Abstract
BACKGROUND
Well-annotated clinical samples are valuable resources for biomarker discovery and validation. Multiplex and integrated methods that simultaneously measure multiple analytes and generate integrated information about these analytes from a single measurement are desirable because these methods help conserve precious samples. We developed a magnetic bead–based system for multiplex and integrated glycoprotein quantification by immunoassays and glycan detection by lectin immunosorbent assays (LISAs).
METHODS
Magnetic beads coupled with antibodies were used for capturing proteins of interest. Biotinylated antibodies in combination with streptavidin-labeled phycoerythrin were used for protein quantification. In the LISAs, biotinylated detection antibodies were replaced by biotinylated lectins for glycan detection.
RESULTS
Using tissue inhibitor of metallopeptidase 1 (TIMP-1), tissue plasminogen activator, membrane metallo-endopeptidase, and dipeptidyl peptidase-IV (DPP-4) as models, we found that the multiplex integrated system was comparable to single immunoassays in protein quantification and LISAs in glycan detection. The merits of this system were demonstrated when applied to well-annotated prostate cancer tissues for validation of biomarkers in aggressive prostate cancer. Because of the system’s multiplex ability, we used only 300 ng of tissue protein for the integrated detection of glycans in these proteins. Fucosylated TIMP-1 and DPP-4 offered improved performance over the proteins in distinguishing aggressive and nonaggressive prostate cancer.
CONCLUSIONS
The multiplex and integrated system conserves samples and is a useful tool for validation of glycoproteins and their glycoforms as biomarkers.
The differential detection of glycosylated proteins is important to human health because changes in glycosylation are associated with many human diseases (1–6). Carbohydrate cancer antigen CA19-9 is increased in patients with colorectal cancer (5). Prostate-specific antigen, the biomarker for early detection of prostate cancer, has decreased sialylation in the serum of patients with prostate cancer (6). In patients with liver diseases, glycoproteins with increased fucosylation have been reported as candidate biomarkers (4, 7–9). Detection of disease-related changes of glycan structures in proteins provides biomarkers with better disease specificity. Such an improvement has been demonstrated by α-fetoprotein (AFP),2 a marker for hepatocellular carcinoma, and AFP-L3, a core-fucosylated glycoform of AFP (10). Because the enzyme fucosyltransferase Fut8 is overexpressed, detection of AFP-L3 provides better diagnostic specificity for hepatocellular carcinoma than detection of AFP alone (11).
For analyses of glycoproteins and their glycoforms, analytical methods integrating glycoprotein quantification and glycan detection are desired. Mass spectrometry (MS)-based approaches for the integrated analyses are emerging (12–14). Because the MS-based approaches usually require glycoproteins in relatively large quantities (nanograms to micrograms), they are impractical for routine analyses of well-annotated clinical samples that often have limited quantities. Multiplex magnetic bead–based immunoassays have been widely used for routine analyses of well-annotated clinical samples for biomarker discovery and validation for several reasons. These immunoassays require little sample preparation and are easy to perform. The assays are high throughput and can analyze a large number of samples in a short period of time. Lastly, the multiplex ability of these assays for simultaneous measurement of proteins in a single sample results in substantial saving of samples. Building on these advantages, we developed a multiplex magnetic bead–based system that integrates protein quantification with glycan detection.
Tissue inhibitor of metallopeptidase 1 (TIMP-1), tissue plasminogen activator (tPA), membrane metallo-endopeptidase (MME), and dipeptidyl peptidase-IV (DPP-4) are glycoproteins previously discovered using MS-based proteomic analyses as candidate biomarkers in prostate cancer (15–16). Glycan structures of TIMP-1 from cell lines and plasma of patients with cancer have been well characterized and are mostly core fucosylated bi-, tri-, or tetraantennary (17–18). Glycan structures of DPP-4, MME, and tPA are less well characterized than those of TIMP-1. Although glycan profiles of MME were evaluated by lectin microarray (19), the detailed glycan structures remain to be determined. Glycan structures of human recombinant tPA produced by Chinese hamster ovary cells have been reported to be core fucosylated and mono-, di-, and trisialylated (20–21). Glycan structures of rat kidney DPP-4 are a mixture of high-mannose–type sugar chains and mono-, bi-, tri-, and tetraantennary complex–type sugar chains (22–23).
