b |
| Adra1d-Mm01328600_m1 | 0.1371 ± 0.0199 | Adrenergic receptor, α1d |
| Adra2a-Mm00845383_s1 | 11.0508 ± 1.3903 | Adrenergic receptor, α2a |
| Adra2b-Mm00477390_s1 | 0.0880 ± 0.0207 | Adrenergic receptor, α2b |
| Adra2c-Mm00431686_s1 | 0.9547 ± 0.0725 | Adrenergic receptor, α2c |
| Adrb1-Mm00431701_s1 | 1.9190 ± 0.0825 | Adrenergic receptor, β1 |
| Adrb2-Mm02524224_s1 | 0.1308 ± 0.0270 | Adrenergic receptor, β2 |
| Adrb3-Mm00442669_m1 | 0.0591 ± 0.0068 | Adrenergic receptor, β3 |
| Cannabinoid receptors (2) | ||
| Cnr1-Mm00432621_s1 | 26.9570 ± 0.4546 | Cannabinoid receptor 1 (brain) |
| Cnr2-Mm02620087_s1 | 0.0306 ± 0.0091 | Cannabinoid receptor 2 (macrophage) |
| Dopamine receptors (4) | ||
| Drd1a-Mm02620146_s1 | 0.6679 ± 0.0648 | Dopamine receptor D1A |
| Drd2-Mm00438541_m1 | 3.1332 ± 0.1559 | Dopamine receptor D2 |
| Drd3-Mm00432887_m1 | 0.5728 ± 0.0507 | Dopamine receptor D3 |
| Drd5-Mm00658653_s1 | 0.1003 ± 0.0160 | Dopamine receptor D5 |
| GABAB receptors (2) | ||
| Gabbr1-Mm00444578_m1 | 171.931 ± 21.1734 | γ-Aminobutyric acid (GABAB) receptor 1 |
| Gabbr2-Mm01352561_m1 | 72.9596 ± 13.7708 | γ-Aminobutyric acid (GABAB) receptor 2 |
| Metabotropic glutamate receptors (6) | ||
| Grm1-Mm00810219_m1 | 3.7511 ± 1.0295 | Glutamate receptor, metabotropic 1 |
| Grm3-Mm00725298_m1 | 9.9115 ± 1.5151 | Glutamate receptor, metabotropic 3 |
| Grm4-Mm01306128_m1 | 1.8876 ± 0.1222 | Glutamate receptor, metabotropic 4 |
| Grm5-Mm00690332_m1 | 27.2579 ± 0.8546 | Glutamate receptor, metabotropic 5 |
| Grm7-Mm01189424_m1 | 7.0332 ± 2.2733 | Glutamate receptor, metabotropic 7 |
| Grm8-Mm00433840_m1 | 2.4910 ± 0.3229 | Glutamate receptor, metabotropic 8 |
| Serotonin (5-hydroxytryptamine) receptors (12) | ||
| Htr1a-Mm00434106_s1 | 2.1189 ± 0.1757 | 5-Hydroxytryptamine (serotonin) receptor 1A |
| Htr1b-Mm00439377_s1 | 14.4398 ± 0.3809 | 5-Hydroxytryptamine (serotonin) receptor 1B |
| Htr1d-Mm00434115_s1 | 1.1242 ± 0.2572 | 5-Hydroxytryptamine (serotonin) receptor 1D |
| Htr1f-Mm02619863_s1 | 0.0847 ± 0.0126 | 5-Hydroxytryptamine (serotonin) receptor 1F |
| Htr2a-Mm00555764_m1 | 7.7976 ± 0.3493 | 5-Hydroxytryptamine (serotonin) receptor 2A |
| Htr2b-Mm00434123_m1 | 0.0208 ± 0.0071 | 5-Hydroxytryptamine (serotonin) receptor 2B |
| Htr2c-Mm00434127_m1 | 2.0104 ± 0.2314 | 5-Hydroxytryptamine (serotonin) receptor 2C |
| Htr4-Mm00434129_m1 | 2.7883 ± 0.4173 | 5 Hydroxytryptamine (serotonin) receptor 4 |
| Htr5a-Mm00434132_m1 | 4.1376 ± 0.7950 | 5-Hydroxytryptamine (serotonin) receptor 5A |
| Htr5b-Mm00439389_m1 | 0.7102 ± 0.1409 | 5-Hydroxytryptamine (serotonin) receptor 5B |
| Htr6-Mm00445320_m1 | 1.0866 ± 0.0790 | 5-Hydroxytryptamine (serotonin) receptor 6 |
| Htr7-Mm00434133_m1 | 1.8060 ± 0.1417 | 5-Hydroxytryptamine (serotonin) receptor 7 |
Table 2.
Classification and Relative Quantity of Neuropeptide Receptors in the Arcuate Nucleus (26)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Galanin receptors (3) | ||
| Galr1-Mm00433515_m1 | 1.3932 ± 0.1538 | Galanin receptor 1 |
| Galr2-Mm00726392_s1 | 0.4943 ± 0.0593 | Galanin receptor 2 |
| Galr3-Mm00443617_m1 | 0.4117 ± 0.0274 | Galanin receptor 3 |
| Bradykinin receptors (2) | ||
| Bdkrb1-Mm00432059_s1 | 0.2190 ± 0.0112 | Bradykinin receptor B1 |
| Bdkrb2-Mm00437788_s1 | 0.2436 ± 0.0214 | Bradykinin receptor, β2 |
| Kisspeptin receptor (1) | ||
| Kiss1r-Mm00475046_m1 | 1.6223 ± 0.1173 | Kisspeptin receptor, KISS1 receptor |
| Neuromedin B receptor (1) | ||
| Nmbr-Mm00435147_m1 | 1.0793 ± 0.0780 | Neuromedin B receptor |
| Neuromedin U receptors (2) | ||
| Nmur1-Mm00515885_m1 | 0.0857 ± 0.0158 | Neuromedin U receptor 1 |
| Nmur2-Mm00600704_m1 | 2.2792 ± 0.1854 | Neuromedin U receptor 2 |
| Neuropeptide FF/neuropeptide AF receptors (2) | ||
| Npffr1-Mm01176033_m1 | 2.6919 ± 0.3299 | Neuropeptide FF receptor 1 |
| Npffr2-Mm00500040_m1 | 0.8695 ± 0.0396 | Neuropeptide FF receptor 2 |
| Neuropeptide W/neuropeptide B receptors (1) | ||
| Npbwr1-Mm02621088_s1 | 0.1830 ± 0.0137 | Neuropeptide B/W receptor 1 |
| Neuropeptide Y receptors (3) | ||
| Npy1r-Mm00650798_g1 | 4.5690 ± 0.7657 | Neuropeptide Y receptor Y1 |
| Npy2r-Mm01956783_s1 | 1.8491 ± 0.0785 | Neuropeptide Y receptor Y2 |
| Npy5r-Mm02620267_s1 | 0.1625 ± 0.0201 | Neuropeptide Y receptor Y5 |
| Neurotensin receptors (2) | ||
| Ntsr1-Mm00444459_m1 | 4.4989 ± 0.3538 | Neurotensin receptor 1 |
| Ntsr2-Mm00435426_m1 | 39.7503 ± 4.6948 | Neurotensin receptor 2 |
| Opioid receptors (4) | ||
| Oprm1-Mm01188089_m1 | 7.0238 ± 0.0742 | Opioid receptor, μ1 |
| Oprd1-Mm00443063_m1 | 1.5361 ± 0.2264 | Opioid receptor, δ1 |
| Oprk1-Mm01230885_m1 | 2.1007 ± 0.4578 | Opioid receptor, κ1 |
| Oprl1-Mm00440563_m1 | 17.7796 ± 1.4257 | Opioid receptor-like 1 |
| Orexin receptors (2) | ||
| Hcrtr1-Mm01185778_m1 | 1.1484 ± 0.1203 | Hypocretin (orexin) receptor 1 |
| Hcrtr2-Mm01179312_m1 | 1.9816 ± 0.0953 | Hypocretin (orexin) receptor 2 |
| Tachykinin receptors (3) | ||
| Tacr1-Mm00436892_m1 | 2.6346 ± 0.3096 | Tachykinin receptor 1 |
| Tacr2-Mm00436898_m1 | 0.0667 ± 0.0139 | Tachykinin receptor 2 |
| Tacr3-Mm00445346_m1 | 5.1109 ± 0.5237 | Tachykinin receptor 3 |
Table 3.
Classification and Relative Quantity of Hormone Receptors in the Arcuate Nucleus (31)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Calcitonin receptors (2) | ||
| Calcr-Mm00432271_m1 | 19.5005 ± 3.0788 | Calcitonin receptor |
| Calcr1-Mm00516986_m1 | 1.3965 ± 0.1857 | Calcitonin receptor-like |
| Cholecystokinin receptors (2) | ||
| Cckar-Mm00438060_m1 | 2.0683 ± 0.1362 | Cholecystokinin A receptor |
| Cckbr-Mm00432329_m1 | 6.0393 ± 0.7506 | Cholecystokinin B receptor |
| Corticotropin-releasing factor receptors (2) | ||
| Crhr1-Mm00432670_m1 | 0.8884 ± 0.1048 | Corticotropin-releasing factor receptor 1 |
| Crhr2-Mm00438303_m1 | 0.0211 ± 0.0074 | Corticotropin-releasing factor receptor 2 |
| Estrogen (G protein–coupled) receptor (1) | ||
| Gpr30-Mm02620446_s1 | 0.2393 ± 0.0171 | GPCR 30 |
| Ghrelin receptor (1) | ||
| Ghsr-Mm00616415_m1 | 2.8523 ± 0.2406 | Ghrelin receptor (growth hormone secretagogue receptor) |
| Glucagon receptor family (4) | ||
| Ghrhr-Mm01326479_m1 | 0.0087 ± 0.0019 | Growth hormone–releasing hormone receptor |
| Gipr-Mm01316351_g1 | 0.0991 ± 0.0039 | Gastric inhibitory polypeptide receptor |
| Glp1r-Mm00445292_m1 | 7.0520 ± 0.3008 | Glucagon-like peptide 1 receptor |
| Glp2r-Mm00558835_m1 | 0.0328 ± 0.0063 | Glucagon-like peptide 2 receptor |
| Gonadotrophin-releasing hormone receptors (1) | ||
| Gnrhr-Mm00439143_m1 | 0.0169 ± 0.0040 | Gonadotrophin-releasing hormone receptor |
| Luteinizing hormone/choriogonadotropin receptor (1) | ||
| Lhcgr-Mm00442931_m1 | 0.2576 ± 0.0518 | Luteinizing hormone/choriogonadotropin receptor |
| Melanin-concentrating hormone receptor (1) | ||
| Mchr1-Mm00653044_m1 | 0.7887 ± 0.0747 | Melanin-concentrating hormone receptor 1 |
| Melanocortin receptors (4) | ||
| Mc1r-Mm00434851_s1 | 0.0397 ± 0.0092 | Melanocortin 1 receptor |
| Mc3r-Mm00434876_s1 | 7.7624 ± 0.2813 | Melanocortin 3 receptor |
| Mc4r-Mm00457483_s1 | 0.9801 ± 0.0467 | Melanocortin 4 receptor |
| Mc5r-Mm00442970_m1 | 0.1284 ± 0.0118 | Melanocortin 5 receptor |
| Parathyroid hormone receptor (2) | ||
| Pthr1-Mm00441046_m1 | 0.6688 ± 0.1087 | Parathyroid hormone receptor 1 |
| Pthr2-Mm00653029_m1 | 0.7876 ± 0.1297 | Parathyroid hormone receptor 2 |
| Prokineticin receptors (2) | ||
| Prokr1-Mm00517546_m1 | 0.6927 ± 0.0960 | Prokineticin receptor 1 |
| Prokr2-Mm00769571_m1 | 1.4569 ± 0.2388 | Prokineticin receptor 2 |
| Prolactin-releasing peptide receptor (1) | ||
| Prlhr-Mm01266991_s1 | 0.6843 ± 0.0669 | Prolactin-releasing peptide receptor |
| Somatostatin receptors (5) | ||
| Sstr1-Mm00436679_s1 | 11.1765 ± 1.4531 | Somatostatin receptor 1 |
| Sstr2-Mm00436685_g1 | 0.1265 ± 0.0432 | Somatostatin receptor 2 |
| Sstr3-Mm00436695_s1 | 4.4996 ± 0.3993 | Somatostatin receptor 3 |
| Sstr4-Mm00436710_s1 | 4.2868 ± 0.3161 | Somatostatin receptor 4 |
| Sstr5-Mm01307775_s1 | 0.9652 ± 0.0484 | Somatostatin receptor 5 |
| TSH receptor (1) | ||
| Tshr-Mm00442027_m1 | 3.0282 ± 0.4828 | TSH receptor |
| TRH receptor (1) | ||
| Trhr-Mm00443262_m1 | 2.2997 ± 0.1972 | TRH receptor |
Table 4.
Classification and Relative Quantity of Angiogenesis and Vasoactive Receptors in the Arcuate Nucleus (10)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Angiotensin receptors (3) | ||
| Agtr1a-Mm01957722_s1 | 0.1780 ± 0.0168 | Angiotensin II receptor, type 1a |
| Agtr2-Mm01341373_m1 | 0.2825 ± 0.0271 | Angiotensin II receptor, type 2 |
| Agtrl1-Mm00442191_s1 | 0.2595 ± 0.0522 | Angiotensin receptor-like 1 |
| Endothelin receptors (2) | ||
| Ednra-Mm01243722_m1 | 1.4502 ± 0.1071 | Endothelin receptor type A |
| Ednrb-Mm00432989_m1 | 21.9352 ± 2.9980 | Endothelin receptor type B |
| Vasoactive intestinal peptide– and pituitary adenylate cyclase–activating polypeptide receptors (3) | ||
| Adcyap1r1-Mm00431683_m1 | 131.2449 ± 8.9925 | Adenylate cyclase activating polypeptide 1 receptor 1 |
| Vipr1-Mm00449214_m1 | 0.0971 ± 0.0154 | Vasoactive intestinal peptide receptor 1 |
| Vipr2-Mm00437316_m1 | 0.6042 ± 0.0708 | Vasoactive intestinal peptide receptor 2 |
| Vasopressin and oxytocin receptors (2) | ||
| Avpr1a-Mm00444092_m1 | 1.6037 ± 0.0892 | Arginine vasopressin receptor 1A |
| Oxtr-Mm01182684_m1 | 1.4708 ± 0.1439 | Oxytocin receptor |
Table 5.
Classification and Relative Quantity of Inflammatory and Immune System Receptors in the Arcuate Nucleus (16)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Anaphylatoxin receptors (2) | ||
| C3ar1-Mm02620006_s1 | 0.2775 ± 0.0535 | Complement component 3a receptor 1 |
| C5ar1-Mm00500292_s1 | 0.3223 ± 0.0492 | Complement component 5a receptor 1 |
| Chemokine receptors (11) | ||
| Ccr1-Mm00438260_s1 | 0.0413 ± 0.0098 | Chemokine (C-C motif) receptor 1 |
| Ccr2-Mm99999051_gH | 0.5748 ± 0.0536 | Chemokine (C-C motif) receptor 2 |
| Ccr3-Mm00515543_s1 | 0.0130 ± 0.0050 | Chemokine (C-C motif) receptor 3 |
| Ccr5-Mm01963251_s1 | 0.1934 ± 0.0798 | Chemokine (C-C motif) receptor 5 |
| Ccr7-Mm01301785_m1 | 0.0083 ± 0.0013 | Chemokine (C-C motif) receptor 7 |
| Ccr9-Mm02620030_s1 | 0.2877 ± 0.0258 | Chemokine (C-C motif) receptor 9 |
| Cxcr3-Mm99999054_s1 | 0.0795 ± 0.0145 | Chemokine (C-X-C motif) receptor 3 |
| Cxcr4-Mm99999055_m1 | 0.1071 ± 0.1026 | Chemokine (C-X-C motif) receptor 4 |
| Cxcr6-Mm02620517_s1 | 0.2566 ± 0.0138 | Chemokine (C-X-C motif) receptor 6 |
| Cx3cr1-Mm02620111_s1 | 6.9515 ± 0.1138 | Chemokine (C-X3-C) receptor 1 |
| Xcr1-Mm00442206_s1 | 0.0177 ± 0.0026 | Chemokine (C motif) receptor 1 |
| Histamine receptors (3) | ||
| Hrh1-Mm00434002_s1 | 8.4888 ± 0.5633 | Histamine receptor H1 |
| Hrh2-Mm00434009_s1 | 0.0709 ± 0.0132 | Histamine receptor H2 |
| Hrh3-Mm00446706_m1 | 18.1531 ± 1.8979 | Histamine receptor H3 |
Table 6.
Classification and Relative Quantity of Lipid Messenger Receptors in the Arcuate Nucleus (20)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Leukotriene receptors (4) | ||
| Ltb4r1-Mm02619879_s1 | 0.0321 ± 0.0091 | Leukotriene B4 receptor 1 |
| Ltb4r2-Mm00498491_s1 | 0.1165 ± 0.0073 | Leukotriene B4 receptor 2 |
| Cysltr1-Mm02620326_s1 | 0.0753 ± 0.0124 | Cysteinyl leukotriene receptor 1 |
| Cysltr2-Mm02620584_s1 | 0.0866 ± 0.0142 | Cysteinyl leukotriene receptor 2 |
| Lysophospholipid (LPA) receptors (4) | ||
| Edg2-Mm00439145_m1 | 7.1370 ± 1.2251 | LPA1, endothelial differentiation, lysophosphatidic acid GPRC 2 |
| Edg4-Mm00469562_m1 | 0.9551 ± 0.0425 | LPA2, endothelial differentiation, lysophosphatidic acid GPRC, 4 |
| Edg7-Mm00469694_m1 | 0.1421 ± 0.0261 | LPA3, endothelial differentiation, lysophosphatidic acid GPRC, 7 |
| P2ry5-Mm00613058_s1 | 2.6972 ± 0.3282 | LPA6, purinergic receptor P2Y, GPRC, 5 |
| Lysophospholipid (S1P) receptors (4) | ||
| Edg1-Mm02619656_s1 | 2.8238 ± 1.1109 | S1P1, endothelial differentiation sphingolipid GPRC, 1 |
| Edg3-Mm02620181_s1 | 0.1047 ± 0.0077 | S1P3, endothelial differentiation, sphingolipid GPRC, 3 |
| Edg6-Mm00468695_s1 | 0.4419 ± 0.0976 | S1P4, endothelial differentiation, GPRC, 6 |
| Edg8-Mm02620565_s1 | 0.4196 ± 0.0341 | S1P5, endothelial differentiation, sphingolipid GPRC, 8 |
| Prostanoid receptors (8) | ||
| Ptgdr-Mm00436050_m1 | 0.0608 ± 0.0176 | Prostaglandin D receptor |
| Gpr44-Mm00438315_s1 | 0.0730 ± 0.0108 | DP2, GPCR 44 |
| Ptger1-Mm00443098_g1 | 0.2651 ± 0.0285 | Prostaglandin E receptor1 (subtype EP1) |
| Ptger2-Mm00436051_m1 | 0.3236 ± 0.0433 | Prostaglandin E receptor2 (subtype EP2) |
| Ptger3-Mm01316856_m1 | 0.8174 ± 0.0324 | Prostaglandin E receptor3 (subtype EP3) |
| Ptger4-Mm00436053_m1 | 0.7047 ± 0.0704 | Prostaglandin E receptor4 (subtype EP4) |
| Ptgfr-Mm00436055_m1 | 0.1216 ± 0.0274 | Prostaglandin F receptor |
| Tbxa2r-Mm00436917_m1 | 0.0674 ± 0.0068 | Thromboxane A2 receptor |
Table 7.
Classification and Relative Quantity of Growth Factor Receptors in the Arcuate Nucleus (12)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Frizzled receptors (12) | ||
| Fzd1-Mm00445405_s1 | 2.2868 ± 0.1984 | Frizzled homolog 1 (Drosophila) |
| Fzd2-Mm02524776_s1 | 0.4988 ± 0.0380 | Frizzled homolog 2 (Drosophila) |
| Fzd3-Mm00445423_m1 | 19.0983 ± 1.9531 | Frizzled homolog 3 (Drosophila) |
| Fzd4-Mm00433382_m1 | 3.0181 ± 0.4399 | Frizzled homolog 4 (Drosophila) |
| Fzd5-Mm00445623_s1 | 13.4249 ± 0.5211 | Frizzled homolog 5 (Drosophila) |
| Fzd6-Mm00433387_m1 | 3.2283 ± 0.2427 | Frizzled homolog 6 (Drosophila) |
| Fzd7-Mm00433409_s1 | 2.0544 ± 0.1326 | Frizzled homolog 7 (Drosophila) |
| Fzd8-Mm00433419_s1 | 1.2603 ± 0.1365 | Frizzled homolog 8 (Drosophila) |
| Fzd9-Mm02621136_s1 | 0.6752 ± 0.0861 | Frizzled homolog 9 (Drosophila) |
| Fzd10-Mm00558396_s1 | 2.0051 ± 0.1161 | Frizzled homolog 10 (Drosophila) |
| Smo-Mm01162710_m1 | 6.7230 ± 0.3111 | Smoothened homolog (Drosophila) |
| Frzb-Mm00441378_m1 | 6.4396 ± 1.0771 | Frizzled-related protein |
Table 8.