Despite the previously published information on the glycan structures of TIMP-1, DPP-4, MME, and tPA, whether glycoforms of these proteins could serve as biomarkers to distinguish aggressive (AG) and non-aggressive (NAG) prostate cancers remains to be determined. In this study, we applied a multiplex integrated system to analyze TIMP-1, tPA, MME, and DPP-4 in tissues, as well as their glycoforms, as candidate biomarkers for AG prostate cancer. Molecular mechanisms of aberrant glycosylation have indicated increased β1–6 branching of N-glycans and α1–2 fucosylation in prostate cancer progression. Increases in β1–6 branching of N-glycans, which result from overexpression of GlcNAc-TV (UDP-GlcNAc:Manα1–6Manβ-R β1–6-N-acetylglucos-aminyltransferase V) in cancer progression (24), have been reported to be associated with lymph node metastasis in breast carcinoma and promotion of tumor growth and metastasis in many cancers (25–27). Fucosylation in cancer progression has been best studied in hepatocellular carcinoma, which results in overexpression of the enzyme fucosyltransferase Fut8 (11). Recently, quantitative real-time reverse-transcription PCR analysis of glycosyltransferases in normal tissue, tissue from prostate cancer patients, and prostate cancer cell lines indicated that cancer cells have higher mRNA concentrations of Fut1 [fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)], which encodes fucosyltransferase, an enzyme responsible for α1–2 fucosylation (28). Because of the indications of increased β1–6 branching of N-glycans and α1–2 fucosylation as aberrant glycosylation in prostate cancer progression, we used the lectins phytohemagglutinin-L (PHA-L) and Ulex europaeus agglutinin (UEA), which preferentially recognize the β1–6 branched N-glycans and the α1–2 fucosylated N-glycans, respectively, in the multiplex lectin immunosorbent assays (LISAs).
Methods
REAGENTS AND CELL CULTURE
We purchased DPP-4, MME, and TIMP-1 capture and biotinylated detection antibodies and their recombinant proteins from R&D Systems. tPA capture and biotinylated detection antibodies were from Abcam. Detailed information for the antibodies and recombinant proteins are described in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol59/issue1. Magnetic beads, amine coupling kits, and cytokine assay kits were purchased from Bio-Rad Laboratories. Biotinylated UEA, PHA-L, Aleuria aurantia lectin (AAL), and Vicia villosa agglutinin (VVA) were from Vector Laboratories. Cells from the human prostate cancer cell lines PC3 and DU145 were purchased from ATCC and were cultured according to the manufacturer’s instructions.
CLINICAL SAMPLES
The 29 tissue samples analyzed in this study were from men with prostate cancer. Informed consent was obtained under protocols that were institutional review board approved and Health Insurance Portability and Accountability Act compliant. The tissue samples surgically removed were flash frozen, embedded in OCT (optimal cutting temperature) media, and stored at −80 °C till cryostat microdissection. Twenty-one of the tissue samples were NAG prostate cancer defined by their pathological Gleason scores of 6; 8 samples were AG prostate cancer defined by their pathological Gleason scores of 8, 9, or 10. Both NAG and AG prostate cancer tissues were cryostatically microdissected from primary tissues to enrich the tumor content (29). The microdissected tissues were then lysed in radioimmunoprecipitation assay buffer on ice. We determined protein concentrations of the lysed tissue samples by using the bicinchoninic acid assay (Thermo Scientific) normalized to 1 mg/mL. The tissue samples were further diluted 50 times with PBS (13.7 mmol/L NaCl, 2.7 mmol/L KCl, 10 mmol/L Na2HPO4, 2.0 mmol/L KH2PO4, pH 7.4) to a protein concentration of 20 μg/mL and stored at −80 °C until use.
MAGNETIC BEAD–BASED INTEGRATED SYSTEM FOR TIMP-1
The TIMP-1 capture antibody was coupled to magnetic beads with the Bio-Rad amine-coupling kit used according to the manufacturer’s instructions. Detailed procedures of the TIMP-1 immunoassays are described in the online Data Supplement. For TIMP-1 LISAs, 2 μg/mL biotinylated TIMP-1 detection antibody used in the immunoassay was replaced with 20 μg/mL of biotinylated UEA, PHA-L, AAL, or VVA. The TIMP-1 immunoassay and TIMP-1 LISAs were used to analyze TIMP1 in the culture media of PC3 and DU145 cells. Four samples were prepared by 4-fold serial dilutions of the neat culture media. We established a calibration curve using 8 calibrators of 100, 25, 6.25, 1.56, 0.39, 0.1, 0.025, and 0 ng/mL of TIMP-1 and used this curve to determine TIMP-1 protein concentrations.
MULTIPLEX INTEGRATED SYSTEM FOR TPA, DPP-4, MME, AND TIMP-1
As done for TIMP-1, we developed single immunoassays for tPA, DPP-4, and MME. The magnetic beads used for coupling the detection antibodies could be distinguished by their distinctive emission fluorescence and therefore could be mixed together. For the multiplex immunoassay, the beads used for capture and the antibodies for detection were prepared by mixing the 2500 couple beads and the detection antibodies used in the single assays. Single-antigen and single-detection cross-reactivity studies were performed to evaluate the specificity of the capture and detection antibodies. The single-antigen study was conducted by testing the individual antigen in the presence of multiplexed capture beads and detection antibodies. The single-detection study was conducted by testing the individual detection antibodies in the presence of multiplexed antigens and capture beads. For the multiplex LISAs, the same mixture of beads used in the multiplex immunoassay was used for capture, and 20 μg/mL of biotinylated UEA, PHA-L, AAL, or VVA was used for detection.
Six serum samples prepared by supplementing a pooled serum (Sigma-Aldrich) with recombinant TIMP-1, tPA, DPP-4, and MME were used for method comparisons of the multiplex and single immunoassays. For the multiplex immunoassay, we established 1 calibration curve using 8 calibrators of 100, 25, 6.25, 1.56, 0.39, 0.1, 0.025, and 0 ng/mL recombinant tPA, MME, DPP-4, and TIMP-1, respectively. For the single immunoassays, 4 calibration curves were established using 8 calibrators of 100, 25, 6.25, 1.56, 0.39, 0.1, 0.025, and 0 ng/mL of recombinant tPA, MME, DPP-4, or TIMP-1. The same calibrators were used for comparisons of the multiplex and single LISAs.