Classification and Relative Quantity of Other G Protein-Coupled Receptors in the Arcuate Nucleus (24)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Bile acid receptor (1) | ||
| Gpbar1-Mm00558112_s1 | 0.2721 ± 0.0230 | GPCR bile acid receptor 1 |
| Calcium-sensing receptors (1) | ||
| Casr-Mm00443375_m1 | 0.7649 ± 0.1186 | Calcium-sensing receptor |
| Formylpeptide receptors (2) | ||
| Fpr1-Mm00442803_s1 | 0.2505 ± 0.0353 | Formyl peptide receptor 1 |
| Fpr-rs2-Mm00484464_s1 | 0.0297 ± 0.0034 | Formyl peptide receptor, related sequence 2 |
| Hydroxycarboxylic acid (HCA) receptors (1) | ||
| Gpr81-Mm00558586_s1 | 0.0663 ± 0.0140 | HCA1, GPCR 81 |
| Melatonin receptors (1) | ||
| Mtnr1a-Mm00434999_m1 | 0.1422 ± 0.0330 | Melatonin receptor 1A |
| Opsins and rhodopsin (1) | ||
| Rho-Mm00520345_m1 | 0.1942 ± 0.0267 | Rhodopsin, a GPCR, extremely sensitive to light |
| Others (1) | ||
| Crcp-Mm01197736_m | 16.5122 ± 0.1912 | Calcitonin gene-related peptide-receptor component protein |
| Pancreatic polypeptide receptor (1) | ||
| Ppyr1-Mm00435894_s1 | 0.3744 ± 0.0595 | Pancreatic polypeptide receptor 1 |
| Platelet-activating factor receptor (1) | ||
| Ptafr-Mm02621061_m1 | 0.5617 ± 0.0502 | Platelet-activating factor receptor |
| Protease-activated receptors (3) | ||
| F2r-Mm00438851_m1 | 4.1429 ± 0.2484 | Protease-activated receptor 1, coagulation factor II (thrombin) receptor |
| F2rl1-Mm00433160_m1 | 0.0650 ± 0.0139 | Protease-activated receptor 2, coagulation factor II (thrombin) receptor-like 1 |
| F2rl2-Mm00438852_m1 | 1.5260 ± 0.2496 | Protease-activated receptor 3, coagulation factor II (thrombin) receptor-like 2 |
| Purinergic (P2Y) receptors (7) | ||
| P2ry1-Mm00435471_m1 | 0.2795 ± 0.0263 | Purinergic receptor P2Y, G protein coupled 1 |
| P2ry2-Mm02619978_s1 | 0.0587 ± 0.0182 | Purinergic receptor P2Y, G protein coupled 2 |
| P2ry4-Mm00445136_s1 | 0.0151 ± 0.0040 | Pyrimidinergic receptor P2Y, G protein coupled, 4 |
| P2ry6-Mm02620937_s1 | 0.1369 ± 0.0147 | Pyrimidinergic receptor P2Y, G protein coupled, 6 |
| P2ry12-Mm01950543_s1 | 0.0479 ± 0.0046 | Purinergic receptor P2Y, G protein coupled 12 |
| P2ry13-Mm01951265_s1 | 0.2298 ± 0.0156 | Purinergic receptor P2Y, G protein coupled 13 |
| P2ry14-Mm02035793_s1 | 0.4214 ± 0.0080 | Purinergic receptor P2Y, G protein coupled, 14 |
| Relaxin family peptide receptors (2) | ||
| Rxfp3-Mm00618735_s1 | 2.9747 ± 0.4442 | Relaxin family peptide receptor 3 |
| Rxfp4-Mm00731536_s1 | 0.0427 ± 0.0076 | Relaxin family peptide receptor 4 |
| Secretin receptor (1) | ||
| Sctr-Mm01290790_m1 | 0.5935 ± 0.0945 | Secretin receptor |
| Transferrin receptors (1) | ||
| Tfrc-Mm00441941_m1 | 10.0531 ± 1.1201 | Transferrin receptor |
Table 9.
Classification and Relative Quantity of Orphan GPCR Genes in the Arcuate Nucleus (109)
| Classification Detector | Mean ± SEM | Annotation |
|---|---|---|
| Class A orphans (66) | ||
| Ccrl2-Mm00516914_g1 | 0.4606 ± 0.0427 | Chemokine (C-C motif) receptor-like 2 |
| Cmklr1-Mm02619757_s1 | 0.2342 ± 0.0666 | Chemokine-like receptor 1 |
| Gpr1-Mm02620665_s1 | 0.1688 ± 0.0195 | GPCR 1 |
| Gpr3-Mm00433719_s1 | 0.4976 ± 0.0296 | GPCR 3 |
| Gpr4-Mm00558777_s1 | 1.3546 ± 0.2395 | GPCR 4 |
| Gpr6-Mm01701705_s1 | 2.5490 ± 0.3355 | GPCR 6 |
| Gpr12-Mm02343661_s1 | 0.8334 ± 0.0373 | GPCR 12 |
| AI853548, Gpr17-Mm02619401_s1 | 2.4195 ± 0.2651 | GPCR 17, expressed sequence AI853548 |
| Gpr18-Mm02620895_s1 | 0.4013 ± 0.0209 | GPCR 18 |
| Gpr19-Mm02619790_s1 | 0.4952 ± 0.0380 | GPCR 19 |
| Gpr20-Mm02620726_s1 | 0.0260 ± 0.0018 | GPCR 20 |
| Gpr21-Mm02620869_s1 | 0.5886 ± 0.0561 | GPCR 21 |
| Gpr22-Mm01291487_m1 | 2.2926 ± 0.3833 | GPCR 22 |
| Gpr23-Mm02620784_s1 | 1.1411 ± 0.1069 | GPCR 23 |
| N/A-Mm02621666_s1 | 0.7404 ± 0.0617 | GPR25, GPCR 25 |
| Gpr26-Mm01165717_m1 | 6.3768 ± 0.3539 | GPCR 26 |
| Gpr27-Mm01962535_s1 | 0.0326 ± 0.0118 | GPCR 27 |
| Gpr34-Mm02620221_s1 | 0.3407 ± 0.0442 | GPCR 34 |
| Gpr35-Mm01973686_s1 | 0.1457 ± 0.0256 | GPCR 35 |
| Gpr45-Mm00446398_s1 | 5.5220 ± 0.4795 | GPCR 45 |
| Gpr50-Mm00439147_m1 | 19.5940 ± 2.0275 | GPCR 50 |
| Gpr55-Mm02621622_s1 | 0.8677 ± 0.0713 | GPCR 55 |
| Gpr61-Mm00558495_s1 | 6.7480 ± 1.1120 | GPCR 61 |
| Gpr62-Mm02621101_s1 | 2.0888 ± 0.1987 | GPCR 62 |
| Gpr63-Mm00446159_s1 | 1.0420 ± 0.0699 | GPCR 63 |
| Gpr65-Mm02619732_s1 | 0.0749 ± 0.0168 | GPCR 65 |
| Gpr68-Mm00558545_s1 | 0.2831 ± 0.0234 | GPCR 68 |
| Gpr75-Mm00558537_s1 | 11.2089 ± 1.5338 | GPCR 75 |
| Gpr82-Mm00558788_s1 | 0.5621 ± 0.0572 | GPCR 82 |
| Gpr83-Mm00439103_m1 | 8.5449 ± 0.5664 | GPCR 83 |
| Gpr84-Mm02620530_s1 | 0.2354 ± 0.0216 | GPCR 84 |
| Gpr85-Mm00460767_s1 | 10.7271 ± 1.2817 | GPCR 85 |
| Gpr87-Mm00519013_s1 | 0.3322 ± 0.0396 | GPCR 87 |
| Gpr88-Mm02620353_s1 | 0.0466 ± 0.0060 | GPCR 88 |
| Gpr101-Mm01296083_m1 | 33.7668 ± 2.1204 | GPCR 101 |
| Gpr119-Mm00731497_s1 | 0.0742 ± 0.0166 | GPCR 119 |
| Gpr120-Mm00725193_m1 | 0.0205 ± 0.0037 | GPCR 120 |
| Gpr132-Mm02620285_s1 | 0.0178 ± 0.0017 | GPCR 132 |
| Gpr135-Mm00731507_s1 | 5.6510 ± 0.7205 | GPCR 135 |
| Gpr139-Mm01280440_m1 | 1.2749 ± 0.1576 | GPCR 139 |
| Gpr141-Mm00731518_s1 | 0.0074 ± 0.0020 | GPCR 141 |
| Gpr146-Mm01951835_s1 | 2.8621 ± 0.2153 | GPCR 146 |
| Gpr149-Mm00805216_m1 | 3.1200 ± 0.3126 | GPCR 149 |
| Gpr150-Mm02344527_s1 | 0.1833 ± 0.0152 | GPCR 150 |
| Gpr151-Mm00808987_s1 | 0.2428 ± 0.0177 | GPCR 151 |
| Gpr152-Mm02620975_s1 | 0.0469 ± 0.0039 | GPCR 152 |
| Gpr153-Mm00805218_m1 | 16.4656 ± 0.9116 | GPCR 153 |
| Gpr160-Mm02621016_s1 | 0.1267 ± 0.0347 | GPCR 160 |
| Gpr161-Mm01291057_m1 | 2.5561 ± 0.3186 | GPCR 161 |
| Gpr162-Mm00496577_m1 | 13.3626 ± 0.2260 | GPCR 162 |
| Gpr171-Mm02620738_s1 | 0.4417 ± 0.0133 | GPCR 171 |
| Gpr173-Mm02620389_s1 | 1.4268 ± 0.1131 | GPCR 173 |
| Gpr174-Mm01238430_m1 | 0.0084 ± 0.0018 | GPCR 174 |
| Gpr176-Mm01277657_m1 | 3.6780 ± 0.0754 | GPCR 176 |
| Admr-Mm01946034_s1 | 0.1446 ± 0.0251 | GPR182, adrenomedullin receptor |
| Ebi2-Mm02620906_s1 | 0.7011 ± 0.0617 | GPR183, Epstein-Barr virus–induced gene 2 |
| Lgr4-Mm00554385_m1 | 6.6814 ± 0.1825 | Leucine-rich repeat–containing GPCR 4 |
| Lgr5-Mm00438890_m1 | 1.3949 ± 0.2244 | Leucine-rich repeat–containing GPCR 5 |
| Lgr6-Mm01291336_m1 | 3.9080 ± 0.2838 | Leucine-rich repeat–containing GPCR 6 |
| Lgr7-Mm01220214_m1 | 0.3115 ± 0.0512 | Leucine-rich repeat–containing GPCR 7 |
| Lgr8-Mm00446529_m1 | 0.2990 ± 0.0400 | Leucine-rich repeat–containing GPCR 8 |
| Mas1-Mm00434823_s1 | 0.1029 ± 0.0234 | MAS1 oncogene, a GPCR, binds the angiotensin II metabolite angiotensin- (1–7) |
| Mrgpra3-Mm02620679_s1 | 0.0123 ± 0.0028 | MAS-related GPR, member A3 |
| Mrgpre-Mm02620836_s1 | 2.5059 ± 0.3675 | MAS-related GPR, member E |
| Mrgprf-Mm02620618_s1 | 0.0250 ± 0.0040 | MAS-related GPR, member F |
| Oxgr1-Mm01960674_s1 | 0.0936 ± 0.0094 | Oxoglutarate (α-ketoglutarate) receptor 1 |
| Class B orphans (23) | ||
| Bai1-Mm00558144_m1 | 52.0334 ± 2.8904 | Brain-specific angiogenesis inhibitor 1, aphagocytic receptor |
| Bai2-Mm00557365_m1 | 4.9234 ± 0.5216 | Brain-specific angiogenesis inhibitor 2, orphan receptor |