DATA ANALYSIS
We established calibration curves for protein quantification using the 5-parameter nonlinear regression model in Bio-Plex Manager™ 6.0. Protein concentrations were calculated by using the calibration curves and reported by Bio-Plex Manager™ 6.0. We used Graphpad Prism 5.04 for linear regression and statistical analysis.
Results
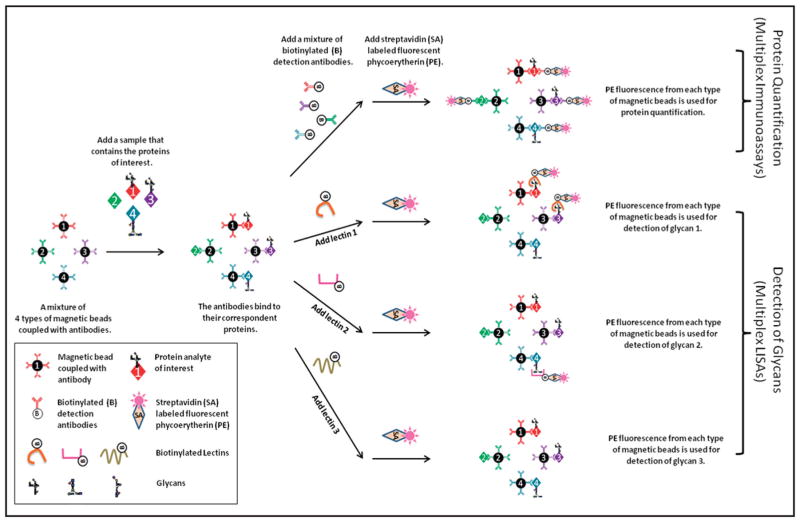
Magnetic beads with different internal fluorescent molecules and different emitted fluorescences (30) have been used for multiplex immunoassays that allow simultaneous measurement of multiple proteins in a single sample (31–32). Building on the multiplex-capability of magnetic beads, we developed a multiplex system integrating protein quantification and glycan detection (Fig. 1). This multiplex system used a standard multiplex immunoassay in which, for the detection of proteins, biotinylated detection antibodies were mixed together. However, for the detection of glycans, biotinylated lectins could not be mixed together because the same glycans could be found on different proteins. As a result, a single lectin was used for the detection of glycans on multiple proteins. This approach of using multiplexed detection of glycans in the magnetic bead system is similar to the detection of glycans in planar array systems such as antibody arrays (33). It is easier, however, to prepare the beads in bulk and reconfigure them to measure a new set of analytes (34), and this method involves fluid-phase kinetics, which is is faster than the solid-phase kinetics of planar arrays and therefore has shorter reaction times (35).
Fig. 1. The magnetic bead–based system that integrates protein quantification with glycan detection.
For protein quantification, magnetic beads coupled with antibodies were used for capturing proteins of interest, and biotinylated antibodies in combination with streptavidin-labeled phycoerythrin were used for detection. Multiplex glycan detection was achieved by running the assay multiple times, each with a different lectin.
MAGNETIC BEAD–BASED INTEGRATED SYSTEM FOR TIMP-1
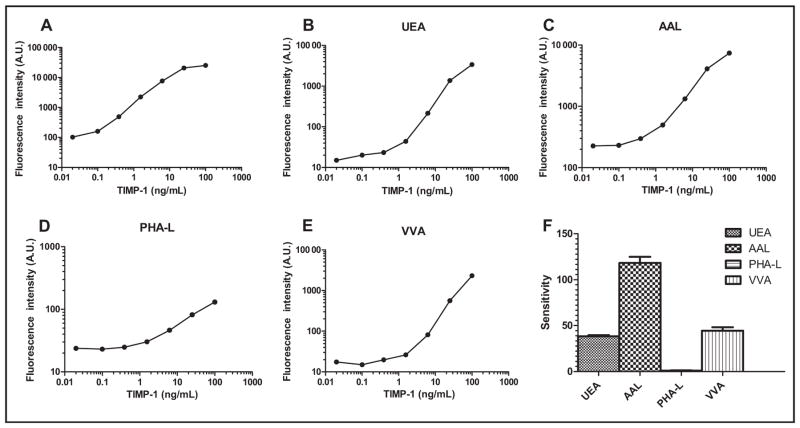
Using TIMP-1 as a model protein, we demonstrated the feasibility of the integrated system for protein quantification and glycan detection. First, we developed a magnetic bead–based immunoassay for TIMP-1 protein. The calibration curve we established using recombinant TIMP-1 protein showed increased fluorescence signals with increasing TIMP-1 concentrations (Fig. 2A). Once the immunoassay was developed, we selected lectins for the development of TIMP-1 LISAs. Antibodies are glycoproteins, and it is known that in LISAs glycans on antibodies may bind to detection lectins, potentially causing high background noise (6, 33, 36). We tried 2 approaches to reduce the background: enzymatic digestion of the glycans off the antibodies and derivatization of the glycans with dipeptides to block glycan–lectin binding (37). Although both approaches were successful in reducing the background for the majority of detection lectins tested (see online Supplemental Figs. 1 and 2), the reduction was not universal. Through these experiments we also observed that not all of the lectins caused high background. We decided that instead of modifying glycans on the capture antibodies, we would circumvent the background issues by selecting the lectins to which the glycans on the antibodies did not bind. In addition, we determined the lectin-binding profile of recombinant TIMP-1 protein using tyramide signal amplification (36). Among the lectins to which TIMP-1 bound, we selected UEA, AAL, PHA-L, and VVA for the development of the LISAs.
Fig. 2. (A) Establishment of the integrated system for protein quantification and glycan detection in TIMP-1: magnetic bead–based immunoassay for TIMP-1 protein; (B–E), dose–response curves of UEA, AAL, PHA-L, and VVA immunosorbent assays for TIMP-1; F, glycan profile of the recombinant TIMP-1.
The glycan profile was established using the sensitivity of the dose–response curves, calculated as changes of fluorescence intensity per TIMP-1 concentration. A.U., arbitrary units.
Dose–response curves of TIMP-1 LISAs (Fig. 2, B–E) showed increased fluorescence signals in UEA, AAL, PHA-L, and VVA with increasing TIMP-1 concentrations. Sensitivity of the curves, calculated as changes of fluorescence intensity per TIMP-1 concentration, was used to establish the glycan profile of the recombinant TIMP-1 [Fig. 2F; mean (SD), 118 (7) for AAL; 44 (4) for VVA; 38 (1) for UEA; and 1.0 (0.2) for PHA-L]. The y intercept of 25 in the TIMP-1 PHA-L assay (Fig. 2D) was likely due to the PHA-L–associated glycans on the TIMP-1 capture antibody. The glycan structures of the same recombinant TIMP-1 were analyzed by Thaysen-Andersen et al. (17), who showed, with the exception of a few core structures, predominantly core-fucosylated glycans to which AAL preferentially binds. This finding is consistent with the glycan profile established by the multiplex LISAs, as evidenced by the highest sensitivity TIMP-1 AAL assay of 118 (7). In agreement with Thaysen-Anderson et al. (17), the TIMP-1 LISAs showed that the recombinant TIMP-1 also contained terminal α1–2 fucosylated glycans associated with UEA, and tri- and tetraantennary core-fucosylated glycans associated with PHA-L.
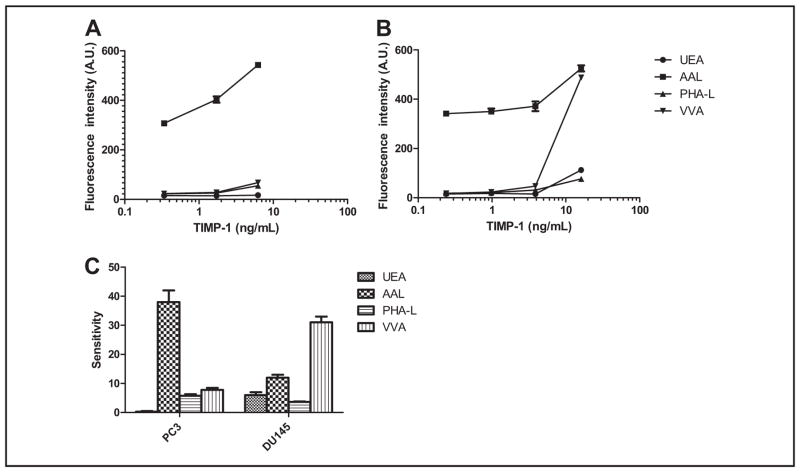
TIMP-1 was present in culture media of both PC3 and DU145 cells. Lectin microarray profiling indicated that TIMP-1 from these 2 sources had different glycans (see online Supplemental Fig. 3). Consistent with the lectin microarray profiling results, TIMP-1 in DU145 medium showed more preferential binding to VVA than TIMP-1 in PC3 medium [shown in Fig. 3C; mean (SD) 31 (2) vs 7.8 (0.7)]. In Fig. 3, A and B, AAL LISAs had background signals in the 300–400 range, which was likely due to the AAL-associated glycans on the TIMP-1 capture antibody (38). Using TIMP-1 from PC3 and DU145 cell culture media, we demonstrated that the magnetic bead–based system integrating protein quantification and glycan detection was capable of sensitive detection of proteins with differential glycan structures. To validate the lectin specificity of UEA toward α1–2–linked fucose, we used TIMP-1 as the model protein. Forty-five minute treatment of TIMP-1 with α1–2 fucosidase, an enzyme that hydrolyzes the α1–2 linked fucose that UEA preferentially recognizes, decreased the fluorescence signals of TIMP-1 measured by the UEA LISA at various concentrations compared to those without the treatment (see online Supplemental Fig. 4). These results supported the specificity of UEA toward α1–2 linked fucose.
Fig. 3. Dose–response curves of the LISAs for TIMP-1 from culture media of PC3 cells (A) and DU145 cells (B).