| Bai3-Mm00657451_m1 | 15.5840 ± 0.7051 | Brain-specific angiogenesis inhibitor 3, orphan receptor |
| Cd97-Mm00516248_m1 | 1.8654 ± 0.0688 | CD97 antigen, member of the epidermal growth factor (EGF)-TM7 family, new GPCR |
| Celsr1-Mm00464808_m1 | 3.1861 ± 0.4955 | Cadherin EGF LAG 7-pass G-type receptor 1 |
| Celsr2-Mm00457515_m1 | 1.9425 ± 0.0908 | EGF, latrophilin 7TM domain–containing 1 |
| Emr1-Mm00802529_m1 | 0.4630 ± 0.0233 | EGF-like module–containing, mucin-like, hormone receptor–like sequence 1 |
| Gpr56-Mm00817704_m1 | 52.3475 ± 1.5049 | GPCR 56 |
| Gpr64-Mm00724545_m1 | 0.3733 ± 0.0369 | GPCR 64 |
| Gpr97-Mm00499385_m1 | 0.0645 ± 0.0157 | GPCR 97 |
| Gpr111-Mm01281279_m1 | 0.1909 ± 0.0170 | GPCR 111 |
| Gpr114-Mm01247578_m1 | 0.0099 ± 0.0025 | GPCR 114 |
| Gpr115-Mm01279261_m1 | 0.0418 ± 0.0056 | GPCR 115 |
| Gpr116-Mm01269030_m1 | 3.9893 ± 0.3116 | GPCR 116 |
| Gpr123-Mm00624275_m1 | 42.3620 ± 4.4316 | GPCR 123 |
| Gpr124-Mm00475185_m1 | 1.5261 ± 0.2603 | GPCR 124 |
| Gpr125-Mm01211383_g1 | 5.8560 ± 0.4562 | GPCR 125 |
| Gpr133-Mm01203407_m1 | 0.0444 ± 0.0032 | GPCR 133 |
| Mass1-Mm00475232_m1 | 2.3356 ± 0.2509 | GPR98, monogenic, audiogenic seizure susceptibility 1 |
| Lphn1-Mm01247150_m1 | 58.5283 ± 4.2677 | Latrophilin 1, a family of secretin-like GPCRs, α-latrotoxin (LTX) receptor |
| Lphn2-Mm01320597_m1 | 16.4134 ± 0.9633 | Latrophilin 2, a family of secretin-like GPCRs, but not bind LTX |
| Lphn3-Mm01216694_m1 | 32.8496 ± 2.0521 | Latrophilin 3, a family of secretin-like GPCRs, but not bind LTX |
| Class C orphans (4) | ||
| Gpr156-Mm00462845_m1 | 0.3794 ± 0.0648 | GPCR 156 |
| Gprc5a-Mm00724888_m1 | 0.0747 ± 0.0093 | GPCR, family C, group 5, member A |
| Gprc5b-Mm00458150_m1 | 30.5954 ± 1.9440 | GPCR, family C, group 5, member B |
| Gprc5c-Mm00548863_m1 | 0.7821 ± 0.1231 | GPCR, family C, group 5, member |
| Non-signaling 7TM chemokine-binding proteins (2) | ||
| Ccrl1-Mm02620636_s1 | 0.1953 ± 0.0102 | Chemokine (C-C motif) receptor-like 1 |
| Darc-Mm02620117_s1 | 0.2885 ± 0.0531 | Duffy blood group, chemokine receptor |
| Other 7TM proteins (14) | ||
| Cmkor1-Mm02619632_s1 | 0.0726 ± 0.0121 | Chemokine orphan receptor 1 |
| Gpr77-Mm01267981_s1 | 0.1266 ± 0.0151 | GPCR 77 |
| Gpr92-Mm02621109_s1 | 0.3266 ± 0.0374 | GPCR 92 |
| Gpr103-Mm01294559_m1 | 1.8488 ± 0.0771 | GPCR 103 |
| Gpr108-Mm00712856_m1 | 9.7793 ± 1.4049 | GPCR 108 |
| Gpr109a-Mm02620500_s1 | 0.0107 ± 0.0018 | GPCR 109A |
| Gpr137-Mm01231752_g1 | 27.4696 ± 0.6611 | GPCR 137 |
| Gpr137b-Mm00652182_m1 | 11.4195 ± 1.0647 | GPCR 137B |
| Gpr143-Mm00440553_m1 | 0.0241 ± 0.0046 | GPCR 143 |
| Gpr154-Mm00558817_m1 | 1.4863 ± 0.2508 | GPCR 154 |
| Gpr175-Mm01183739_m1 | 1.3931 ± 0.1338 | GPCR 175 |
| Grm2-Mm01235831_m1 | 1.9818 ± 0.2209 | GPCR, family C, group 1, member B |
| Tm7sf3-Mm01255186_m1 | 7.5160 ± 0.2914 | TM7 superfamily member 3 |
| N/A-Mm02621695_s1 | 0.5950 ± 0.0693 | mCG6879 Celera annotation, hypothetical protein |
Overall GPCR expression profile in hypothalamic arcuate nucleus
The known GPCR genes that were expressed in the arcuate nucleus (183) were divided into 8 functional groups and the orphan GPCRs (109) into 5 orphan groups as shown in Tables 1 to 9 and in Figure 2. For the known GPCRs, these included 44 neurotransmitter receptors, 26 neuropeptide receptors, 31 hormone receptors, 10 receptors related to angiogenesis and vasoactivity, 16 receptors involved with inflammation and immune systems, 20 lipid messenger receptors, 12 growth factor receptors, and 24 other known GPCRs (Figure 2 and Tables 1 to 8). Of the 109 orphan GPCRs, 66 belong to class A (rhodopsin), 23 to class B (adhesion), 4 to class C (other GPCRs), 2 to nonsignaling 7-transmembrane (TM) chemokine-binding proteins, and 14 to other 7TM protein genes (Figure 2 and Table 9).
Verification of low-density array results using individual TaqMan GPCR assays
To verify the quantitative results of the GPCR low-density array we chose 4 GPCRs that were found to be expressed in high to low levels (GABAB-R2 [Gabbr2], μ-opioid receptor [Oprm1], 5HT receptor 2c [Htr2c], and CRH receptor 2 [Crhr2], as well as β-actin as an internal control), to be analyzed in individual TaqMan qPCR assays. As expected, all 4 GPCR genes were expressed in the arcuate nucleus in the following order: Gabbr2 ≫ Oprm1 > Htr2c > Crhr2, which was similar to the rank order of expression of these genes as revealed by the TaqMan low-density array (R2 = 0.9944; P < .005). Therefore, the relative expression of these 4 GPCRs was similar using the 2 different assay methods in 2 different PCR systems (ABI HT 7900 and ABI 7500), supporting the accuracy and reliability of the GPCR low-density array analysis.
Expression of the glucagon receptor family
The glucagon receptor family of GPCRs includes GHRH- receptor (Ghrhr), gastric inhibitory polypeptide receptor (Gipr), Glp1r, and Glp2r. Of these, Glp1r mRNA was found to be highly expressed (relative expression [RE], 7.05 ± 0.30) in the arcuate nucleus of POMC-EGFP mice, whereas the mRNA levels of the other receptors in this family were barely detectable (Figure 3 and Table 3). Glp1r vs Glp2r in the arcuate nucleus was also analyzed in wild-type C57 mice (n = 3) using qPCR (SYBR Green method) with similar results; ie, Glp1r mRNA was highly expressed (RE, 10.07 ± 0.60), whereas Glp2r mRNA could hardly be detected (RE, 0.01 ± 0.01).
Figure 3.

Glucagon receptor family mRNA expression in the arcuate nucleus. Bar graphs illustrating the relative mRNA expression of the glucagon receptor family as measured by GPCR array.
Glp1r vs Glp2r in arcuate nucleus POMC neurons
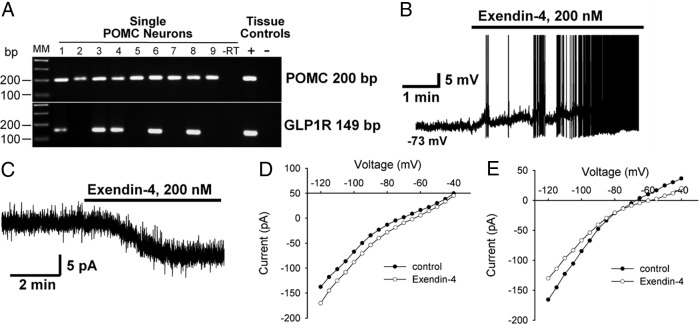
Our finding that the GLP1R was highly expressed in the arcuate nucleus is consistent with earlier observations that this receptor is involved in inhibition of feeding (38). Therefore, we measured the expression and function of GLP1R in arcuate POMC neurons. The studies revealed that Glp1r mRNA was expressed in at least 50% of POMC neurons in both males and females (Figure 4A), but this transcript could not be detected in NPY/agouti-related peptide (NPY/AgRP) neurons (data not shown). Consistent with the GPCR array measurements of low expression of Glp2r mRNA (RE, 0.03 ± 0.01) in the arcuate nucleus, Glp2r transcripts could not be detected in POMC neurons. We next examined the effects of a selective GLP1R agonist, exendin-4, on POMC neurons (39, 40). As shown in Figure 4B, exendin-4 (200 nM) induced a depolarization (5.3 ± 1.8 mV, n = 5) and increased firing in 3 of 5 cells (the other 2 cells did not respond). To examine whether exendin-4 directly activated POMC neurons, whole-cell current was measured in the presence of a synaptic blockade cocktail of drugs [tetrodotoxin, 1 μM; picrotoxin, 100 μM; and CNQX (6-cyano-7-nitroquinoxaline-2,3-dione, 20 μM)]. As shown in Figure 4C, exendin-4 induced an inward current in 70% (n = 16) of POMC neurons with a mean amplitude of 5.2 ± 0.6 pA. The current-voltage plots revealed that the GLP1R agonist activated a nonselective cation conductance along with inhibiting an inwardly rectifying potassium conductance (Figure 4, D and E, respectively).
Figure 4.
GLP1R expression and function in arcuate POMC neurons. A, Representative gel illustrating the mRNA expression of Glp1r in arcuate POMC neurons. Thirty-six neurons were analyzed from each of 3 females and 3 males, and at least 50% of POMC neurons were positive for Glp1r mRNA. MM, molecular marker; −RT, cell reacted without RT; + and −, tissue RNA reacted with and without RT. B, Whole-cell current-clamp recording showing that the GLP1R agonist exendin-4 (200 nM) induced a depolarization and increased firing in POMC neurons. Resting membrane potential was −73 mV. C, In the voltage clamp and in the presence of synaptic blockade, exendin-4 induced an inward current. D and E, current-voltage plots revealed that the inward current was generated by the activation of a nonselective cation current (D, reversal at −40 mV) and inhibition of an inwardly rectifying K+ current (E, reversal at −80 mV).
Discussion
With a TaqMan low-density array assay, we investigated the mRNA expression of 353 nonolfactory GPCRs and found that most of these genes were expressed in the mouse arcuate nucleus, including 183 known and 109 orphan GPCRs. The known GPCR genes expressed were classified into 8 functional groups, including neurotransmitter/neuropeptide receptors, hormone receptors, angiogenesis/vasoactive receptors, inflammatory/immune system receptors, lipid messenger receptors, and growth factor receptors. The orphan genes expressed were grouped into 5 structure-related classes including class A (rhodopsin), class B (adhesion), class C (other GPCRs), nonsignaling 7TM chemokine-binding proteins, and other 7TM proteins. Therefore, the arcuate nucleus of the hypothalamus is richly endowed with both known and orphan GPCRs.
Based on the analysis of independent biological samples run on replicate plates, the arcuate GPCR expression profile was consistent across the different plates. Similarly, individual qPCR assays of selected genes, ranging from high to very low expression levels in the array, exhibited the same relative expression of these genes in individual real-time PCR analysis. Of the genes tested individually (Gabbr2, Oprm1, Htr2c, and Crhr2), Gabbr2 was the most highly expressed, followed by Oprm1, Htr2c, and finally Crhr2, which was barely detectable, essentially as noted in the array analysis. The very low expression of Crhr2 (RE, 0.02 ± 0.01) in the mouse arcuate nucleus is also consistent with a previous report on the expression of this receptor gene (41). Therefore, the quantitative array profile of GPCRs in the arcuate nucleus appears to be consistent and should represent a reliable measure of the expression levels of known and orphan GPCRs. Moreover, the PCR array analysis is congruent with the electrophysiological analysis of the GABAB, μ-opioid, and 5HT receptor 2C–mediated responses in hypothalamic arcuate neurons (18, 24–27, 42).
Known GPCRs
Many of the known neurotransmitter GPCRs, which were presently identified as being moderate to highly expressed, have been documented in the arcuate nucleus, eg, adrenergic receptors α1a, α1b, α2a, and β1 and serotonergic receptors 5HT1B, 5HT1A, 5HT2A, and 5HT2C, all of whose expression and functions in the arcuate nucleus are relatively well known (43–48). Less is known about the quantitative expression of many of the other known GPCRs that were currently identified in the arcuate nucleus of adult female mice (Tables 1–8), eg, the glucagon receptor family, tachykinin receptors, RFamide receptors, calcitonin receptors, relaxin receptors, and frizzled receptors.
The arcuate nucleus of the hypothalamus is composed of multiple neuronal types that control many homeostatic functions of which the control of energy homeostasis and reproduction have been the most extensively investigated (7, 49, 50). For example, neurons expressing NPY/AgRP stimulate feeding but are also involved in the control of reproduction via modulation of GnRH neuronal excitability (51). Importantly, the α2 adrenergic receptor appears to be involved in the NPY-induced feeding behavior and may also act in part to modulate the NPY-induced LH surge in rodents (52, 53). Several NPY receptors including Y1, Y2, and Y5 are all expressed in the arcuate nucleus (54, 55) with highest expression of Y1 (RE, 4.57 ± 0.76) and lowest expression of Y5 mRNA (RE, 0.16 ± 0.02) (Table 2). NPY neurons project to and synapse on dopamine and POMC cells in the arcuate nucleus (56, 57), an indication that NPY receptors are expressed on these neurons. The gut hormone ghrelin also stimulates feeding, and, not surprisingly, we found that the ghrelin receptor (GH secretagogue receptor) is expressed in the arcuate nucleus in relatively high quantities (RE, 2.85 ± 0.24).
Neurons expressing POMC/cocaine- and amphetamine-regulated transcript (CART) inhibit feeding and may also be involved in the control of GnRH neurons (58, 59). Although much is known about POMC peptides and their receptors, the CART receptor(s) has not yet been identified and may be among the orphan GPCRs identified in the arcuate nucleus (Table 9). The POMC peptide α-melanocyte-stimulating hormone (α-MSH), acting primarily through the melanocortin receptors 3 and 4 (MC3R and MC4R), has received much attention for its role in the control of energy homeostasis (13, 60, 61). MC4R deficiencies (mutations) lead to severe obesity in human and animal models, and MC3R mutation appears also to be associated with obesity (61, 62). Currently, we have identified 4 melanocortin receptors (MC1, MC3, MC4, and MC5) in the arcuate nucleus, of which the Mc3r transcript was by far the most highly expressed (RE, 7.76 ± 0.28) followed by much lower expression of the Mc4r transcript (RE, 0.98 ± 0.05). These results are consistent with previous reports documenting the mRNA expression of melanocortin receptors in the hypothalamus (61, 63–67). The functional significance of the low expression of Mc1r and Mc5r mRNAs within the arcuate nucleus is uncertain because these receptors are believed to be important in the periphery, eg, for skin pigmentation (MC1R) and sebaceous lipid production (MC5R) (67).
The proglucagon peptide, which is processed to glucagon in the pancreas, is processed to GLP1 and GLP2 in the brain and in the gut (for review, see ref. 68). These peptides act at the GLP1R and GLP2R, respectively (68, 69). We found that Glp1r but not Glp2r mRNA was highly expressed in the arcuate nucleus, and Glp1r mRNA (not Glp2r) was specifically identified in male and female POMC neurons. Importantly, a selective and potent agonist (exendin-4) for GLP1R (39) depolarized and increased the firing of the anorexigenic POMC neurons (Figure 4). Moreover, in the presence of synaptic blockade, the GLP1R agonist induced an inward (depolarizing) current in most POMC neurons via activation of nonselective cation conductance. Collectively, these findings suggest that functional GLP1Rs are expressed in POMC neurons, and their activation leads to increased excitation of POMC neurons. These actions of this postprandial-released gut hormone augment actions of insulin in POMC neurons (70). This could be a mechanism by which GLP1 and its agonist (exendin-4) attenuate feeding (38, 71). In comparison to Glp1r mRNA, Glp2r mRNA was expressed at many-fold lower levels in the arcuate nucleus (Figure 3). This finding is consistent with previous in situ hybridization studies, which localized Glp2r mRNA primarily outside the arcuate nucleus within the dorsomedial hypothalamus (72). In addition, intracerebroventricular injection of GLP2 induced cFos expression in the dorsomedial hypothalamus but not in the arcuate nucleus (72), a further indication that GLP2Rs may not be highly expressed or active in the arcuate nucleus. It is therefore interesting that it has been reported that Glp2r mRNA and protein are highly expressed in arcuate neurons including POMC neurons (73). The reason for the discrepancy is unclear but could be related to different animal models and experimental approaches. Although central nervous system GLP2Rs appear to be involved in the regulation of food intake in rodents, the cellular targets are not known but may involve functional interaction (cross-talk) between the GLP1- and GLP2-responsive neurons given that the anorectic effect of GLP2 was potentiated in GLP1R knockout animals (74).
GPR54, the kisspeptin 1 (KISS1) receptor, was found to be moderately expressed (RE, 1.6 ± 0.12) in the mouse arcuate nucleus (Table 2). In contrast, using a transgenic GPR54 LacZ knock-in mouse model, GPR54 mRNA was not detected in the arcuate nucleus (75). However, KISS1 has been found to excite POMC neurons in the arcuate nucleus, an indication that the kisspeptin receptor is expressed and may play a role in the control of feeding (16). Neurons expressing KISS1 are localized in the arcuate nucleus and coexpress the tachykinin, neurokinin (NK) B, as well as the opioid peptide, dynorphin (76, 77). NKB is the ligand for the tachykinin receptor 3 (Tacr3), which was one of the high-expressing GPCRs in our analysis (RE, 5.11 ± 0.52), and Tacr3 mRNA is expressed in KISS1/NKB neurons (77, 78). Both KISS1, NKB, and their respective receptors have been shown to be essential for sexual development and reproductive function in human and animal models (79–84). In addition, we found that Tacr1 (substance P receptor) mRNA was expressed in moderate levels (RE, 2.63 ± 0.31), whereas the mRNA expression of the Tacr2 (ligand, neurokinin A) was quite low (RE, 0.07 ± 0.01) in the mouse arcuate nucleus. Immunoreactive substance P has been identified in the rhesus macaque infundibular nucleus (85), and substance P mRNA is also expressed in the mouse arcuate nucleus (O. K. Rønnekleiv, unpublished observations). The current findings are consistent with a recent publication illustrating that NKB, NKA, and substance P all induced membrane depolarization in mouse arcuate KISS1 neurons (86). We found that κ-opioid receptor mRNA was moderately expressed (RE, 2.10 ± 0.46) within the arcuate nucleus. As stated above, the arcuate KISS1 neurons coexpress dynorphin, the ligand for the κ-opioid receptors. In addition, these neurons express κ-opioid receptor mRNA, and the mRNA expression levels of both dynorphin and its receptor are inhibited by estradiol (77). Interestingly, based on a series of neuroanatomical and neuroendocrine analyses, a model has been presented showing that NKB and dynorphin may act autosynaptically on arcuate KISS1 neurons to shape pulsatile secretion of GnRH (77).
We found moderate expression of the relaxin family peptide receptor 3 (Rxfp3) in the arcuate nucleus. This receptor has been identified as the receptor for the relaxin-3 (RLN-3) peptides (87). Both RLN-3 and its receptor are widely distributed in the mouse brain, and centrally administered RLN-3 induces cFos expression in different brain regions including the supraoptic nucleus and the arcuate nucleus (88–90). It is therefore, not surprising that centrally administered RLN-3 facilitates both water and food intake (88, 89). However, overall very little is known about the mechanism by which RLN-3 and its receptor induce feeding.