C, comparison of glycan profiles of TIMP-1 from PC3 and DU145. Protein concentrations of TIMP-1 in these samples were quantified by TIMP-1 immunoassay and plotted against the signals detected in these samples by LISAs. The plotted results are shown as the dose–response curves. A.U., arbitrary units.
MULTIPLEX MAGNETIC BEAD–BASED INTEGRATED SYSTEM FOR TPA, DPP-4, MME, AND TIMP-1
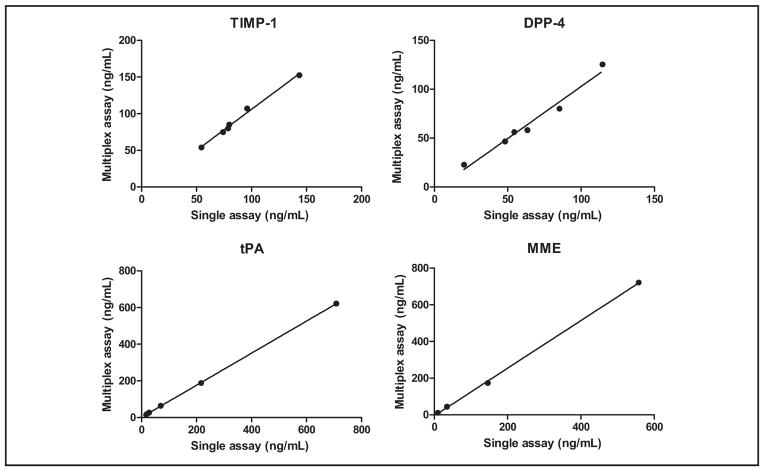
tPA, DPP-4, and MME, in addition to TIMP-1, were included as model proteins to demonstrate the feasibility of the integrated system for multiplexing. As done for TIMP-1, magnetic bead–based single immunoassays were developed for tPA, DPP-4, and MME. We examined the multiplex ability of the 4 single immunoassays by performing single-antigen and single-detection cross-reactivity studies, the results of which showed that the degree of cross-reactivity across the 4 immunoassays was <2% (see online Supplemental Fig. 5). By mixing the magnetic beads and the detection antibodies used in the single assays, we developed the multiplex immunoassay and evaluated it by comparing it to the single immunoassays for measuring concentrations of tPA, DPP-4, MME, and TIMP-1 in serum samples. Linear regression analyses showed that the multiplex-to–single assay comparisons had slopes of 0.93 (R2 =0.91), 1.05 (R2 =0.97), 0.87 (R2 =1.00), and 1.30 (R2 =1.00) for DPP-4, TIMP-1, tPA, and MME, respectively (Fig. 4; also see online Supplemental Table 1). These results indicated that the multiplex immunoassay was comparable to the single assays in protein quantification.
Fig. 4.
Comparison of the multiplex immunoassays and single immunoassays for measurement of DPP-4, TIMP-1, tPA, and MME in serum.
Next, by replacing the mixture of detection antibodies in the multiplex immunoassay with UEA, AAL, PHA-L, or VVA, we developed the multiplex LISAs. We found that, similar to TIMP-1 antibody, the capture antibodies for tPA, DPP-4, and MME did not cause background binding to the selected lectins. We evaluated the performances of these assays by comparing them to the performances of the respective single LISAs. Both the multiplex and single LISAs gave low signals for the recombinant MME (data not shown), indicating that the recombinant MME had few or no glycan structures that were associated with UEA, AAL, PHA-L, or VVA. The glycan profiles established for recombinant tPA, DPP-4, and TIMP-1 by the single assays were similar to the profiles established by the multiplex assays (Fig. 5). With the use of tPA, DPP-4, MME, and TIMP-1, the protein quantification and glycan detection on the multiplex integrated system was comparable to those of respective single immunoassays and single LISAs.
Fig. 5. Comparison of the multiplex LISAs and single LISAs for detection of glycan structures of recombinant TIMP-1, DPP-4, and tPA.
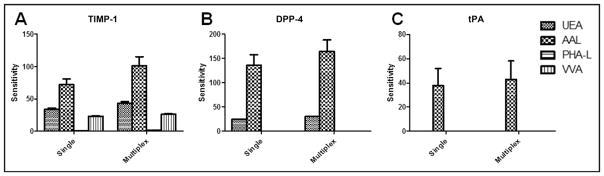
The recombinant MME had few or no glycan structures associated with UEA, AAL, PHA-L, or VVA, and therefore its glycan profile was not included.
APPLICATION OF THE MULTIPLEX INTEGRATED ASSAY FOR EVALUATION OF BIOMARKERS IN AG PROSTATE CANCER
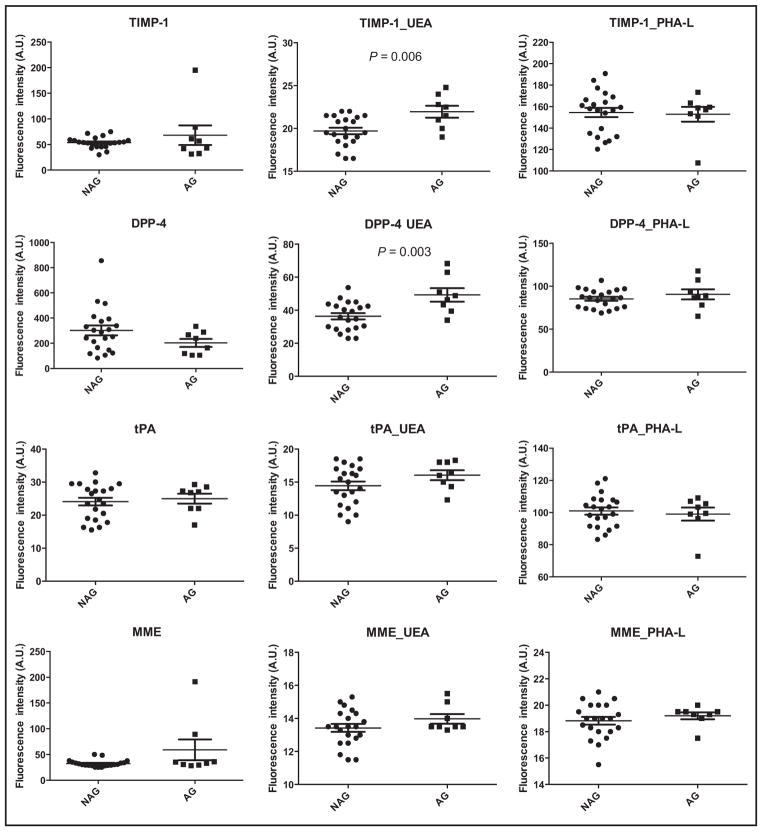
We applied the system to analyze 21 NAG tissues and 8 AG prostate cancer tissues to evaluate tPA, DPP-4, MME, and TIMP-1 and their UEA- and PHA-L–associated glycoforms as biomarkers in AG prostate cancer. Because of the system’s sensitive, multiplex, and integrated merits, we were able to carry out the evaluation using 300 ng of tissue samples. Neither tPA nor MME, nor their glycoforms detected by UEA or PHA-L, could be used to distinguish AG cancer (Fig. 6). tPA is a tissue type plasminogen activator. The primary role of tPA is the generation of plasmin for fibrinolysis in blood vessels (39). The initial study in which we discovered tPA as a candidate biomarker for prostate cancer was performed with serum samples (16). This use of serum might explain the lack of differences of tPA concentrations in AG and NAG prostate tissues. MME is a cell surface metallopeptidase. Expression of MME has been reported to be lost in several human tumors, including androgen-independent prostate cancer. Our data showed no difference in MME protein concentrations between AG and NAG prostate cancer tissues obtained from patients with androgen-dependent cancer.
Fig. 6.
Expression of TIMP-1, DPP-4, tPA, and MME and their UEA- and PHA-L-associated glycoforms in NAG and AG prostate cancer. A.U., arbitrary units.
TIMP-1 has been shown to be decreased in prostate cancer tissues compared to noncancer tissues (15). Our data showed that TIMP-1 protein concentrations did not differ between AG and NAG prostate cancer tissues. We were able to make the distinction, however, on the basis of the detection of UEA-associated fucosylated TIMP-1 (Fig. 6, P =0.006). DPP-4 is a cell surface serine protease associated with cancer suppression. DPP-4 also blocks fibroblast growth factor 2 signaling, altering the cell adhesion properties and malignant phenotype of prostate cancer cells (40). Unlike DPP-4, the detection of UEA-associated fucosylated DPP-4 showed a statistically significant (P =0.003) increase in AG cancer. A preliminary experiment that used 10 NAG and 6 AG prostate tissues (see online Supplemental Fig. 6) showed the same patterns as those in Fig. 6, indicating that the changes observed were consistent. In addition, in this experiment we tested the amount of tissue to be used in the system and found that 1500 ng of tissue gave the same patterns for distinguishing AG and NAG prostate cancer as 300 ng of tissue (see online Supplemental Fig. 7), highlighting the capabilities of this system in saving precious human samples.
Discussion
We were unable to distinguish between AG and NAG cancer with the detection of TIMP-1 and DPP-4, but we were able to make the distinction with the detection of fucosylated TIMP-1 and DPP-4. The results of the study highlight 2 merits of the multiplex and integrated system. First, because of its advantage in multiplexing, we used only 300 ng of tissue in protein for the validation and we saved quantities of the precious samples. Second, because of the advantages of integrated detection of glycoforms of proteins, we discovered that the use of fucosylated TIMP-1 and DPP-4 was associated with improved performance over the use of proteins in distinguishing AG and NAG prostate cancer.
Although recombinant proteins are widely used as standards in protein quantification, whether they are appropriate as standards for the detection of protein glycans, in our opinion, depends on the contexts in which they are being used. If used as standards for the quantification of protein glycoforms, such as the percentage of AFP-L3, highly purified single glycoforms of recombinant proteins should be used. The recombinant glycoproteins produced through current protein engineering technology would not be appropriate, because they usually exist in heterogeneous, rather than single, glycoforms. On the other hand, if the recombinant glycoproteins were to be used only as model proteins to demonstrate the feasibility of an analytical system, such as the integrated system in this study, they should be appropriate.