Calcitonin receptor (Calcr) mRNA was found to be highly expressed (RE, 19.50 ± 3.08) in the arcuate nucleus. A related peptide receptor, calcitonin receptor-like (Calcrl), was also detected, but at a much lower level. A prior immunocytochemical analysis also found a high level of immunoreactive calcitonin receptor in the rat arcuate nucleus as well as in numerous other brain regions (91). Calcitonins are a family of peptides, including calcitonin, aminoprocalcitonin, calcitonin gene–related peptide, amylin, and adrenomedullin, which can act on Calcr, Calcrl, or on heterodimers of these receptors with single transmembrane proteins (92, 93). Activation of Calcr or Calcrl affects primarily calcium homeostasis (92). In addition, the calcitonin-calcitonin receptor system may be involved in regulating energy homeostasis (93). However, the specific role of the calcitonins and their receptors in the arcuate nucleus is not known and warrants further investigation.
Of interest is that 10 growth factor receptors of the frizzled family were found to be expressed in the arcuate nucleus and the majority at quite high levels. In particular, the frizzled homolog 3 and 5 receptors were highly expressed (RE, 19.10 ± 1.95 and 13.42 ± 0.52, respectively) in this brain region in support of previous publications (94, 95). This family of receptors is known primarily to be critically involved in fetal development. However, evidence suggests that these receptors, which are activated by the Wingless-Int (WNT) family of lipoglycoproteins, are important in the adult brain and other organs (96). The WNTs comprise a large family of highly conserved growth factors that upon binding to frizzled receptors cause accumulation of β-catenin in the cytoplasm, which translocates to the nucleus and alters transcription of target genes (97). WNTs through binding to frizzled receptors can also signal via pathways involving calcium/calmodulin-dependent kinase II and protein kinase C or GTP-binding proteins, leading to activation of phospholipase C and phosphodiesterase (98). However, the function and signaling of frizzled receptors within the arcuate nucleus in adult animals appear to be unexplored.
In the low-expressing GPCR group, we identified GPR30 (RE, 0.24 ± 0.13), also known as G protein–coupled estrogen receptor 1 (GPER1) (99–102). Considerable evidence indicates that GPR30 is an estrogen receptor (ER) at the plasma membrane and signals independent of ERα and ERβ (99). GPR30 stimulates calcium oscillations and GnRH release in immature rhesus monkey GnRH neuronal cultures (103, 104); however, a physiological role for GPR30 in the arcuate nucleus of adult animals has not been identified (105, 106).
Orphan GPCRs
We arbitrarily identified these orphan genes as follows: high-expressing (>5-fold), including GPR50, GPR56, GPR75, GPR85, GPR153, and GPR brain-specific angiogenesis inhibitor (Bai) 1 and Bai3; moderate-expressing (between 1- and 5-fold), including GPR6, GPR17, GPR23, GPR176, and Bai2; and low-expressing (<1-fold), the largest group of orphan GPCRs including GPR1, GPR3, and GPR12.
GPR50, a member of the rhodopsin GPCR family, shares significant sequence homology with the two melatonin receptors (MT1 and MT2) but does not bind melatonin (107) and is, therefore, still considered an orphan GPCR. We found that this transcript is one of the high expressing genes (RE, 19.59 ± 2.03) in the arcuate nucleus. Previous in situ hybridization analysis had already revealed that GPR50 mRNA is expressed in the ventromedial hypothalamus including the arcuate nucleus (107). Immunoreactive GPR50 neurons have been identified in the dorsomedial hypothalamus but have been more difficult to detect in the arcuate nucleus (108, 109). GPR50 is believed to be involved in adaptive thermogenesis in mammals (110).
GPR75, also of the rhodopsin family of orphan GPCRs, is highly expressed (RE, 11.21 ± 1.53) in the arcuate nucleus. This receptor is a novel chemokine receptor, which is expressed in the brain, spinal cord, and in the pancreas (111, 112). Although GPR75 has limited homology (12%–16%) with chemokine receptors, it is activated by the chemokine (C-C motif) ligand 5 (CCL5; also called RANTES [regulated upon activation, normal T-cell expressed and secreted]) in pancreatic β cells, which causes intracellular calcium release and increased insulin secretion (112, 113). In the hippocampal cell line HT22, which endogenously expresses GPR75, CCL5 reduced the neurotoxicity of amyloid-β peptides by activating phospholipase C and phosphoinositide 3-kinase (113). The role of GPR75 in the arcuate nucleus has not been investigated but could be involved in anti-inflammatory responses associated with a high-fat diet (114).
GPR56, a member of the adhesion GPCR family, is one of the highest expressing (RE, 52.35 ± 1.50) orphan GPCRs in the arcuate nucleus. This transcript appears to be important for the development of the cortex because mutations in GPR56 cause bilateral frontoparietal polymicrogyria (115, 116). Possible ligands for GPR56 are collagen, type III and/or transglutaminase 2 (115–117). A role for GPR56 in the arcuate nucleus needs to be elucidated.
Bai1 to Bai3 of the adhesion GPCR family were also highly expressed in the arcuate nucleus with Bai1 ≫ Bai3 ≫ Bai2 (RE, 52.03 ± 2.89, 15.58 ± 0.71, and 4.92 ± 0.52, respectively). These receptors are involved not only in angiogenesis and phagocytosis but may also play important roles in neurogenesis, synaptogenesis, and regulation of spine morphology (118–120). Importantly, it has been found that Bai1 is enriched in postsynaptic density in mouse brain and signals through Gα12/13 to activate the Rho pathway as shown in human embryonic kidney 293 cells (119). The role of Bai transcripts (proteins) within the arcuate nucleus needs to be investigated but could be associated with spine formation and synaptic remodeling within this important nucleus (118, 120).
In summary, this quantitative analysis of GPCR expression in the arcuate nucleus should serve as a resource on which to further elucidate the functional significance of known GPCRs as well as help to deorphanize the orphan GPCRs. The expression of these orphans within the arcuate nucleus argues for their role in important homeostatic functions, such as metabolism and energy homeostasis, as well as temperature regulation and reproduction.
Acknowledgments
We thank Martha A. Bosch, Marina V. Rulevskaya, and Uyen-Vy Navarro for their excellent technical support.
This work was supported by the National Science Foundation Eager IOS Grant 110822. C.CN. was supported by National Institutes of Health Training Grant T32 HD007133.
Current address for Y.F.: Hangzhou Derlead Bio-Tech Co. Ltd, Hangzhou Tianhe High Tech Park, Hangzhou, Zhejiang Province, China 310052.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ABI
- Applied Biosystems
- aCSF
- artificial cerebrospinal fluid
- Bai
- brain-specific angiogenesis inhibitor
- CART
- cocaine- and amphetamine-regulated transcript
- CCL5
- chemokine (C-C motif) ligand 5
- EGFP
- enhanced green fluorescent protein
- ER
- estrogen receptor
- GABA
- γ-aminobutyric acid
- GLP
- glucagon-like peptide
- GLP1R
- glucagon-like peptide 1 receptor
- GLP2R
- glucagon-like peptide 2 receptor
- GPCR
- G protein–coupled receptor
- 5HT
- 5-hydroxytryptamine (serotonin)
- KISS1
- kisspeptin
- MC
- melanocortin
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- NKB
- neurokinin B
- NPY
- neuropeptide Y
- nt
- nucleotide
- OVX
- ovariectomized
- POMC
- proopiomelanocortin
- qPCR
- quantitative PCR
- RE
- relative expression
- RLN-3
- relaxin-3
- WNT
- Wingless-Int.
References
- 1. Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–685 [DOI] [PubMed] [Google Scholar]
- 2. Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11:998–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lyons DJ, Horjales-Araujo E, Broberger C. Synchronized network oscillations in rat tuberoinfundibular dopamine neurons: switch to tonic discharge by thyrotropin-releasing hormone. Neuron. 2010;65:217–229 [DOI] [PubMed] [Google Scholar]
- 4. Lehman MN, Ladha Z, Coolen LM, et al. Neuronal plasticity and seasonal reproduction in sheep. Eur J Neurosci. 2010;32:2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohkura S, Takase K, Matsuyama S, et al. Gonadotrophin-releasing hormone pulse generator activity in the hypothalamus of the goat. J Neuroendocrinol. 2009;21:813–821 [DOI] [PubMed] [Google Scholar]
- 6. Rønnekleiv OK, Bosch MA, Zhang C. 17β-Oestradiol regulation of gonadotrophin-releasing hormone neuronal excitability. J Neuroendocrinol. 2011;24:122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelly MJ, Rønnekleiv OK. Membrane-initiated actions of estradiol that regulate reproduction, energy balance and body temperature. Front Neuroendocrinol. 2012;33:376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Micevych PE, Kelly MJ. Membrane estrogen receptor regulation of hypothalamic function. Neuroendocrinology. 2012;96:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kelly MJ, Zhang C, Qiu J, Rønnekleiv OK. Pacemaking kisspeptin neurons. Exp Physiol. 2013;98:1535–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gearing M, Terasawa E. Suppression of luteinizing hormone release by the α1-adrenergic receptor antagonist prazosin in the ovariectomized female rhesus monkey. Am J Primatol. 1991;25:23–33 [DOI] [PubMed] [Google Scholar]
- 11. Kelly MJ, Qiu J, Wagner EJ, Rønnekleiv OK. Rapid effects of estrogen on G protein-coupled receptor activation of potassium channels in the central nervous system (CNS). J Steroid Biochem Mol Biol. 2002;83:187–193 [DOI] [PubMed] [Google Scholar]
- 12. Ibrahim N, Bosch MA, Smart JL, et al. Hypothalamic proopiomelanocortin neurons are glucose-responsive and express KATP channels. Endocrinology. 2003;144:1331–1340 [DOI] [PubMed] [Google Scholar]
- 13. Cone RD. Anatomy and regulation of the central melanocortin system. Nat Neurosci. 2005;8:571–578 [DOI] [PubMed] [Google Scholar]
- 14. Qiu J, Bosch MA, Tobias SC, et al. A G-protein-coupled estrogen receptor is involved in hypothalamic control of energy homeostasis. J Neurosci. 2006;26:5649–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu J, Xue C, Bosch MA, et al. Serotonin 5-hydroxytryptamine2c receptor signaling in hypothalamic POMC neurons: role in energy homeostasis in females. Mol Pharmacol. 2007;72:885–896 [DOI] [PubMed] [Google Scholar]