Antibody characteristics such as affinity and specificity can be used to determine the performance of immunoassays (32). To select the best antibody pair to develop the magnetic bead immunoassays for TIMP-1, MME, DPP-4, or tPA, we reviewed the information provided by antibody vendors on the specificity of the antibodies. A monoclonal antibody is preferred for use as the capture antibody because it is potentially more specific than polyclonal antibodies. In addition, when experimental conditions are kept constant, results from monoclonal antibodies are highly reproducible because of the homogeneity of monoclonal antibodies. Commercially available biotinylated antibodies for the detection antibody are preferred over nonbiotinylated antibodies. Antibodies generated using proteins as immunogens are preferred over polypeptides. Furthermore, we used indirect LISAs to perform in-house testing of the affinity and specificity of the selected antibodies toward the recombinant protein. Once the immunoassays were developed, we used the same capture antibodies for the development of LISAs.
In summary, we developed a magnetic bead–based multiplex system that allows for sensitive, multiplex, integrated detection of glycoproteins and their glycoforms. Although TIMP-1 and DPP-4 could not be used for distinguishing AG and NAG cancer, their fucosylated glycoforms could. This result highlights the importance of the integrated detection of glycoproteins and their glycoforms. Through integrated detection, we might be able to discover glycoforms of proteins with improved performance as biomarkers.
Supplementary Material
Acknowledgments
Research Funding: NIH/NCI/EDRN, National Cancer Institute Early Detection and Research Network grant U24CA115102 and National Cancer Institute Early Detection and Research Network grant U01CA152813.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: AFP, α-fetoprotein; AFP-L3, core-fucosylated glycoform of AFP; MS, mass spectrometry; TIMP-1, tissue inhibitor of metallopeptidase 1; tPA, tissue plasminogen activator; MME, membrane metallo-endopeptidase; DPP-4, dipeptidyl peptidase-IV; AG, aggressive; NAG, nonaggressive; PHA-L, phytohemagglutinin-L; UEA, Ulex europaeus agglutinin; LISA, lectin immunosorbent assay; AAL, Aleuria aurantia lectin; VVA, Vicia villosa-B-4 agglutinin.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
Patents: Glycosylation patterns for the diagnosis of prostate cancer.
References
- 1.Meany DL, Chan DW. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin Proteomics. 2011;8:7–20. doi: 10.1186/1559-0275-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lau KS, Dennis JW. N-Glycans in cancer progression. Glycobiology. 2008;18:750– 60. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- 3.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855– 67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Mehta A, Block TM. Fucosylated glycoproteins as markers of liver disease. Dis Markers. 2008;25:259– 65. doi: 10.1155/2008/264594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narimatsu H, Iwasaki H, Nakayama F, Ikehara Y, Kudo T, Nishihara S, et al. Lewis and secretor gene dosages affect CA19-9 and DU-PAN-2 serum levels in normal individuals and colorectal cancer patients. Cancer Res. 1998;58:512– 8. [PubMed] [Google Scholar]
- 6.Meany DL, Zhang Z, Sokoll LJ, Zhang H, Chan DW. Glycoproteomics for prostate cancer detection: changes in serum PSA glycosylation patterns. J Proteome Res. 2009;8:613–9. doi: 10.1021/pr8007539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, et al. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–15. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, et al. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:1914–21. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, et al. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43:1007–12. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Oka H, Saito A, Ito K, Kumada T, Satomura S, Kasugai H, et al. Multicenter prospective analysis of newly diagnosed hepatocellular carcinoma with respect to the percentage of Lens culinaris agglutinin-reactive alpha-fetoprotein. J Gastroenterol Hepatol. 2001;16:1378– 83. doi: 10.1046/j.1440-1746.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson WL, Du MQ, Johnson PJ, Williams R. Fucosyltransferases: differential plasma and tissue alterations in hepatocellular carcinoma and cirrhosis. Hepatology. 1991;13:683–8. [PubMed] [Google Scholar]
- 12.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal Chem. 2001;73:4530– 6. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 13.Stalnaker SH, Hashmi S, Lim JM, Aoki K, Porterfield M, Gutierrez-Sanchez G, et al. Site mapping and characterization of O-glycan structures on alpha-dystroglycan isolated from rabbit skeletal muscle. J Biol Chem. 2010;285:24882–91. doi: 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leymarie N, Zaia J. Effective use of mass spectrometry for glycan and glycopeptide structural analysis. Anal Chem. 2012;84:3040– 8. doi: 10.1021/ac3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu AY, Zhang H, Sorensen CM, Diamond DL. Analysis of prostate cancer by proteomics using tissue specimens. J Urol. 2005;173:73–8. doi: 10.1097/01.ju.0000146543.33543.a3. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Sokoll LJ, Rush J, Meany D, Zou N, Chan DW, Zhang H. Targeted detection of prostate cancer proteins in serum using heavy-isotope-labeled-peptide standards and MALDI-TOF/TOF. Proteomics Clin Appl. 2009;3:597–608. [Google Scholar]
- 17.Thaysen-Andersen M, Thogersen IB, Nielsen HJ, Lademann U, Brunner N, Enghild JJ, Hojrup P. Rapid and individual-specific glycoprofiling of the low abundance N-glycosylated protein tissue inhibitor of metalloproteinases-1. Mol Cell Proteomics. 2007;6:638– 47. doi: 10.1074/mcp.M600407-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Thaysen-Andersen M, Thogersen IB, Lademann U, Offenberg H, Giessing AM, Enghild JJ, et al. Investigating the biomarker potential of glycoproteins using comparative glycoprofiling: application to tissue inhibitor of metalloproteinases-1. Biochim Biophys Acta. 2008;1784:455–63. doi: 10.1016/j.bbapap.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Tao SC, Bova GS, Liu AY, Chan DW, Zhu H, Zhang H. Detection and verification of glycosylation patterns of glycoproteins from clinical specimens using lectin microarrays and lectin-based immunosorbent assays. Anal Chem. 2011;83:8509–16. doi: 10.1021/ac201452f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papac DI, Wong A, Jones AJ. Analysis of acidic oligosaccharides and glycopeptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 1996;68:3215–23. doi: 10.1021/ac960324z. [DOI] [PubMed] [Google Scholar]
- 21.Basa LJ, Spellman MW. Analysis of glycoprotein-derived oligosaccharides by high-pH anion-exchange chromatography. J Chromatogr. 1990;499:205–20. doi: 10.1016/s0021-9673(00)96974-7. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita K, Tachibana Y, Matsuda Y, Katunuma N, Kochibe N, Kobata A. Comparative studies of the sugar chains of aminopeptidase N and dipeptidylpeptidase IV purified from rat kidney brush-border membrane. Biochemistry. 1988;27:5565–73. doi: 10.1021/bi00415a026. [DOI] [PubMed] [Google Scholar]
- 23.Schmauser B, Kilian C, Reutter W, Tauber R. Sialoforms of dipeptidylpeptidase IV from rat kidney and liver. Glycobiology. 1999;9:1295–305. doi: 10.1093/glycob/9.12.1295. [DOI] [PubMed] [Google Scholar]
- 24.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 25.Handerson T, Camp R, Harigopal M, Rimm D, Pawelek J. Beta1,6-branched oligosaccharides are increased in lymph node metastases and predict poor outcome in breast carcinoma. Clin Cancer Res. 2005;11:2969–73. doi: 10.1158/1078-0432.CCR-04-2211. [DOI] [PubMed] [Google Scholar]
- 26.Guo HB, Lee I, Kamar M, Pierce M. N-acetylglucosaminyltransferase V expression levels regulate cadherin-associated homotypic cell-cell adhesion and intracellular signaling pathways. J Biol Chem. 2003;278:52412–24. doi: 10.1074/jbc.M308837200. [DOI] [PubMed] [Google Scholar]
- 27.Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–12. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 28.Fukushima K, Satoh T, Baba S, Yamashita K. alpha1,2-Fucosylated and beta-N-acetylgalactosaminylated prostate-specific antigen as an efficient marker of prostatic cancer. Glycobiology. 2010;20:452–60. doi: 10.1093/glycob/cwp197. [DOI] [PubMed] [Google Scholar]
- 29.Tian Y, Bova GS, Zhang H. Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal Chem. 2011;83:7013–9. doi: 10.1021/ac200815q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishhan VV, Khan IH, Luciw PA. Multiplexed microbead immunoassays by flow cytometry for molecular profiling: basic concepts and proteomics applications. Crit Rev Biotechnol. 2009;29:29– 43. doi: 10.1080/07388550802688847. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto K, Yasukawa F, Ito S. Measurement of the sugar-binding specificity of lectins using multiplexed bead-based suspension arrays. Methods Mol Biol. 2007;381:401–9. doi: 10.1007/978-1-59745-303-5_21. [DOI] [PubMed] [Google Scholar]
- 32.Li D, Chiu H, Gupta V, Chan DW. Validation of a multiplex immunoassay for serum angiogenic factors as biomarkers for aggressive prostate cancer. Clin Chim Acta. 2012;413:1506–11. doi: 10.1016/j.cca.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, et al. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437–44. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 34.Nolan JP, Sklar LA. Suspension array technology: evolution of the flat-array paradigm. Trends Biotechnol. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- 35.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5:310–20. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meany DL, Hackler L, Jr, Zhang H, Chan DW. Tyramide signal amplification for antibody-overlay lectin microarray: a strategy to improve the sensitivity of targeted glycan profiling. J Proteome Res. 2011;10:1425–31. doi: 10.1021/pr1010873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Haab BB. Analysis of glycans on serum proteins using antibody microarrays. Methods Mol Biol. 2009;520:39–58. doi: 10.1007/978-1-60327-811-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huhn C, Selman MH, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics. 2009;9:882–913. doi: 10.1002/pmic.200800715. [DOI] [PubMed] [Google Scholar]
- 39.Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57:25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajiyama H, Kikkawa F, Khin E, Shibata K, Ino K, Mizutani S. Dipeptidyl peptidase IV overexpression induces up-regulation of E-cadherin and tissue inhibitors of matrix metalloproteinases, resulting in decreased invasive potential in ovarian carcinoma cells. Cancer Res. 2003;63:2278– 83. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.