- 16. Fu LY, Van den Pol AN. Kisspeptin directly excites anorexigenic proopiomelanocortin neurons but inhibits orexigenic neuropeptide Y cells by an indirect synaptic mechanism. J Neurosci. 2010;30:10205–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pennock RL, Hentges ST. Differential expression and sensitivity of presynaptic and postsynaptic opioid receptors regulating hypothalamic proopiomelanocortin neurons. J Neurosci. 2011;31:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith AW, Bosch MA, Wagner EJ, Rønnekleiv OK, Kelly MJ. The membrane estrogen receptor ligand STX rapidly enhances GABAergic signaling in NPY/AgRP neurons: role in mediating the anorexigenic effects of 17β-estradiol. Am J Physiol Endocrinol Metab. 2013;305:E632–E640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Regard JB, Sato IT, Coughlin SR. Anatomical profiling of g protein-coupled receptor expression. Cell. 2008;135:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allen Institute for Brain Science. Allen Brain Atlas. Seattle, WA: Allen Institute for Brain Science; 2011 [Google Scholar]
- 21. Civelli O. Orphan GPCRs and neuromodulation. Neuron. 2012;76:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Civelli O, Reinscheid RK, Zhang Y, Wang Z, Fredriksson R, Schiöth HB. G protein-coupled receptor deorphanizations. Annu Rev Pharmacol Toxicol. 2013;53:127–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011;51:117–144 [DOI] [PubMed] [Google Scholar]
- 24. Kelly MJ, Loose MD, Rønnekleiv OK. Estrogen suppresses μ-opioid- and GABAB-mediated hyperpolarization of hypothalamic arcuate neurons. J Neurosci. 1992;12:2745–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lagrange AH, Wagner EJ, Rønnekleiv OK, Kelly MJ. Estrogen rapidly attenuates a GABAB response in hypothalamic neurons. Neuroendocrinology. 1996;64:114–123 [DOI] [PubMed] [Google Scholar]
- 26. Wagner EJ, Bosch MA, Kelly MJ, Rønnekleiv OK. A powerful GABAB receptor-mediated inhibition of GABAergic neurons in arcuate nucleus. NeuroReport. 1999;10:2681–2687 [DOI] [PubMed] [Google Scholar]
- 27. Qiu J, Bosch MA, Tobias SC, et al. Rapid signaling of estrogen in hypothalamic neurons involves a novel G-protein-coupled estrogen receptor that activates protein kinase C. J Neurosci. 2003;23:9529–9540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng SX, Bosch MA, Rønnekleiv OK. Mu-opioid receptor mRNA expression in identified hypothalamic neurons. J Comp Neurol. 2005;487:332–344 [DOI] [PubMed] [Google Scholar]
- 29. Bosch MA, Hou J, Fang Y, Kelly MJ, Rønnekleiv OK. 17β-Estradiol regulation of the mRNA expression of T-type calcium channel subunits: role of estrogen receptor α and estrogen receptor β. J Comp Neurol. 2009;512:347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang C, Tonsfeldt KJ, Qiu J, Bosch MA, Kobayashi K, Steiner RA, Kelly MJ, Rønnekleiv OK. Molecular mechanisms that drive estradiol-dependent burst firing of Kiss1 neurons in the rostral periventricular preoptic area. Am J Physiol Endocrinol Metab. 2013;305:E1384–E1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bosch MA, Xue C, Rønnekleiv OK. Kisspeptin expression in guinea pig hypothalamus: effects of 17β-estradiol. J Comp Neurol. 2012;520:2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C, Bosch MA, Rick EA, Kelly MJ, Rønnekleiv OK. 17β-Estradiol regulation of T-type calcium channels in gonadotropin-releasing hormone neurons. J Neurosci. 2009;29:10552–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qiu J, Fang Y, Rønnekleiv OK, Kelly MJ. Leptin excites proopiomelanocortin neurons via activation of TRPC channels. J Neurosci. 2010;30:1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bosch MA, Tonsfeldt KJ, Rønnekleiv OK. mRNA expression of ion channels in GnRH neurons: subtype-specific regulation by 17β-estradiol. Mol Cell Endocrinol. 2013;367:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 36. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001;29:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor S. A MIQE case study—effect of RNA sample quality and reference gene stability on gene expression data. Bio-Rad Tech Bull. 2011;6245 [Google Scholar]
- 38. Turton MD, O'Shea D, Gunn I, et al. A role for glucagon-like peptide-1 in the central regulation of feeding. Nature. 1996;379:69–72 [DOI] [PubMed] [Google Scholar]
- 39. Göke R, Fehmann HC, Linn T, et al. Exendin-4 is a high potency agonist and truncated exendin-(9–39)-amide an antagonist at the glucagon-like peptide 1-(7–36)-amide receptor of insulin-secreting beta-cells. J Biol Chem. 1993;268:19650–19655 [PubMed] [Google Scholar]
- 40. Dalvi PS, Nazarians-Armavil A, Purser MJ, Belsham DD. Glucagon-like peptide-1 receptor agonist, exendin-4, regulates feeding-associated neuropeptides in hypothalamic neurons in vivo and in vitro. Endocrinology. 2012;153:2208–2222 [DOI] [PubMed] [Google Scholar]
- 41. Van Pett K, Viau V, Bittencourt JC, et al. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol. 2000;428:191–212 [DOI] [PubMed] [Google Scholar]
- 42. Lagrange AH, Rønnekleiv OK, Kelly MJ. The potency of μ-opioid hyperpolarization of hypothalamic arcuate neurons is rapidly attenuated by 17β-estradiol. J Neurosci. 1994;14:6196–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Unnerstall JR, Kopajtic TA, Kuhar MJ. Distribution of α2 agonist binding sites in the rat and human central nervous system: analysis of some functional, anatomic correlates of the pharmacologic effects of clonidine and related adrenergic agents. Brain Res. 1984;319:69–101 [DOI] [PubMed] [Google Scholar]
- 44. Condon TP, Rønnekleiv OK, Kelly MJ. Estrogen modulation of the α1-adrenergic response of hypothalamic neurons. Neuroendocrinology 1989;50:51–58 [DOI] [PubMed] [Google Scholar]
- 45. King PR, Gundlach AL, Louis WJ. Quantitative autoradiographic localization in rat brain of α2-adrenergic and non-adrenergic I-receptor binding sites labelled by [3H]rilmenidine. Brain Res. 1995;675:264–278 [DOI] [PubMed] [Google Scholar]
- 46. Acosta-Martinez M, Fiber JM, Brown RD, Etgen AM. Localization of α1B-adrenergic receptor in female rat brain regions involved in stress and neuroendocrine function. Neurochem Int. 1999;35:383–391 [DOI] [PubMed] [Google Scholar]
- 47. Gundlah C, Pecins-Thompson M, Schutzer WE, Bethea CL. Ovarian steroid effects on serotonin 1A, 2A and 2C receptor mRNA in macaque hypothalamus. Mol Brain Res. 1999;63:325–339 [DOI] [PubMed] [Google Scholar]
- 48. Etgen AM. Estrogen regulation of neurotransmitter and growth factor signaling in the brain. In: Pfaff DW, ed. Hormones, Brain and Behavior. 3rd ed San Diego, CA: Academic Press; 2002:381–440 [Google Scholar]
- 49. Kishi T, Elmquist JK. Body weight is regulated by the brain: a link between feeding and emotion. Mol Psychiatry. 2005;10:132–146 [DOI] [PubMed] [Google Scholar]
- 50. Smith SM, True C, Grove KL. The neuroendocrine basis of lactation-induced suppression of GnRH: role of kisspeptin and leptin. Brain Res. 2010;1364:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Acosta-Martinez M, Horton T, Levine JE. Estrogen receptors in neuropeptide Y neurons: at the crossroads of feeding and reproduction. Trends Endocrinol Metab. 2007;18:48–50 [DOI] [PubMed] [Google Scholar]
- 52. Allen LG, Crowley WR, Kalra SP. Interactions between neuropeptide Y and adrenergic systems in the stimulation of luteinizing hormone release in steroid-primed ovariectomized rats. Endocrinology. 1987;121:1953–1959 [DOI] [PubMed] [Google Scholar]
- 53. Clark JT, Gist RS, Kalra SP, Kalra PS. Alpha2-adrenoceptor blockade attenuates feeding behavior induced by neuropeptide Y and epinephrine. Physiol Behav. 1988;43:417–422 [DOI] [PubMed] [Google Scholar]
- 54. Pralong FP, Gonzales C, Voirol MJ, et al. The neuropeptide Y Y1 receptor regulates leptin-mediated control of energy homeostasis and reproductive functions. FASEB J. 2002;16:712–714 [DOI] [PubMed] [Google Scholar]
- 55. Fetissov SO, Byrne LC, Hassani H, Ernfors P, Hökfelt T. Characterization of neuropeptide Y Y2 and Y5 receptor expression in the mouse hypothalamus. J Comp Neurol. 2004;470:256–265 [DOI] [PubMed] [Google Scholar]
- 56. Guy J, Pelletier G. Neuronal interactions between neuropeptide Y (NPY) and catecholaminergic systems in the rat arcuate nucleus as shown by dual immunocytochemistry. Peptides. 1988;9:567–570 [DOI] [PubMed] [Google Scholar]
- 57. Horvath TL, Naftolin F, Kalra SP, Leranth C. Neuropeptide-Y innervation of β-endorphin-containing cells in the rat mediobasal hypothalamus: a light and electron microscopic double immunostaining analysis. Endocrinology. 1992;131:2461–2467 [DOI] [PubMed] [Google Scholar]
- 58. Lagrange AH, Rønnekleiv OK, Kelly MJ. Estradiol-17β and μ-opioid peptides rapidly hyperpolarize GnRH neurons: A cellular mechanism of negative feedback? Endocrinology. 1995;136:2341–2344 [DOI] [PubMed] [Google Scholar]
- 59. Roa J, Herbison AE. Direct regulation of GnRH neuron excitability by arcuate nucleus POMC and NPY neuron neuropeptides in female mice. Endocrinology. 2012;153:5587–5599 [DOI] [PubMed] [Google Scholar]
- 60. Chen AS, Marsh DJ, Trumbauer ME, et al. Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet. 2000;26:97–102 [DOI] [PubMed] [Google Scholar]
- 61. Lee Y-S, Poh LK, Loke KY. A novel melanocortin 3 receptor gene (MC3R) mutation associated with severe obesity. J Clin Endocrinol Metab. 2002;87:1423–1426 [DOI] [PubMed] [Google Scholar]
- 62. Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O'Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–1095 [DOI] [PubMed] [Google Scholar]
- 63. Gantz I, Konda Y, Tashiro T, et al. Molecular cloning of a novel melanocortin receptor. J Biol Chem. 1993;268:8246–8250 [PubMed] [Google Scholar]
- 64. Roselli-Rehfuss L, Mountjoy KG, Robbins LS, et al. Identification of a receptor for γ melanotropin and other proopiomelanocortin peptides in the hypothalamus and limbic system. Proc Natl Acad Sci USA. 1993;90:8856–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mountjoy KG, Mortrud MT, Low MJ, Simerly RB, Cone RD. Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Mol Endocrinol. 1994;8:1298–1308 [DOI] [PubMed] [Google Scholar]
- 66. Beck B. Neuropeptide Y in normal eating and in genetic and dietary-induced obesity. Phil Trans R Soc B Biol Sci. 2006;361:1159–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ramachandrappa S, Gorrigan RJ, Clark AJL, Chan LF. The melanocortin receptors and their accessory proteins. Front Endocrinol (Lausanne). 2013;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev. 2007;87:1409–1439 [DOI] [PubMed] [Google Scholar]
- 69. Munroe DG, Gupta AK, Kooshesh F, et al. Prototypic G protein-coupled receptor for the intestinotrophic factor glucagon-like peptide 2. Proc Natl Acad Sci USA. 1999;96:1569–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qui J, Zhang C, Borgquist A, et al. Insulin excites anorexigenic proopiomelanocortin neurons via activation of canonical transient receptor potential channels. Cell Metab. 2014;19:682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Szayna M, Doyle ME, Betkey JA, et al. Exendin-4 decelerates food intake, weight gain, and fat deposition in Zucker rats. Endocrinology. 2000;141:1936–1941 [DOI] [PubMed] [Google Scholar]
- 72. Tang-Christensen M, Larsen PJ, Thulesen J, Rømer J, Vrana N. The proglucagon-derived peptide, glucagon-like peptide-2, is a neurotransmitter involved in the regulation of food intake. Nat. Med. 2000;6:802–807 [DOI] [PubMed] [Google Scholar]
- 73. Guan X, Shi X, Li X, et al. GLP-2 receptor in POMC neurons suppresses feeding behavior and gastric motility. Am J Physiol Endocrinol Metab. 2012;303:E853–E864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lovshin J, Estall J, Yusta B, Brown TJ, Drucker DJ. Glucagon-like peptide (GLP)-2 action in the murine central nervous system is enhanced by elimination of GLP-1 receptor signaling. J Biol Chem. 2001;276:21489–21499 [DOI] [PubMed] [Google Scholar]
- 75. Herbison AE, de Tassigny Xd, Doran J, Colledge WH. Distribution and postnatal development of Gpr54 gene expression in mouse brain and gonadotropin-releasing hormone neurons. Endocrinology. 2010;151:312–321 [DOI] [PubMed] [Google Scholar]
- 76. Goodman RL, Lehman MN, Smith JT, et al. Kisspeptin neurons in the arcuate nucleus of the ewe express both dynorphin A and neurokinin B. Endocrinology. 2007;148:5752–5760 [DOI] [PubMed] [Google Scholar]
- 77. Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci. 2009;29:11859–11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Navarro VM, Gottsch ML, Wu M, et al. Regulation of NKB pathways and their roles in the control of Kiss1 neurons in the arcuate nucleus of the male mouse. Endocrinology. 2011;152:4265–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627 [DOI] [PubMed] [Google Scholar]
- 81. d'Anglemont de Tassigny X, Fagg LA, Dixon JP, et al. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci USA. 2007;104:10714–10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Topaloglu AK, Reimann F, Guclu M, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guran T, Tolhurst G, Bereket A, et al. Hypogonadotropic hypogonadism due to a novel missense mutation in the first extracellular loop of the neurokinin B receptor. J Clinl Endocrinol Metab. 2009;94:3633–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Topaloglu AK, Tello JA, Kotan LD, et al. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med. 2012;366:629–635 [DOI] [PubMed] [Google Scholar]
- 85. Rønnekleiv OK, Kelly MJ, Eskay RL. Distribution of immunoreactive substance P neurons in the hypothalamus and pituitary of the rhesus monkey. J Comp Neurol. 1984;224:51–59 [DOI] [PubMed] [Google Scholar]
- 86. de Croft S, Boehm U, Herbison AE. Neurokinin B activates arcuate kisspeptin neurons through multiple tachykinin receptors in the male mouse. Endocrinology. 2013;154:2750–2760 [DOI] [PubMed] [Google Scholar]
- 87. Halls ML, van der Westhuizen ET, Bathgate RA, Summers RJ. Relaxin family peptide receptors—former orphans reunite with their parent ligands to activate multiple signalling pathways. Br J Pharmacol. 2007;150:677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. McGowan BM, Stanley SA, White NE, et al. Hypothalamic mapping of orexigenic action and Fos-like immunoreactivity following relaxin-3 administration in male Wistar rats. Am J Physiol Endocrinol Metab. 2007;292:E913–E919 [DOI] [PubMed] [Google Scholar]
- 89. Otsubo H, Onaka T, Suzuki H, et al. Centrally administered relaxin-3 induces Fos expression in the osmosensitive areas in rat brain and facilitates water intake. Peptides. 2010;31:1124–1130 [DOI] [PubMed] [Google Scholar]
- 90. Smith CM, Shen PJ, Banerjee A, et al. Distribution of relaxin-3 and RXFP3 within arousal, stress, affective, and cognitive circuits of mouse brain. J Comp Neurol. 2010;518:4016–4045 [DOI] [PubMed] [Google Scholar]
- 91. Becskei C, Riediger T, Zünd D, Wookey P, Lutz TA. Immunohistochemical mapping of calcitonin receptors in the adult rat brain. Brain Res. 2004;1030:221–233 [DOI] [PubMed] [Google Scholar]
- 92. Lerner UH. Deletions of genes encoding calcitonin/α-CGRP, amylin and calcitonin receptor have given new and unexpected insights into the function of calcitonin receptors and calcitonin receptor-like receptors in bone. J Musculoskelet Neuronal Interact. 2006;6:87–95 [PubMed] [Google Scholar]
- 93. Tavares E, Maldonado R, Miñano FJ. Aminoprocalcitonin-mediated suppression of feeding involves the hypothalamic melanocortin system. Am J Physiol Endocrinol Metab. 2013;304:E1251–E1262 [DOI] [PubMed] [Google Scholar]
- 94. Yamada M, Iwabuchi T, Takahashi K, et al. Identification and expression of frizzled-3 protein in rat frontal cortex after antidepressant and electroconvulsive treatment. J Pharmacol Sci. 2005;99:239–246 [DOI] [PubMed] [Google Scholar]
- 95. Liu C, Wang Y, Smallwood PM, Nathans J. An essential role for Frizzled5 in neuronal survival in the parafascicular nucleus of the thalamus. J Neurosci. 2008;28:5641–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Schulte G, Bryja V. The frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:518–525 [DOI] [PubMed] [Google Scholar]
- 97. Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433 [DOI] [PubMed] [Google Scholar]
- 98. Kohn AD, Moon RT. Wnt and calcium signaling: β-catenin-independent pathways. Cell Calcium. 2005;38:439–446 [DOI] [PubMed] [Google Scholar]
- 99. Filardo EJ, Thomas P. GPR30: a seven-transmembrane-spanning estrogen receptor that triggers EGF release. Trends Endocrinol Metab. 2005;16:362–367 [DOI] [PubMed] [Google Scholar]
- 100. Funakoshi T, Yanai A, Shinoda K, Kawano MM, Mizukami Y. G protein-coupled receptor 30 is an estrogen receptor in the plasma membrane. Biochem Biophys Res Commun. 2006;346:904–910 [DOI] [PubMed] [Google Scholar]
- 101. Filardo E, Quinn J, Pang Y, et al. Activation of the novel estrogen receptor G protein-coupled receptor 30 (GPR30) at the plasma membrane. Endocrinology. 2007;148:3236–3245 [DOI] [PubMed] [Google Scholar]
- 102. Filardo EJ, Thomas P. Minireview: G protein-coupled estrogen receptor-1, GPER-1: its mechanism of action and role in female reproductive cancer, renal and vascular physiology. Endocrinology. 2012;153:2953–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Noel SD, Keen KL, Baumann DI, Filardo EJ, Terasawa E. Involvement of G protein-coupled receptor 30 (GPR30) in rapid action of estrogen in primate LHRH neurons. Mol Endocrinol. 2009;23:349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kenealy BP, Keen KL, Rønnekleiv OK, Terasawa E. STX, a novel nonsteroidal estrogenic compound, induces rapid action in primate GnRH neuronal calcium dynamics and peptide release. Endocrinology. 2011;152:182–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Qiu J, Rønnekleiv OK, Kelly MJ. Modulation of hypothalamic neuronal activity through a novel G-protein-coupled estrogen membrane receptor. Steroids. 2008;73:985–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Micevych P, Dewing P. Membrane-initiated estradiol signaling regulating sexual receptivity. Front Endocrinol (Lausanne). 2011;2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Reppert SM, Weaver DR, Edisawa T, Mahle CD, Kolakowski LF., Jr Cloning of a melatonin-related receptor from human pituitary. FEBS Lett. 1996;386:219–224 [DOI] [PubMed] [Google Scholar]
- 108. Sidibe A, Mullier A, Chen P, et al. Expression of the orphan GPR50 protein in rodent and human dorsomedial hypothalamus, tanycytes and median eminence. J Pineal Res. 2010;48:263–269 [DOI] [PubMed] [Google Scholar]
- 109. Batailler M, Mullier A, Sidibe A, et al. Neuroanatomical distribution of the orphan GPR50 receptor in adult sheep and rodent brains. J Neuroendocrinol. 2012;24:798–808 [DOI] [PubMed] [Google Scholar]
- 110. Bechtold DA, Sidibe A, Saer BR, et al. A role for the melatonin-related receptor GPR50 in leptin signaling, adaptive thermogenesis, and torpor. Curr Biol. 2012;22:70–77 [DOI] [PubMed] [Google Scholar]
- 111. Tarttelin EE, Kirschner LS, Bellingham J, et al. Cloning and characterization of a novel orphan G-protein-coupled receptor localized to human chromosome 2p16. Biochem Biophys Res Commun. 1999;260:174–180 [DOI] [PubMed] [Google Scholar]
- 112. Liu B, Hassan Z, Amisten S, et al. The novel chemokine receptor, G-protein-coupled receptor 75, is expressed by islets and is coupled to stimulation of insulin secretion and improved glucose homeostasis. Diabetologia. 2013;56:2467–2476 [DOI] [PubMed] [Google Scholar]
- 113. Ignatov A, Robert J, Gregory-Evans C, Schaller HC. RANTES stimulates Ca2+ mobilization and inositol trisphosphate (IP3) formation in cells transfected with G protein-coupled receptor 75. Br J Pharmacol. 2006;149:490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Thaler JP, Schwartz MW. Minireview: Inflammation and obesity pathogenesis: the hypothalamus heats up. Endocrinology. 2010;151:4109–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci USA. 2011;108:12925–12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Singer K, Luo R, Jeong SJ, Piao X. GPR56 and the developing cerebral cortex: cells, matrix, and neuronal migration. Mol Neurobiol. 2013;47:186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Davenport AP, Alexander SP, Sharman JL, et al. International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands. Pharmacol Rev. 2013;65:967–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Duman JG, Tzeng CP, Tu YK, et al. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J Neurosci. 2013;33:6964–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Stephenson JR, Paavola KJ, Schaefer SA, Kaur B, Van Meir EG, Hall RA. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J Biol Chem. 2013;288:22248–22256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Stephenson JR, Purcell RH, Hall RA. The BAI subfamily of adhesion GPCRs: synaptic regulation and beyond. Trends Pharmacol Sci. 2014;35:208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]